Abstract
This report shows that loss of heterozygosity at the mannose 6-phosphate/insulin-like growth factor II receptor (M6P/IGF2R) locus occurred in 5/8 (63%) dysplastic liver lesions and 11/18 (61%) hepatocellular carcinomas (HCCs) associated with the high risk factors of hepatitis virus infection and liver cirrhosis. Mutations in the remaining allele were detected in 6/11 (55%) HCCs, including deletions in a polydeoxyguanosine region known to be a target of microsatellite instability. M6P/IGF2R allele loss was also found in cirrhotic tissue of clonal origin adjacent to these dysplastic lesions and HCCs, demonstrating that M6P/IGF2R inactivation occurs early in liver carcinogenesis. In conclusion, HCCs frequently develop from clonal expansions of phenotypically normal, M6P/IGF2R-mutated hepatocytes, providing further support for the idea that M6P/IGF2R functions as a liver tumor-suppressor gene.
Hepatocellular carcinoma (HCC) is the most common type of malignancy in sub-Saharan Africa, Southeast Asia, Japan, and much of China and is the most common cause of cancer death worldwide (1). Etiological risk factors for HCC formation include hepatitis virus (HV) infection, alcohol consumption, and dietary exposure to aflatoxin B1 (1, 2). The p53 tumor suppressor gene is a target for mutation in HCCs, especially in humans exposed to aflatoxin B1, and its loss has generally been shown to occur late in transformation (3). The early molecular events involved in multistage liver carcinogenesis, however, still remain to be elucidated.
The mannose 6-phosphate/insulin-like growth factor-II receptor (M6P/IGF2R) plays a critical role in regulating cell growth by facilitating the activation of the growth inhibitor transforming growth factor β (TGFβ) (4, 5) and inactivating the growth stimulator insulin-like growth factor-II (IGF2) (6). Loss of this receptor is therefore predicted to both increase cell proliferation and reduce apoptosis, consistent with the M6P/IGF2R gene functioning as a tumor suppressor. We have recently shown that M6P/IGF2R is mutated in HCCs developing in patients without HV infection or liver cirrhosis (7, 8); however, the majority of HCCs arise in patients with these clinical conditions (1). The results of this study demonstrate that M6P/IGF2R is also frequently mutated in HCCs associated with the high risk factors of HV infection and cirrhosis, and that M6P/IGF2R gene inactivation occurs early in liver carcinogenesis.
MATERIALS AND METHODS
Patients.
Frozen tissue sections (n = 6) or paraffin-embedded tissue sections (n = 31) from 27 patients with histopathologically confirmed HCC or liver dysplasia (S. Finkelstein) were obtained from the University of Pittsburgh and the University of Miami. All patients had a history of HV infection and/or cirrhosis. These patients had undergone hepatic resection or transplantation for the treatment of their disease.
Tissue Microdissection and Loss of Heterozygosity (LOH) Analysis.
The microdissection and polymerase chain reaction (PCR) conditions employed were described previously (7, 9). Liver stroma was used as the normal tissue. Purified PCR products were digested with 10 units of EcoRI, and then labeled with [α-33P]ATP (2,000 Ci/mmol, 10 mCi/ml; 1 Ci = 37 GBq) using 2.5 units of the large (Klenow) fragment of DNA polymerase I. Labeled products were analyzed for LOH as previously described (7, 9). Due to the potential of contaminating the tumor tissue sample with normal stroma, allele loss in informative patients was defined as a >50% decrease in the ratio of the two alleles in the target tissue versus that in the surrounding normal stromal tissue. This was quantified using a densitometer. Statistical comparisons were performed with the Fisher exact test (10).
Direct Sequencing of PCR Products.
Genomic DNA was amplified and sequenced as previously described (8, 9). Since infidelity of Taq DNA polymerase can introduce errors during PCR, a number of precautions were taken. Normal and mutant templates were amplified in two or more independent PCRs, and mutants were confirmed by direct sequencing in both directions.
Primer pairs were as follows:
Identification of exon 27 mutations.
First-round primer pair: 5′-GAAAATGTGAATGCGTGTGTGG-3′ and 5′-ACTCATGTTTCTGACTCAAGGG-3′. Second-round primer pair: 5′-GCGTGTGTGGTTGCAGTTGCCC-3′ and 5′-CCTTTGCAACAAAAGGAAAACG-3′.
