Abstract
For a large number of T cell-mediated immunopathologies, the disease-related antigens are not yet identified. Identification of T cell epitopes is of crucial importance for the development of immune-intervention strategies. We show that CD4+ T cell epitopes can be defined by using a new system for synthesis and screening of synthetic peptide libraries. These libraries are designed to bind to the HLA class II restriction molecule of the CD4+ T cell clone of interest. The screening is based on three selection rounds using partial release of 14-mer peptides from synthesis beads and subsequent sequencing of the remaining peptide attached to the bead. With this approach, two peptides were identified that stimulate the β cell-reactive CD4+ T cell clone 1c10, which was isolated from a newly diagnosed insulin-dependent diabetes mellitus patient. After performing amino acid-substitution studies and protein database searches, a Haemophilus influenzae TonB-derived peptide was identified that stimulates clone 1c10. The relevance of this finding for the pathogenesis of insulin-dependent diabetes mellitus is currently under investigation. We conclude that this system is capable of determining epitopes for (autoreactive) CD4+ T cell clones with previously unknown peptide specificity. This offers the possibility to define (auto)antigens by searching protein databases and/or to induce tolerance by using the peptide sequences identified. In addition the peptides might be used as leads to develop T cell receptor antagonists or anergy-inducing compounds.
Keywords: insulin-dependent diabetes mellitus
A large number of T cell-mediated diseases, exemplified by the autoimmune disease insulin-dependent diabetes mellitus (IDDM), are thought to be caused by the activation of CD4+ T cells by antigen-derived peptides in the context of HLA class II (1, 2). For many T cell-mediated diseases, it is not known which antigen(s) cause(s) the activation of pathogenic CD4+ T cells. Identification of antigens and epitopes in T cell-mediated diseases might enable immune intervention strategies with respect to treatment or prevention of disease. Even in the absence of an identified antigen source, peptides that stimulate pathogenic CD4+ T cells may be useful in the development of primary immune intervention approaches via the induction of tolerance or the application of altered peptide ligands (3, 4).
Various methods for the identification of T cell epitopes have been described. Most of them are based on identification of the source protein of an epitope by using molecular biology approaches (5–7) or peptide elution from HLA molecules followed by fractionation and sequence analysis (8, 9). These methods are time consuming or require large amounts of biological material. Alternative, fast synthetic peptide library approaches have been shown to be useful in the identification of T cell epitopes only for CD8+ T cells (10, 11).
Herein we show that CD4+ T cell epitopes can be identified by using a newly developed system for construction and screening synthetic peptide libraries. We applied this generally applicable system to identify epitopes for CD4+ T cell clones from a recent onset IDDM patient (12, 13). IDDM is caused by the destruction of β cells on the pancreatic islet (2). A number of autoantibodies that are associated with the development of IDDM have been characterized and can be used for predicting its development (14). Although a number of candidate autoantigens are recognized by antibodies and/or T cells (GAD65, GAD67, insulin, ICA69, hsp65, and a 38-kDa protein), additional, as yet unidentified, antigens are recognized by diabetogenic CD4+ T cell clones (14).
The CD4+ T cell clones we used in this study are β cell-reactive and DR1-restricted (12, 13). Although DR1 is not associated with the highest risk for IDDM (15), we chose for DR1 because of the availability of a positive control clone for the validation of the procedure. An epitope for this clone, 1c6, has been identified by using a β cell subtraction library (6). Clone 1c6 and a clone with unknown peptide specificity (clone 1c10) were tested for proliferation with pools of the peptide library.
MATERIALS AND METHODS
Peptide Synthesis.
Synthetic peptides were made on an Abimed 422 multiple peptide synthesizer (Abimed, Langenfeld, Germany) at the 10-μmol scale (16, 17). TentagelS AC resins (18, 19) (Rapp, Tübingen, Germany) were used in combination with fluorenylmethoxycarbonyl-protected amino acids carrying trifluoroacetic acid (TFA)-labile side-chain-protecting groups where needed (20). Acylations were carried out with a 6-fold excess amino acid by using (benzotriazolyl)-N-oxytripyrrolidinophosphonium hexafluorophosphate/N-methylmorpholine activation in N-methylpyrrolidinone (17, 21). Deprotection was performed with piperidine/dimethylacetamide, 1:4 (vol/vol). Coupling and deprotection times were increased as synthesis proceeded, starting with a 45-min period for coupling and three 3-min periods for deprotection. Cleavage of the peptides and removal of the side-chain-protecting groups was performed with TFA/water, 19:1 (vol/vol) for 2.5 h. For Trp-containing peptides, ethanethiol (5%) was added to the cleavage mixture (22). Peptides were isolated and purified by repeated ether/pentane (1:1, vol/vol) precipitations, dissolved in 10% acetic acid or water, and lyophilized.
Synthetic Peptide Library Design and Synthesis.