Identification of exon 28 mutations.
First-round primer pair: 5′-AGTTTGACAGCCTAGGGACCCG-3′ and 5′-TCCCAGCAGCCTGAGGGTGGGG-3′. Second-round primer pair: AGTTTGACAGCCTAGGGACCCG-3′ and 5′-AGACCCCGCTGAGGGCCGTCGG-3′.
Identification of exon 31 mutations.
First-round primer pair: 5′-GCCGTGCCCTCCAGAAGCAGCC-3′ and 5′-CACTTGGCTCTCGCTGCAGGTG-3′. Second-round primer pair: 5′-CGCGTGTCTGCTGGGTGGCTCC-3′ and 5′-GGATGGTGGTTGACTTTTTCCG-3′.
Immunohistochemical Staining for M6P/IGF2R.
Frozen sections were fixed in Omnifix II (An-Con Genetics, Melville, NY) and immunoperoxidase stained with diaminobenzidine as previously described (8). In all cases, nonimmune rabbit IgG was used as a negative control on serial sections (data not shown). The sections were counterstained with hematoxylin.
RESULTS AND DISCUSSION
The human M6P/IGF2R gene maps to chromosome 6q26 (11). It has both a polymorphic dinucleotide repeat sequence and a tetranucleotide deletion/insertion polymorphism in the 3′ untranslated region of the gene that are useful in determining LOH at this locus by PCR technology (12). We used these polymorphisms to first screen for M6P/IGF2R heterozygosity in 27 patients associated with HV infection and/or liver cirrhosis (Fig. 1). Eighteen of the 27 (67%) patients were informative (heterozygous) for these polymorphisms, a frequency consistent with that previously reported (7, 9, 12). Summary data for the M6P/IGF2R-informative patients are provided in Table 1.
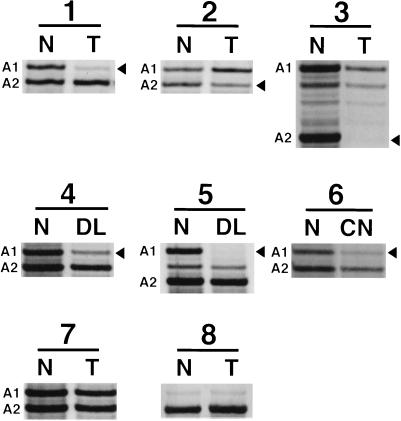
Figure 1.
LOH at the M6P/IGF2R locus in human liver lesions. Patients 1–3 are informative with LOH in HCCs; patients 4 and 5 are informative with LOH in premalignant dysplastic lesions; patient 6 is informative with LOH in a cirrhotic nodule; patient 7 is informative without LOH in a HCC; and patient 8 is noninformative with a HCC. Alleles are defined as A1 and A2 for the informative patients. Arrowheads mark the alleles lost; faint bands are due to contaminating normal stromal tissue. N, normal stromal tissue; T, tumor; DL, dysplastic lesion; CN, cirrhotic nodule.
Table 1.
Summary data for the M6P/IGF2R-informative patients
| Case no. | Informative patient no. | Age, yr | Sex | Cirrhosis etiology | Diagnosis | M6P/IGF2R LOH | M6P/IGF2R mutations |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 52 | M | Alcohol | HCC | No | ND |
| 2 | DL | No | ND | ||||
| 3 | DL | Yes | No mutations in exons 27, 28, 31 | ||||
| 4 | 2 | 60 | M | Alcohol | HCC | Yes | G deletion in exon 28 |
| 5 | DL | Yes | No mutations in exons 27, 28, 31 | ||||
| 6 | 3 | 60 | M | HBV | HCC | No | ND |
| 7 | 4 | 64 | F | HCV | HCC | Yes | G·C → T·A in exon 31 |
| 8 | 5 | 68 | M | HAV and HBV | HCC | Yes | No mutations in exons 27, 28, 31 |
| 9 | 6 | 51 | M | Drugs | HCC | No | ND |
| 10 | DL | Yes | No mutations in exons 27, 28, 31 | ||||
| 11 | 7 | 48 | M | HBV and HCV | HCC | No | ND |
| 12 | HCC | Yes | No mutations in exons 27, 28, 31 | ||||
| 13 | 8 | 59 | M | Alcohol | HCC | Yes | No mutations in exons 27, 28, 31 |
| 14 | HCC | Yes | G·C → T·A in exon 31 | ||||
| 15 | DL | Yes | No mutations in exons 27, 28, 31 | ||||
| 16 | 9 | 50 | F | HAV | HCC | Yes | No mutations in exons 27, 28, 31 |
| 17 | 10 | 64 | M | HCV and alcohol | HCC | Yes | G deletion in exon 28 |
| 18 | DL | No | ND | ||||
| 19 | 11 | 49 | M | HCV and alcohol | HCC | Yes | No mutations in exon 27, 28, 31 |
| 20 | 12 | 50 | M | Alcohol | DL | Yes | No mutations in exon 27, 28, 31 |
| 21 | 13 | 60 | F | HCV | DL | No | ND |
| 22 | 14 | 62 | M | HBV | HCC | No | ND |
| 23 | 15 | 58 | M | HCV | HCC | Yes | G deletion in exon 28 |
| 24 | 16 | 76 | M | HCV | HCC | Yes | G·C → C·G in exon 27 |
| 25 | 17 | 56 | F | Alcohol | HCC | No | ND |
| 26 | 18 | 76 | F | Unknown | HCC | No | ND |
DL, dysplastic lesion; HAV, hepatitis A virus; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus, ND, not determined.
Three pathological lesions believed to depict different stages of liver carcinogenesis (i.e., cirrhotic nodules, premalignant dysplastic lesions, and primary HCCs) (13) were analyzed for LOH at the M6P/IGF2R locus (Fig. 1). LOH was observed at the M6P/IGF2R locus in 11/18 (61%) of the HCCs (Table 1). This LOH frequency is not significantly different (P > 0.1) from that previously observed in HCCs that developed in patients without HV infection and cirrhosis (i.e., 63%) (7). Thus, M6P/IGF2R allele loss is a common event in HCCs irrespective of whether the patients are HV-infected and/or have liver cirrhosis.
Mutations should also be present in the remaining allele of those HCCs exhibiting LOH if the M6P/IGF2R is a tumor suppressor gene, as our previous results in both liver (7, 8) and breast (9) tumors indicate. Since the M6P/IGF2R gene is large (i.e., cDNA, 9.1 kb; genomic DNA, >100 kb) (14, 15), we focused our mutational screening on those receptor regions associated with ligand binding (i.e., exons 27, 28, and 31) (6, 16) and previously shown to be mutated in other tumors (8, 9, 17). We found M6P/IGF2R missense mutations and single base deletions in 6/11 (55%) of HCCs with LOH (Tables 1 and 2). None of the observed mutations were found in the corresponding normal stromal tissue.
Table 2.
Mutation profile at the M6P/IGF2R locus in HCCs with LOH
| Region* | Codon change | Amino acid change | Secondary structure alteration† | Mutation frequency |
|---|---|---|---|---|
| Exon 27 | TGT → TCT | Cys-1262 → Ser | β-Sheeting ↓ | 1/11 (9%) |
| Exon 28 | G deletion in poly(G) repeat causing frameshift | 3/11 (27%) | ||
| Exon 31 | GGC → GTC | Gly-1449 → Val | β-Sheeting ↑ | 2/11 (18%) |
| Total 6/11 (55%) | ||||
Corresponding to the mouse gene (15).
The Chou–Fasman algorithm, macdnasis Pro version 3.2, Hitachi Software Engineering of America.
A G⋅C → C⋅G transversion that results in a Cys-1262 → Ser substitution was identified in 1/11 (9%) HCCs with LOH (Tables 1 and 2); the patient had HCV-induced liver cirrhosis (Table 1, case 24, patient 16). This mutation occurred in exon 27 of the M6P/IGF2R gene, which encodes a region of the receptor associated with phosphomannosyl glycoprotein binding (6, 16). The amino acid alteration is predicted by the Chou–Fasman algorithm to decrease β-sheeting. Additionally, Cys-1262 is conserved among human, bovine, rat, and mouse, suggesting a significant functional role for this amino acid.