TentagelS AM [Rapp; particle size, 90 μm; loading, 100 pmol per bead; linker, 5-(4′-aminomethyl-3′,5′-dimethoxyphenoxy)valeric acid (23)] was used to synthesize a random 14-mer library containing a DR1 binding motif (24), by following the synthesis protocol as described above. γ-Aminobutyric acid (GABA) was synthesized between the peptide and the linker to make the rate of cleavage independent of the C-terminal amino acid of the peptide. The design of the library represents a compromise between high dedication for DR1 binding and complete randomness. We expected that identification of epitopes from a completely random library would only be possible for large libraries (on the order of a billion peptides). To reduce this large number of peptides required, we decided to use anchor residues at DR1 anchor positions (p1, p4, and p6). In this way, libraries were enriched for possible DR1 binding peptides and, therefore, also enriched for possible epitopes. Anchor positions were created by Trp, Tyr, or Phe at relative position 1; Leu, Ala, Ile, or Met at relative position 4; and Gly, Ser, Pro, Ala, or Thr at relative position 6, summarized in the following synthesis scheme: XXX(W,Y,F)XX(L,A,I,M)X(G, S,P,A,T)XXXXX-GABA, where X is one of 19 l-amino acids (an natural amino acids except cysteine, which was omitted for synthetic reasons). Compared with the anchor positions mentioned, p9 is very degenerate (amino acids Leu, Ile, Ala, Val, Asn, Phe, Tyr, Gln, Met, Cys, Ser, Arg and probably others are all possible) (24) and was, therefore, treated as a random position. The library synthesis was performed according to the one-bead/one-peptide principle (25) by following a mix and split protocol (26); in one reaction column, one amino acid was coupled in each step and after each step the total amount of resin was mixed and redivided over the appropriate number of reaction columns. Thus, at the end of the synthesis one bead contained only a single peptide. The complexity of the library synthesized was 8 × 106.
Partial Release of Peptides from Synthesis Beads.
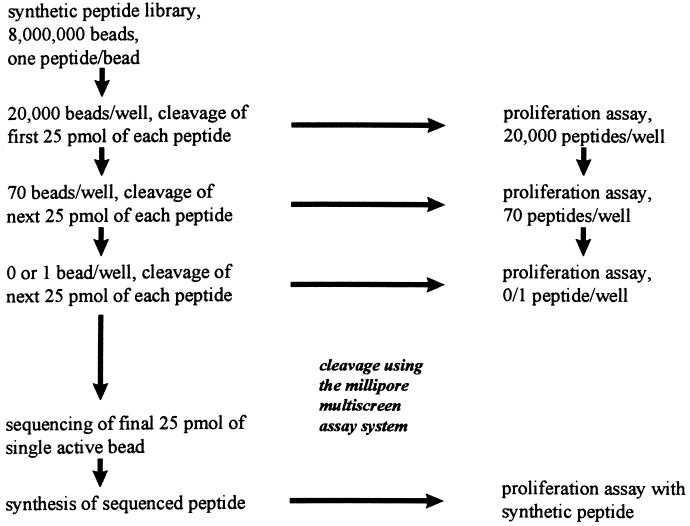
To make stepwise screening of large numbers of peptides possible, we defined conditions for stepwise partial cleavage of C-terminal GABA peptides from TentagelS AM (17) synthesis beads. The use of different linkers and orthogonal cleavage conditions (27, 28) was shown not to be required for partial cleavage. The complete procedure is schematically summarized in Fig. 1. For the first step of the screening, a suspension of 8 × 106 library beads in C2H4Cl2/CH3CN, 82:18 (vol/vol), was dispensed in four multiscreen-PF1 96-well polypropylene plates with 1.0-μm (pore size) hydrophophilic polytetrafluoroethylene (Teflon) membrane filters (Millipore) to 2 × 104 beads per well. The filtration plates were placed on a filtration assembly (Millipore multiscreen assay system). Beads were successively washed twice with CH2Cl2 (200 μl per well) and three times with ether (200 μl per well) and dried overnight in air. To liberate the first quarter of the peptide material from each bead, the beads were incubated for 60 min in TFA/water/CH3CN, 8:1:11 (vol/vol) (150 μl per well), after which the filtrate was collected into 96-well serocluster polypropylene U-bottom plates (Costar). After washing the beads with 75 μl of TFA/water/CH3CN, 8:1:11 (vol/vol), per well in the same U-bottom plate, the filtrate was dried overnight under vacuum. Immediately after filtration, the beads were neutralized by washing twice with 200 μl of 0.5 M Tris⋅HCl, pH 7.5/CH3CN, 1:1 (vol/vol), per well and incubated for 1 h in the same buffer solution. After sequential washings with CH3CN/water, 1:1 (vol/vol), CH3CN, CH2Cl2, and ether (all at 200 μl per well) and drying, the beads were stored at −20°C. To remove all protecting groups, the cleaved and dried peptides were treated with TFA/water/ethanethiol, 18:1:1 (vol/vol) (100 μl per well), for 3 h and dried overnight under vacuum. The peptide mixtures obtained were dissolved in 15 μl of dimethyl sulfoxide/water, 1:2 (vol/vol), diluted, and neutralized with 200 μl of 30 mM sodium phosphate buffer (pH 7.5) per well. This solution was used in a proliferation assay to determine the activity of library pools. In the second step of the screening, the 2 × 104 beads, from which the solubilized peptides were found to be active in the first step of the screening, were dispensed to approximately 70 beads per well, by the same protocol as described above for the first step of the screening. For the cleavage of a second quarter of the peptides from each bead, beads were incubated in TFA/water/CH3CN, 8:1:11 (vol/vol) for 100 min. In the third step of the screening, beads from which the solubilized peptides were found to be active in the second screening were dispensed to 0 or 1 bead per well and beads were incubated 120 min in TFA/water/CH3CN, 8:1:11 (vol/vol), to cleave off a third quarter of the peptide from each bead. The bead responsible for the activity in the third step of the screening was isolated for determining the sequence of the peptide attached to it by Edman sequencing.