Single G deletions were identified in a M6P/IGF2R poly deoxyguanosine [poly(G)] region of exon 28 in 3/11 (27%) HCCs with LOH (Tables 1 and 2, Fig. 2A). One patient had alcohol-induced cirrhosis (Table 1, case 4, patient 2), a second patient had HCV- and alcohol-induced cirrhosis (Table 1, case 17, patient 10), and the third patient had HCV-induced cirrhosis (Table 1, case 23, patient 15). The gene encoding the transforming growth factor β type II receptor was the first tumor suppressor gene demonstrated to be a target for microsatellite instability (MI) in replication/repair error-positive (RER+) colon tumors (18). The poly(G) region in the M6P/IGF2R gene was recently shown to likewise be a target of MI in RER+ endometrial, stomach, and colorectal tumors (17, 19). We did not determine if the HCCs with poly(G) deletions were RER+, but cirrhotic liver tissue associated with HV infection and alcohol consumption has been shown to have MI (20). Therefore, our findings suggest that the poly(G) region of the M6P/IGF2R may be a target of MI also in HCCs developing in patients with these clinical risk factors.
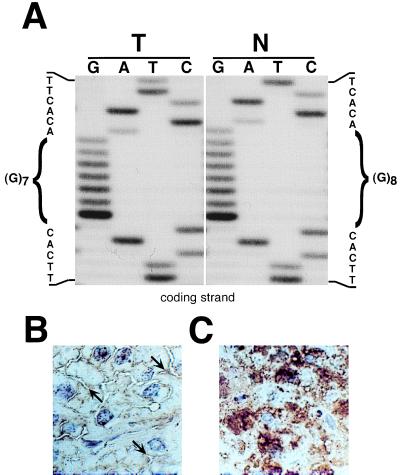
Figure 2.
Deoxyguanosine (G) deletion in a (G)8 repeat of the M6P/IGF2R gene in an HCC with LOH. (A) Direct sequencing of the PCR template derived from tumor (T) and normal stromal tissue (N) shows a single G deletion in exon 28 [based on the mouse gene (15)] of the tumor. (B) HCC cells with LOH and a G deletion are immunohistochemically negative for the M6P/IGF2R protein; however, the extracellular spaces are positively stained (arrows). (Counterstained with hematoxylin; ×275.) (C) Immunohistochemical staining for the M6P/IGF2R in normal hepatocytes demonstrates a high concentration of intracellular receptors. (Counterstained with hematoxylin; ×275.)
These single G deletions cause a frameshift resulting in the substitution of the stop codon TAA for the Leu-1343 codon. Truncation of the M6P/IGF2R protein at this position prevents the synthesis of the IGF2 binding domain, the transmembrane domain, and the intracellular lysosomal trafficking region, suggesting that if this truncated receptor is synthesized, it would be secreted (6, 16). Immunohistochemical staining for the M6P/IGF2R protein in a HCC with both LOH and a G deletion in the remaining allele shows a significant lack of the receptor (Fig. 2B) compared with normal hepatocytes (Fig. 2C). Immunohistochemical staining was also absent in another HCC (Table 1, case 24, patient 16) with inactivated M6P/IGF2R, but present in a HCC with normal M6P/IGF2R (Table 1, case 22, patient 14) (results not shown). These findings, coupled with those previously published (8), indicate that M6P/IGF2R gene inactivation in HCCs may be detectable by immunohistochemistry.
A G⋅C → T⋅A transversion that results in a Gly-1449 → Val substitution was identified in 2/11 (18%) HCCs with LOH (Tables 1 and 2). One of these patients had HCV-induced cirrhosis (Table 1, case 7, patient 4) and the other patient had alcohol-induced cirrhosis (Table 1, case 14, patient 8). This G⋅C → T⋅A transversion occurs in exon 31 of the M6P/IGF2R gene, which encodes a region of the receptor closely associated with both phosphomannosyl glycoprotein and IGF2 ligand binding (6, 16). The amino acid alteration is predicted by the Chou–Fasman algorithm to increase β-sheeting. Gly-1449 is also conserved among human, bovine, rat, and mouse, indicating functional importance of this amino acid. Interestingly, this mutation is identical to that previously identified in HCCs not associated with HV infection or cirrhosis. The overall frequency of this mutation is 13% in HCCs (8). This establishes the G⋅C → T⋅A transversion as a potential M6P/IGF2R mutational “hot spot” in human liver cancer.