Figure 1.
Schematic representation of the screening the library of 8 million beads for stimulating peptides. Beads were divided in four 96-well plates (20,000 peptides per well) and partially cleaved for first-round screening (proliferation assay). Beads of each positive pool from round 1 were divided in three 96-well plates (70 beads per well) and partially cleaved for second round screening (proliferation assay). Beads of each positive pool from round 2 were divided by limiting dilution to 0 or 1 bead per well for the third round screening (proliferation assay). Beads from positive wells from round 3 were sequenced (Edman degradation). Sequenced peptides were synthesized and tested for activity in a proliferation assay.
Proliferation Assay.
CD4+ T cell proliferation assays for testing library fractions and synthetic peptides were performed with 1 × 104 T cells and 5 × 104 irradiated HLA DR1-matched peripheral blood mononuclear cells per well in flat-bottomed 96-well plates in 150 μl of complete Iscove’s modified Dulbecco’s medium containing 10% pooled human serum. Phytohemagglutinin (10 μg/ml), rat insulinoma membranes (10 μg/ml), and interleukin 2 (T cell growth factor, 10% Lymphocult) were used as positive controls for T cell proliferation. [3H]Thymidine (0.5 μCi in 50 μl of RPMI 1640 medium; 1 Ci = 37 GBq) was added after 72 h, cells were harvested (Micro Cell Harvester; Skatron, Sterling, VA), and activity of the T cell DNA was counted after another 18 h (LKB 1205 Betaplate Liquid Scintillation Counter). Library pools were tested in a quantity of 7 μl per well giving final concentrations of 5 pmol/ml for each individual peptide and 0.1% dimethyl sulfoxide.
Bead Sequencing.
A single bead containing a stimulating peptide was isolated from the 96-well multiscreen plate by pipetting under a microscope (magnification, ×40). The pipet tip was emptied in a sequence cartridge of a Hewlett–Packard G1005A protein sequencer by centrifuging 10 s at 103 rpm. Of the 14-mer peptides, at least 13 amino acids could be determined by Edman degradation, until yields became too low, resulting in dropping of the phenylthiohydantoin amino acid absorbance below detection limit.
Swiss Prot Database Searching.
The Swiss Prot database was searched for possible autoantigens by using the Genetic Computer Group software, with the fasta program for homology searches with single sequences or the findpatterns program for searches with patterns based on substitution studies performed with the peptides identified. Based on their homology and origin, peptides were selected for synthesis and testing in a proliferation assay.
RESULTS AND DISCUSSION
Development of the System.
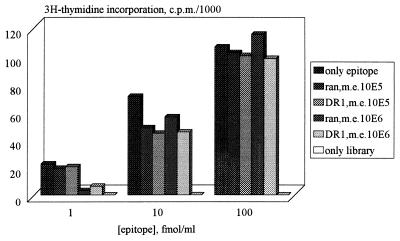
First we defined the maximal complexity of a synthetic peptide library, containing the HLA-DR1 binding motif XXX(W,Y,F)XX(L,I,A,M)X(G,S,P,A,T)XXXXX-GABA, in which the activity of a single peptide can still be measured in a CD4+ T cell proliferation assay as 105-106. For this purpose, clone 1c6, a clone with known peptide, was used (6). This clone was isolated from the same recent-onset IDDM patient as the clone of unknown peptide specificity (1c10), which was used in this study (12, 13), Data for clone 1c6 and its previously identified 14-mer epitope are shown in Fig. 2. Since factors that might affect proliferation, such as competition for HLA binding and antagonism, seem to occur only at large excess of irrelevant peptides, screening of library pools containing 20,000 peptides was shown to be feasible. A C-terminal GABA residue does not influence the response to a CD4+ T cell epitope in a proliferation assay as had been determined before with various known peptide epitopes (data not shown).
Figure 2.
Effect of mixing random library or library containing a DR1 motif with the 1c6 14-mer epitope (PSLWEIEFAKQLAS) on the proliferation of 1c6 to this epitope. ran, Random 14-mer library; DR1, random 14-mer library containing a DR1 motif; m.e., molar excess of library over epitope. In this experiment neither the random library nor the library containing a DR1 motif caused proliferation of clone 1c6. All responses are the means of duplicate tests.