Evidence suggests that dysplastic liver lesions are both clonal and precursors to HCCs (13, 21). To determine whether M6P/IGF2R inactivation occurs early in liver carcinogenesis, premalignant dysplastic lesions were screened for LOH (Fig. 1). Dysplastic lesions were defined as cirrhotic nodules in which hepatocytes showed enlarged pleomorphic nuclei and included both large and small cell types (22). Dysplastic hepatocytes were present in the form of atypical cirrhotic nodules that were large and often poorly demarcated. LOH was detected at the M6P/IGF2R locus in 5/8 (63%) premalignant dysplastic lesions (Table 1). The LOH frequency is not significantly different (P > 0.1) from that for HCCs (61%). This demonstrates that M6P/IGF2R allele inactivation occurs early rather than late in the pathogenesis of liver tumors.
Cirrhotic nodules have generally been regarded as hyperplastic regenerative lesions, but some also arise by clonal expansion (23–25). Therefore, we questioned whether the cirrhotic nodules adjacent to dysplastic lesions and HCCs had M6P/IGF2R allele loss, a genetic change observable only if the cirrhotic nodules were of clonal origin (Figs. 1, 3, and 4). A randomly chosen cirrhotic nodule in each liver tissue section that contained either a dysplastic lesion or an HCC was examined for M6P/IGF2R allele inactivation. M6P/IGF2R allele loss was observed in 9/10 (90%) cirrhotic nodules when the adjacent tumor lesions also had the M6P/IGF2R mutated. Furthermore, the same allele was inactivated in the cirrhotic nodule and tumor lesion. In contrast, the frequency of M6P/IGF2R allele loss (0/3) was significantly lower (P = 0.01) in the cirrhotic nodules when they were adjacent to tumor lesions in which the M6P/IGF2R was not mutated.
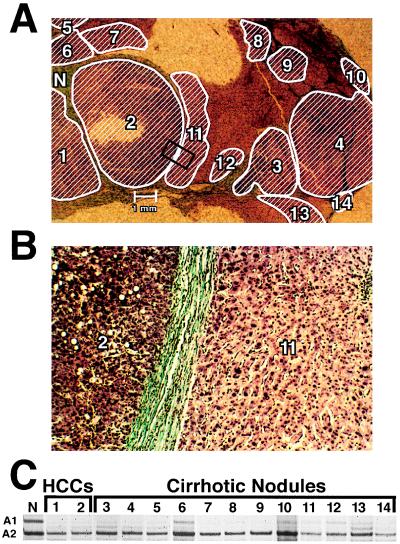
Figure 3.
LOH at the M6P/IGF2R locus in HCCs and adjacent cirrhotic nodules of an informative patient with a history of HCV infection. (A) LOH at the M6P/IGF2R locus is mapped in an HCC and adjacent cirrhotic nodules. (Mason’s stain; ×4.) Area within the rectangle is shown at higher magnification in B. Hatched areas have LOH at the M6P/IGF2R locus. (B) HCC region 2 and cirrhotic nodule 11 within the rectangle in A are shown at higher magnification. (Mason’s stain; ×58.) (C) Autoradiograph shows both allele 1 (A1) and allele 2 (A2) at the M6P/IGF2R locus in normal stromal tissue (N). In contrast, allele A1 is lost in HCC regions 1 and 2 and cirrhotic nodules 3–14; faint bands are due to contaminating normal stromal tissue.
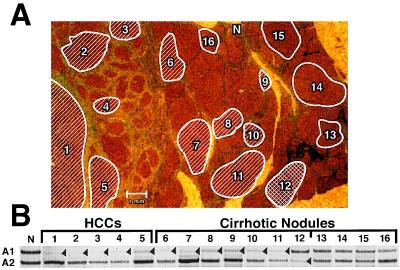
Figure 4.
LOH at the M6P/IGF2R locus in an HCC and adjacent cirrhotic nodules of an informative patient with a history of HBV and HCV infection. (A) LOH at the M6P/IGF2R locus is mapped in an HCC and adjacent cirrhotic nodules. (Mason’s stain; ×8.) Hatched and crosshatched areas have LOH at the M6P/IGF2R locus, while both alleles are present in those enclosed areas without hatch marks. (B) Autoradiograph shows both allele 1 (A1) and allele 2 (A2) at the M6P/IGF2R locus in normal stromal tissue (N). In contrast, allele A1 is lost in HCC regions 1–5 and cirrhotic nodules 6–11. Allele A2 is lost in cirrhotic nodule 12, while cirrhotic nodules 13–16 have both alleles present. Arrowheads mark the lost alleles; faint bands are due to contaminating normal stromal tissue.
To further these studies, the distribution of the cirrhotic nodules with M6P/IGF2R allele loss was mapped in four tissue sections. Two representative cases are shown in Figs. 3 and 4. The patient in Fig. 3 had HCV-induced liver cirrhosis (Table 1, case 7, patient 4). Twelve cirrhotic nodules (Fig. 3A) composed of phenotypically normal hepatocytes (Fig. 3B) adjacent to HCC regions 1 and 2 were assessed for LOH (Fig. 3C). All of the cirrhotic nodules examined had lost allele A1 (Fig. 3 A and C). Thus, each of these 12 cirrhotic nodules is a clonal neoplastic lesion, since both alleles of the M6P/IGF2R would be present if they were hyperplastic regenerative lesions. M6P/IGF2R allele loss in these cirrhotic nodules is also not random, since only allele A1 is missing. This is consistent with the 12 cirrhotic nodules arising from either a single or a very few progenitor cells lacking allele A1. Clonal cirrhotic nodules occurring in clusters, as in this patient, is consistent with the clonal expansion preceding the fibrous septa formation involved in the pathogenesis of cirrhosis (13, 25). The LOH pattern in these cirrhotic nodules is also identical to that of the associated HCC, indicating that the liver tumor developed from this clonal cirrhotic nodular tissue.
A more heterogeneous pattern of M6P/IGF2R allele loss is evident in the liver tissue from the HBV- and HCV-infected patient in Fig. 4 (Table 1, case 12, patient 7). Cirrhotic nodules 6–11 adjacent to HCC regions 1–5 lost allele A1, indicating that these nodules again arose from either a single or very few progenitor cells lacking allele A1. Cirrhotic nodule 12 is also a clonal lesion; however, since allele A2 rather than A1 is lost, it developed from a different precursor cell than cirrhotic nodules 6–11. Cirrhotic nodules 13–16 are distant from the HCC. They have retained both alleles of the M6P/IGF2R gene, demonstrating that these nodules are hyperplastic regenerative lesions rather than clonal neoplastic lesions. These results show that the HCC has the same M6P/IGF2R pattern of allele loss as the adjacent cirrhotic nodules. This again supports the postulate that the liver tumor arose from the clonal, M6P/IGF2R-mutated cirrhotic tissue. These results stress that M6P/IGF2R allele inactivation is a very early event in human liver carcinogenesis, occurring even prior to observable phenotypic changes in the hepatocyte.
When both M6P/IGF2R alleles are mutated in the HCCs, only one allele is inactivated in the adjacent clonal cirrhotic tissues. This implies that inactivation of a single M6P/IGF2R allele may be sufficient to provide a selective growth advantage to the mutated hepatocytes. Alternatively, M6P/IGF2R-mutated hepatocyte growth may be facilitated by HV factors inhibiting the normal M6P/IGF2R protein function in a manner similar to that observed with the HBV X transactivator and p53 (26). Finally, M6P/IGF2R allele inactivation may not alter hepatocyte growth potential, but rather clonal expansion of these mutated hepatocytes may occur simply because they fortuitously survived chronic HV infection and/or alcohol toxicity (27). Irrespective of the mechanism for the clonal growth of these M6P/IGF2R-mutated hepatocytes, the consequence of their proliferation is a greatly enlarged population of premalignant cells in which only a single genetic hit is required to completely inactivate M6P/IGF2R tumor suppressor function.