The DR1 binding properties of the library were validated by pool sequencing (29) and binding studies to soluble DR1 (30) and comparison of the data to the data of a completely random peptide library (data not shown). An additional control to validate the new system was performed by mixing one bead containing the previously identified GABA-linked 14-mer epitope (PSLWEIEFAKQLAS) of clone 1c6 (6) with 20,000 beads of the library. We found that clone 1c6 was able to recognize the pools containing the epitope in the selection rounds. Sequencing of the bead indeed yielded the 14-mer epitope sequence.
Screening of the Library.
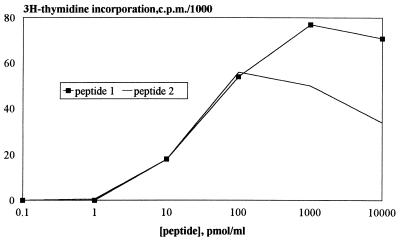
The total library containing the DR1 motif was divided in 384 pools, each containing 20,000 peptides, and each pool was screened with two different clones. Wells were scored positive when stimulation indices were at least 10 and activity was higher than 1,000 cpm. The first step of the screening with the DR1-restricted CD4+ T cell clones yielded two positive pools for clone 1c10, but no positive pools for clone 1c6, which indicates that the complexity of the 14-mer library containing a DR1 binding motif (8 × 106) may not always be large enough to find activity for DR1-restricted clones. Depending on the library design and the HLA restriction of the clones, the outcome might also be different in other experiments. Proceeding with the second and the third step of the screening of the library for clone 1c10, two epitopes were identified: TAMYELLQPAAIV (Pept.-1) and FDSYEIMQGRAVVH (Pept.-2). Dose–response curves of the two defined and synthesized epitopes are shown in Fig. 3. Retrospectively, it was shown that the concentration of the single peptide in the library screening, 5 pmol/ml, indeed induces a response for both identified peptides in a proliferation assays with clone 1c10. Although this response is measured at suboptimal peptide concentration, the response is still strong enough to be detected in a reproducible manner (±100× background). The responses of clone 1c10 to the identified epitopes were indeed DR1-restricted (data not shown).
Figure 3.
Peptide concentration dependency of the 1c10 proliferative responses. Dose–response curves are shown for Pept.-1 (TAMYELLQPAAIV) and Pept.-2 (FDSYEIMQGRAVVH) and responses are the means of duplicate tests.
We conclude that it is possible to identify epitopes from this library by using the newly developed screening protocol. When we compared the two independently identified epitopes for clone 1c10, we found extensive sequence similarity. In addition to the anchor positions defined in the synthesis procedure, we found the following similarity: Glu (relative position 2), Ile/Leu (relative position 3), Gln (relative position 5), and Ala (relative position 8). It has been reported that a T cell clone can respond to considerably distinct peptide epitopes (31, 32). However, our data suggest that this is not a common event. Alternatively, our results may imply that the definition of the anchor residues affects the requirements of the other unspecified peptide positions for T cell recognition.
Minimal Epitope Determination and Database Searching with fasta.
Various length variants of the two newly identified epitopes were tested for recognition in a proliferation assay to determine the minimal epitopes. This was done in a proliferation experiment with all possible 13-, 12-, 11-, and 10-mers derived from the two peptides. Table 1 shows that the minimal sequences required for recognition are MYELLQPAAIV for Pept.-1 and DSYEIMQGRAVV for Pept.-2. It is clear that Pept.-1 requires only a Met at the N-terminal side of p1 (Tyr), whereas Pept.-2 requires two amino acids in this part of the peptide. This might indicate that recognition is dependent on some bulkiness next to p1 that is provided by Met or Asp-Ser but not by a single small residue like Ser. The binding affinities of both minimal epitopes for soluble DR1 were determined (30). The IC50 values for the minimal epitopes of Pept.-1 and Pept.-2 were 10 nM and 20 nM, respectively. Since the IC50 value for the standard DR1-binding hemagglutinin peptide (30) in our assay is 100 nM, the data indicate that both peptides are high-affinity DR1 binders.
Table 1.
Proliferative responses of clone 1c10 to length variants of Pept.-1 and Pept.-2
| Pept.-1 | cpm (×10−3) | Pept.-2 | cpm (×10−3) |
|---|---|---|---|
| TAMYELLQPAAIV | 66 | FDSYEIMQGRAVVH | 111 |
| MYELLQPAAIV | 89 | DSYEIMQGRAVV | 117 |
| AMYELLQPAAI | 52 | FDSYEIMQGRAV | 41 |
| YELLQPAAIV | 0 | SYEIMQGRAVVH | 0 |
| MYELLQPAAI | 55 | SYEIMQGRAVV | 0 |
| MYELLQPAA | 0 | DSYEIMQGRAV | 43 |
| DSYEIMQGRA | 0 |
Peptides were tested at a concentration of 10−5 M. The minimal epitopes determined are in boldface. [3H]Thymidine incorporation is shown as cpm (×10−3). All responses are the means of duplicate tests.