Liver is not the only organ in which regions of normal-appearing tissue have a clonal origin. Entire lobules and large ducts of normal breast tissue have been shown to be derived from a single progenitor cell (28). Furthermore, LOH at various chromosomal locations has been detected in morphologically normal lobules adjacent to breast tumors, implying there are localized regions of breast tissue at increased risk for transformation (29). Multiple head and neck tumors that develop in a localized region may also have a common clonal origin (30). Thus, the clonal expansion of normal-appearing but genetically altered cells has been documented in a number of tissues. This may explain the interesting phenomenon of multiple primary tumors arising in a localized region (i.e., “field cancerization”) initially described in 1953 by Slaughter and colleagues (31). Our results provide strong evidence that M6P/IGF2R plays a role in the formation of these clonal premalignant lesions in the liver. Involvement of M6P/IGF2R in this process in other tissues, such as the breast, is also possible, since this gene is lost frequently at an early stage in breast carcinogenesis (9).
Our finding that M6P/IGF2R inactivation is an early event in liver carcinogenesis may provide new and better approaches for both the diagnosis and treatment of liver cancer. These results may also have an impact on human carcinogen risk assessment based upon rodent studies, because M6P/IGF2R is genomically imprinted in mice (i.e., monoallelic expression) but not in most humans (i.e., biallelic expression) (for review see ref. 32). Inactivation of M6P/IGF2R would require two genetic events in humans but only one in mice. Thus, mice potentially may be more sensitive than humans to those carcinogens in which M6P/IGF2R is mechanistically involved in the transformational process.
In conclusion, this study has shown that the M6P/IGF2R gene is inactivated in both dysplastic liver lesions and HCCs associated with the high risk factors of HV infection and cirrhosis. The M6P/IGF2R gene is also often mutated in cirrhotic nodules of clonal origin adjacent to these lesions. These findings demonstrate that the M6P/IGF2R is lost frequently and early in human liver carcinogenesis, and they provide further evidence that M6P/IGF2R functions as a liver tumor-suppressor gene.
Acknowledgments
We thank R. Reddy and B. Carr for providing us with frozen human liver tumor samples, C. Scott for the polyclonal antibody to the M6P/IGF2R, and F. Kong for technical assistance with the immunohistochemical staining. This research was supported in part by National Institutes of Health Grants CA25951 and ES08823, Sumitomo Chemical Company, and Zeneca Pharmaceuticals.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HCC, hepatocellular carcinoma; HV, hepatitis virus; HBV, hepatitis B virus: HCV, hepatitis C virus; LOH, loss of heterozygosity; MI, microsatellite instability; M6P/IGF2R, mannose 6-phosphate/insulin-like growth factor II receptor.
References
- 1.Lotze M T, Flickinger J C, Carr B I. In: Cancer: Principles & Practice of Oncology. DeVita V T Jr, Hellman S, Rosenberg S A, editors. Philadelphia: Lippincott; 1993. pp. 883–914. [Google Scholar]
- 2.Groopman J D, Wogan G N, Roebuck B D, Kensler T W. Cancer Res. 1994;54:1907s–1911s. [PubMed] [Google Scholar]
- 3.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 4.Dennis P A, Rifkin D B. Proc Natl Acad Sci USA. 1991;88:580–584. doi: 10.1073/pnas.88.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Bleser P J, Jannes P, Van Buul-Offers S C, Hoogerbrugge C M, Van Schravendijk C F H, Niki T, Rogiers V, Van den Brande J L, Wisse E, Geerts A. Hepatology. 1995;21:1429–1437. doi: 10.1002/hep.1840210529. [DOI] [PubMed] [Google Scholar]