We searched the Swiss Prot database with the 9-mer sequences of Pept.-1 and Pept.-2 (relative positions 1 to 9), because class II binding and T cell receptor triggering depend on a 9-mer core sequence (p1–p9). None of the 27 14-mer peptides tested from the top of the list of retrieved sequences were able to stimulate clone 1c10 in a proliferation assay (Table 2). There are several possibilities why this search was not successful. (i) The (auto)antigen might not yet exist in the database. (ii) It has been reported that T cells are able to recognize different epitopes with little or no sequence homology (31, 32). (iii) The input for the database search might not have been sufficiently precise, yielding a hit list too large to be tested. The latter problem seems to be profound since the hit list was shown to consist of a rather heterogeneic peptide population. (iv) The use of other databases (such as the GenBank, Protein Identification Resource, or whole nonredundant database) would have been a possibility. However, as we expected that the use of other databases would also give (a longer) list of heterogeneous peptides, we decided to focus in more detail on the Pept.-1 and Pept.-2 sequences to obtain a more precise search pattern.
Table 2.
Peptides selected with 9-mer
| 14-mer peptide | Organism and protein |
|---|---|
| VTFYELTQPPSVSL | Human Ig chain V–IV region |
| LTIYEMLQNIFAIF | Human interferon β precursor |
| KRKYELGRPAANTK | Human 40S ribosomal protein |
| NPAYEVLQHVKIPT | Variola virus DNA topoisomerase |
| ASLWEALQPEAFSQ | Equine herpes virus type I large tegument protein |
| VPILELENPAAFYR | Human genetic coat protein |
| TAMYKLLKHSRVRA | Epstein–Barr virus transcription activator BRLF1 |
| YGVYEILQPLISFN | Schizosaccharomyces pombe start control protein CDC10 |
| QKAIELLEPAAFLQ | Caenorhabditis elegans 66-kDa hypothetical prot. F47D12.7 |
| GLIWELLQASAFCG | Human cystic fibrosis transm. conductance regulator |
| YNFLELLQHPAFAQ | Equine arteritus virus pol polyprotein |
| IDIYELLKQAAAIT | Haemophilus influenzae hypothetical prot. HI0420 |
| DVLYELLQHILKQR | Human ETS-related protein tel. |
| PEDYELLQIISEDH | Rat guanine nucleotide dissociation stimulator |
| ANGLFLLAPSAFFI | Haemophilus influenzae hypothetical prot. HI0169 |
| KVRVELLHNPAFCS | Human complement C3 Prec. |
| ITGYEVIQPDLIVV | Clostridium thermocellum cell surf. glycoprot.1 prec. |
| KMIVELVQPIAIEK | Astasia longa elongation factor Tu |
| RQRYEILTPNAIPK | Human myosin heavy chain, nonmuscle type B |
| PSQEELLQPAGSEA | Mouse breast cancer type 1 susceptibility protein |
| GVKYELWQPECELT | Trypanosoma brucei variant surface glycoprotein |
| LMTRQLLDPTAIFW | Human olfactory marker protein |
| YGVYEILQPLISFN | Schizosaccharomyces pombe start control protein CDC10 |
| VLSYELLQKAGYLF | Chlamydia trachomatis prolyl-tRNA synthetase |
| INIYEIIKPATANS | Human bone morphogenetic protein 2 prec. |
| LISYELVLSSAILL | Neurospora crassa NADH-ubiquinone oxired. chain 1 |
| KMKVELIQPIAIEK | Euglena gracilis elongation factor Tu |
The 14-mers that were selected from the Swiss Prot protein database by searching with the 9-mer core sequences (relative positions 1–9) of Pept.-1 and Pept.-2 for testing. The 9-mer cores of the 14-mers that share homology with the 9-mer core sequences of Pept.-1 and Pept.-2 are in boldface type.
Testing of Substitution Analogs and Database Searching with findpatterns.
To gain more information on which amino acids are responsible for activity, alanine-scan peptides were tested (Table 3). Only a few Ala substitutions are allowed, indicating a structural restriction at most amino acid positions in the peptides. In addition to that, substitution analogs were tested in which amino acids from one peptide were synthesized into the other peptide. Results of this experiment are shown in the lower part of Table 3. As not all exchanges are permitted, we confirm that the requirements for one amino acid position do not only depend on that particular position but were shown to be also influenced by the amino acids surrounding that position. This has implications for homology searches in protein databases.
Table 3.
Proliferative responses of 1c10 toward Ala-substltuted and exchange variants of Pept.-1 and Pept.-2
| Pept.-1 | cpm (×10−3) | Pept.-2 | cpm (×10−3) |
|---|---|---|---|
| MYELLQPAAIV | 89 | DSYEIMQGRAVV | 117 |
| AYELLQPAAIV | 0 | ASYEIMQGRAVV | 0 |
| MAELLQPAAIV | 1 | DAYEIMQGRAVV | 68 |
| MYALLQPAAIV | 0 | DSAEIMQGRAVV | 0 |
| MYEALQPAAIV | 23 | DSYAIMQGRAVV | 1 |
| MYELAQPAAIV | 0 | DSYEAMQGRAVV | 0 |
| MYELLAPAAIV | 0 | DSYEIAQGRAVV | 1 |
| MYELLQAAAIV | 0 | DSYEIMAGRAVV | 1 |
| MYELLQPAAAV | 96 | DSYEIMQARAVV | 0 |
| MYELLQPAAIA | 87 | DSYEIMQGAAVV | 51 |
| DSYEIMQGRAAV | 129 | ||
| DSYEIMQGRAVA | 125 | ||
| DSYELLQPAAIV | 104 | MYEIMQGRAVV | 0 |
| MYEILQPAAIV | 93 | DSYELMQGRAVV | 66 |
| MYELMQPAAIV | 0 | DSYEILQGRAVV | 97 |
| MYELLQGAAIV | 0 | DSYEIMQPRAVV | 0 |
| MYELLQPRAIV | 0 | DSYEIMQGRAIV | 108 |
| MYELLQPAAVV | 82 |
Peptides were tested at a concentration of 10−5 M. Minimal epitopes and the substituted amino acids are in boldface type. [3H]Thymidine incorporation is reported in cpm (×10−3). All responses are the means of duplicate tests.
We performed a second Swiss Prot database search with a 6-mer pattern that is based on relative positions 1–6 of the two identified epitopes and the substitution studies performed after the first database homology search. The six positions at which Ala substitution affected recognition were taken into account (p1–p6). To define the new search pattern, at these six positions, the amino acids from Pept.-1 and Pept.-2 and their conservative substitutions were included. The search pattern was thus defined as (Trp, Tyr, Phe, Leu, Ile, Met)(Asp, Glu)(Leu, Ile, Met, Val)(Leu, Ile, Met, Val)(Gln, Asn)(Gly, Pro). We tested a set of 48 14-mer peptides obtained from a findpatterns search of the Swiss Prot database with the pattern mentioned above (Table 4). Activity was observed in a proliferation assay for one of the 48 peptides, namely, the Haemophilus influenzae TonB-derived peptide SISMELLQGMVLEE. A duplicate test of this peptide (10 μM) gave 12 × 103 cmp (mean).
Table 4.
Peptides selected with 6-mer
| 14-mer peptide | Organism and protein |
|---|---|
| SFILDLMNGGDLHY | Human β-adrenergic receptor kinase 1 |
| PLPIDMLQGIIGAK | Human erythrocyte band 7 integral protein |
| VSLFDIINPEIITR | Human cholesteryl ester transfer protein prec. |
| LSGYDILQGYPKDI | Human neutrophil collagenase prec. |
| GSFFELLNPFALLC | Haemophilus influenzae cytochrome oxidase |
| DRVIEVVQGAYRAI | Human immunodeficiency virus type 1 envelope |
| GHLLELLNPSEVFG | Human lanosterol synthase |
| YIVMELVQGGDFLT | Human protooncogene tyrosine protein kinase |
| KKRYEVVNGKLTNT | Human β-1,3-galactosyl-O-glycosyl-glycoprot. |
| CLIYEMIQGHSPFK | Human G-protein-coupled receptor kinase |
| KNNIDVINGFGKFV | Haemophilus influenzae glutathione reductase |
| QAAIDLINGKVNRV | Influenza A virus hemagglutinin protein |
| IPALDLINGQVVRL | Haemophilus influenzae phosphorib. form. am. ca. |
| KNNIDVINGEGKFV | Haemophilus influenzae glutathione reductase |
| QAAIDLINGKVNRV | Influenza A virus hemagglutinin protein |
| AIPIEIMQGTGEEL | Human hexokinase type I |
| HRFFELVNGPLFDH | Herpes simplex virus processing and transport |
| RKFFELVNGPLFAH | Pseudorabies virus strain |
| HKFFELVNGPLFNH | Varicella-zoster virus strain Dumas |
| KTVLELMNPEAQLP | Human RAS-GTPase-activating-like protein |
| WQVFDILNGKPYEP | Human nuclear-factor Nκ-B P105 subunit |
| KYSLDVVNGLLFLH | Human mos protooncogene S/T-protein kinase |
| DLGFEVLQPLQSGS | Pseudorabies virus strain K |
| GYWIEILNPNKMAT | Human lactoylglutathione lyase |
| DDVLEIVQGPDFPT | Haemophilus influezae topoisomerase IV subunit A |
| SVTLDIVQGIESAE | Human PMEL1 protein prec. |
| TPVLEVINPRAVVI | Simian foamy virus pol polyprotein |
| LTTIDILNGGRQAL | Haemophilus influenzae phosphorib. form. gl. am. synth. |
| REELELVQGASNEF | Haemophilus influenzae peptide chain release fact. 3 |
| PDPIEVVNGFLIVG | Human parainfluenza 2 virus RNA polymerase |
| YSPMDLINPRSAVS | Haemophilus influenzae meth.-tRNA synth. |
| KALIEVLQPLIAEH | Human tryptophanyl-tRNA synth. |
| IHKFDMIQGLAEHN | Human transf. growth factor β prec. |
| KETLDMINPVQQQN | Human nucleoylsin TIA-1 |
| SISMELLQGMVLEE | Haemophilus influenzae tonB protein |
| VRLFEILQGKYAYV | Epstein–Barr virus strain B virion protein |
| VHLFEILQGKYAYV | Herpesvirus saimiri strain 11 virion gene 43 prot. |
| YDHFDIVNGKECCY | Vaccinia virus strain Copenhagen protein B3 |
| SGHLDMVNGFFDQF | Rat 31.4-kDa water channel protein |
| YGHLEVTQPQLNSQ | Mouse β-glucuronidase prec. |
| AGGLELLQGGPTRP | Mouse CCAAT enhancer binding prot. δ |
| QLLLDILQPQQNGR | Rabbit phosphorylase B kinase β regulat. |
| NSQIEVVNGKLKDS | Rat Kupffer cell receptor |
| QRDLELVQGFMPHF | Mouse plasminogen activator inh.-1 prec. |
| RKDFEMNQGMKTKL | Rat mitochondrial intermediate peptidase prec. |
| KPLFEILNGDSNPI | Mouse mammary tumor virus |
| LSTLEMLQGANCVL | Mouse downreg. prot. of IL-2 receptor |
| PGSLELLQPGESKT | Mouse TNF receptor ass. factor 2 |
The 14-mers that were selected from the SwissProt protein database by searching with a 6-mer pattern for testing. The cores that share full sequence homology with the search pattern are in boldface type.