- 6.Kornfeld S. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 7.De Souza A T, Hankins G R, Washington M K, Fine R L, Orton T C, Jirtle R L. Oncogene. 1995;10:1725–1729. [PubMed] [Google Scholar]
- 8.De Souza A T, Hankins G R, Washington M K, Orton T C, Jirtle R L. Nat Genet. 1995;11:447–449. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 9.Hankins G R, De Souza A T, Bentley R C, Patel M R, Marks J R, Iglehart J D, Jirtle R L. Oncogene. 1996;12:2003–2009. [PubMed] [Google Scholar]
- 10.Brownlee K A. Statistical Theory and Methodology in Science and Engineering. New York: Wiley; 1967. pp. 163–166. [Google Scholar]
- 11.Rao P H, Murty V V V S, Gaidano G, Hauptschein R, Dalla-Favera R, Chaganti R S K. Cytogenet Cell Genet. 1994;66:272–273. doi: 10.1159/000133710. [DOI] [PubMed] [Google Scholar]
- 12.Hol F A, Geurds M P, Hamel B C, Mariman E C. Hum Mol Genet. 1992;1:347. doi: 10.1093/hmg/1.5.347. [DOI] [PubMed] [Google Scholar]
- 13.Theise N D. Semin Liver Dis. 1995;15:360–371. doi: 10.1055/s-2007-1007287. [DOI] [PubMed] [Google Scholar]
- 14.Morgan D O, Edman J C, Standring D N, Fried V A, Smith M C, Roth R A, Rutter W J. Nature (London) 1987;329:301–307. doi: 10.1038/329301a0. [DOI] [PubMed] [Google Scholar]
- 15.Szebenyi G, Rotwein P. Genomics. 1994;19:120–129. doi: 10.1006/geno.1994.1021. [DOI] [PubMed] [Google Scholar]
- 16.Dahms N M. Biochem Soc Trans. 1996;24:136–141. doi: 10.1042/bst0240136. [DOI] [PubMed] [Google Scholar]
- 17.Souza R F, Appel R, Yin J, Wang S, Smolinski K N, Abraham J M, Zou T-T, Shi Y-Q, Lei J, Cottrell J, Cymes K, Biden K, Simms L, Leggett B, Lynch P M, Frazier M, Powell S M, Harpaz N, Sugimura H, Young J, Meltzer S J. Nat Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]
- 18.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Willson J K V. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang H, Shiwaku H O, Hagiwara H, Miura K, Abe T, Kato Y, Ohtani H, Shiiba K, Souza R F, Meltzer S J, Horii A. Cancer Res. 1997;57:1851–1854. [PubMed] [Google Scholar]
- 20.Salvucci M, Lemoine A, Azoulay D, Sebagh M, Bismuth H, Reynes M, May E, Debuire B. Oncogene. 1996;13:2681–2685. [PubMed] [Google Scholar]
- 21.Aihara T, Noguchi S, Sasaki Y, Nakano H, Monden M, Imaoka S. Gastroenterology. 1996;111:455–461. doi: 10.1053/gast.1996.v111.pm8690212. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Okita K, Harada T, Kodama T, Numa Y, Takemoto T, Takahashi T. Cancer. 1983;51:2197–2205. doi: 10.1002/1097-0142(19830615)51:12<2197::aid-cncr2820511208>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Shafritz D A, Shouval D, Sherman H I, Hadziyannis S J, Kew M C. N Engl J Med. 1981;305:1067–1073. doi: 10.1056/NEJM198110293051807. [DOI] [PubMed] [Google Scholar]
- 24.Yasui H, Hino O, Ohtake K, Machinami R, Kitagawa T. Cancer Res. 1992;52:6810–6814. [PubMed] [Google Scholar]
- 25.Aihara T, Noguchi S, Sasaki Y, Nakano H, Imaoka S. Gastroenterology. 1994;107:1805–1811. doi: 10.1016/0016-5085(94)90824-9. [DOI] [PubMed] [Google Scholar]
- 26.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J, Harris C C. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knudson A G. J Cancer Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai Y C, Lu Y, Nichols P W, Zlotnikov G, Jones P A, Smith H S. Cancer Res. 1996;56:402–404. [PubMed] [Google Scholar]
- 29.Deng G, Lu Y, Zlotnikov G, Thor A D, Smith H S. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 30.Bedi G C, Westra W H, Gabrielson E, Koch W, Sidransky D. Cancer Res. 1996;56:2484–2487. [PubMed] [Google Scholar]
- 31.Slaughter D P, Southwick H W, Smejkal W. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 32.De Souza A T, Yamada T, Mills J J, Jirtle R L. FASEB J. 1997;11:60–67. doi: 10.1096/fasebj.11.1.9034167. [DOI] [PubMed] [Google Scholar]