Concluding Remarks.
The data presented above show that CD4+ T cell clone epitopes can be identified relatively easy and fast by using this newly developed screening system based on partial peptide cleavage and bead sequencing.
Performing substitution studies with the identified peptides as lead sequences enabled us to define a pattern for the peptide restriction of clone 1c10. Using this pattern in a findpatterns Swiss Prot database search yielded a Haemophilus influenzae TonB-derived peptide that was able to stimulate clone 1c10. Although there is no epidemiological indication that Haemophilus influenzae is involved in the induction of IDDM, its possible role is currently being investigated. Next to the TonB peptide, other epitopes derived from a database search might be identified by screening other databases or by performing more detailed substitution studies that lead to different patterns.
Apart from using peptides identified from the library for database searches, they can also be used for tolerance induction (33) or the design of antagonists (34, 35) or anergy inducing peptides (36, 37). The latter might also be possible by making use of peptide library strategies, which enables screening of large numbers of potentially interesting peptides. These possibilities are part of current research.
Thus, the method introduced is applicable for identifying epitopes of (autoreactive) CD4+ T cell clones and offers the potential to modulate CD4+ T cell responses based on the peptides identified, with or without knowledge about (auto)antigens.
Acknowledgments
We thank Dr. F. Koning, Dr. H. Bruning, and Dr. S. Schoenberger for critically reading the manuscript. This work was supported by the University of Leiden, the Macropa Foundation, and the Diabetes Fonds Nederland.
ABBREVIATIONS
- IDDM
insulin-dependent diabetes mellitus
- TFA
trifluoroacetic acid
- GABA
γ-aminobutyric acid
References
- 1.Gianani R, Sarvetnick N. Proc Natl Acad Sci USA. 1996;93:2257–2259. doi: 10.1073/pnas.93.6.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson M A, Maclaren N K. N Engl J Med. 1994;331:1428–1436. doi: 10.1056/NEJM199411243312107. [DOI] [PubMed] [Google Scholar]
- 3.Wucherpfennig K W, Strominger J L. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walden P. Curr Opin Immunol. 1996;8:68–74. doi: 10.1016/s0952-7915(96)80107-5. [DOI] [PubMed] [Google Scholar]
- 5.Boon T, Cerottini B, Van den Eynde P, Van der Bruggen P, Van Pel A. Annu Rev Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 6.Neophytou P I, Roep B O, Arden S D, Muir E M, Duinkerken G, Kallan A, De Vries R R P, Hutton J C. Proc Natl Acad Sci USA. 1996;93:2014–2018. doi: 10.1073/pnas.93.5.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavin M A, Dere B, Grandea A G, III, Hogquist K A, Bevan M J. Eur J Immunol. 1994;24:2124–2133. doi: 10.1002/eji.1830240929. [DOI] [PubMed] [Google Scholar]
- 8.Udaka K, Tsomides T J, Eisen H N. Cell. 1992;69:989–998. doi: 10.1016/0092-8674(92)90617-l. [DOI] [PubMed] [Google Scholar]
- 9.Hunt D F, Henderson R A, Shabnowitz J, Sakaguchi K, Michel H, Selilir N, Cox A, Apella E, Engelhard V H. Science. 1992;255:1261–1263. [Google Scholar]
- 10.Gundlach B J, Wiesmüller K-H, Junt T, Kienle S, Jung G, Walden P. J Immunol Methods. 1996;192:149–155. doi: 10.1016/0022-1759(96)00040-3. [DOI] [PubMed] [Google Scholar]
- 11.Blake J, Johnston J V, Hellström K E, Marquardt H, Chen L. J Exp Med. 1996;184:121–130. doi: 10.1084/jem.184.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Vliet E, Roep B O, Meulenbroek L, Bruining G J, De Vries R R P. Eur J Immunol. 1989;19:213–217. doi: 10.1002/eji.1830190136. [DOI] [PubMed] [Google Scholar]
- 13.Roep B O, Arden S D, De Vries R R P, Hutton J C. Nature (London) 1990;345:632–634. doi: 10.1038/345632a0. [DOI] [PubMed] [Google Scholar]
- 14.Roep B O. Diabetes. 1996;45:1147–1156. doi: 10.2337/diab.45.9.1147. [DOI] [PubMed] [Google Scholar]
- 15.She J-X. Immunol Today. 1996;17:323–329. doi: 10.1016/0167-5699(96)10014-1. [DOI] [PubMed] [Google Scholar]
- 16.Gausepohl H, Kraft M, Boulin C, Frank R W. In: Peptides 1990. Giralt E, Andreu D, editors. Leiden, The Netherlands: ESCOM; 1990. pp. 206–207. [Google Scholar]
- 17.Gausepohl H, Kraft M, Boulin C, Frank R W. In: Peptides: Chemistry, Structure and Biology. Rivier J E, Marshall G R, editors. Leiden, The Netherlands: ESCOM; 1990. pp. 1003–1004. [Google Scholar]
- 18.Rapp W, Zhang L, Bayer E. In: Innovation and Perspectives in Solid Phase Peptide Synthesis. Epton R, editor. Birmingham, U.K.: SPCC; 1990. pp. 205–210. [Google Scholar]
- 19.Sheppard R C, Williams B J. Int J Pept Protein Res. 1982;20:451–454. doi: 10.1111/j.1399-3011.1982.tb03067.x. [DOI] [PubMed] [Google Scholar]
- 20.Fields G B, Noble R L. Int J Pept Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 21.Coste J, Dufour M-N, Nguyen D, Castro B. In: Peptides: Chemistry, Structure and Biology. Rivier J E, Marshall G R, editors. Leiden, The Netherlands: ESCOM; 1990. pp. 885–888. [Google Scholar]
- 22.Pearson D A, Blanchette M, Baker M L, Guindon C A. Tetrahedron Lett. 1989;30:2739–2742. [Google Scholar]
- 23.Albericio F, Barany G. Int J Pept Protein Res. 1987;30:206–216. doi: 10.1111/j.1399-3011.1987.tb03328.x. [DOI] [PubMed] [Google Scholar]
- 24.Rammensee H-G, Friede T, Stevanovic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 25.Lam K S, Salmon S E, Hersh E M, Hruby V J, Kazmierski W M, Knapp R J. Nature (London) 1991;354:82–86. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 26.Lebl M, Krchnak V, Sepetov N F, Seligman B, Strop P, Felder S. Biopolymers. 1995;37:177–198. doi: 10.1002/bip.360370303. [DOI] [PubMed] [Google Scholar]
- 27.Lebl M, Patek M, Kocis P, Krchnak V, Hruby V J, Salmon S E, Lam K S. Int J Pept Protein Res. 1993;41:201–203. doi: 10.1111/j.1399-3011.1993.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 28.Salmon S E, Lam K S, Lebl M, Kandola A, Khattri P S, Wade S, Patek M, Kocis P, Krchnak V, Thorpe D, Felder S. Proc Natl Acad Sci USA. 1993;90:11708–11712. doi: 10.1073/pnas.90.24.11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Nature (London) 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 30.Geluk A, Van Meijgaarden K E, Drijfhout J W, Ottenhoff T H M. Mol Immunol. 1995;32:975–981. doi: 10.1016/0161-5890(95)00058-m. [DOI] [PubMed] [Google Scholar]
- 31.Nanda K N, Arzoo K K, Geysen H M, Sette A, Sercasz E E. J Exp Med. 1995;182:531–539. doi: 10.1084/jem.182.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quaratino S, Thorpe C J, Travers P J, Londei M. Proc Natl Acad Sci USA. 1995;92:10398–10402. doi: 10.1073/pnas.92.22.10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruisbeek A M, Armsen D. Curr Opin Immunol. 1996;8:233–244. doi: 10.1016/s0952-7915(96)80062-8. [DOI] [PubMed] [Google Scholar]
- 34.De Magistris M T, Alexander J, Coggeshall M, Altman A, Gaeta F C A, Grey H M, Sette A. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 35.Ruppert J, Alexander J, Snoke K, Coggeshall M, Herbert E, McKenzie D, Grey H M, Sette A. Proc Natl Acad Sci USA. 1993;90:2671–2675. doi: 10.1073/pnas.90.7.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloan-Lancaster J, Evavold B D, Allen P M. Nature (London) 1993;363:156–159. doi: 10.1038/363156a0. [DOI] [PubMed] [Google Scholar]
- 37.Sloan-Lancaster J, Allen P M. Curr Opin Immunol. 1995;7:103–109. doi: 10.1016/0952-7915(95)80035-2. [DOI] [PubMed] [Google Scholar]