Abstract
This study was originally conceived to test in a rigorous way the specificity of three major approaches to high-throughput array-based DNA methylation analysis: (1) MeDIP, or methylated DNA immunoprecipitation, an example of antibody-mediated methyl-specific fractionation; (2) HELP, or HpaII tiny fragment enrichment by ligation-mediated PCR, an example of differential amplification of methylated DNA; and (3) fractionation by McrBC, an enzyme that cuts most methylated DNA. These results were validated using 1466 Illumina methylation probes on the GoldenGate methylation assay and further resolved discrepancies among the methods through quantitative methylation pyrosequencing analysis. While all three methods provide useful information, there were significant limitations to each, specifically bias toward CpG islands in MeDIP, relatively incomplete coverage in HELP, and location imprecision in McrBC. However, we found that with an original array design strategy using tiling arrays and statistical procedures that average information from neighboring genomic locations, much improved specificity and sensitivity could be achieved, e.g., ∼100% sensitivity at 90% specificity with McrBC. We term this approach “comprehensive high-throughput arrays for relative methylation” (CHARM). While this approach was applied to McrBC analysis, the array design and computational algorithms are fractionation method-independent and make this a simple, general, relatively inexpensive tool suitable for genome-wide analysis, and in which individual samples can be assayed reliably at very high density, allowing locus-level genome-wide epigenetic discrimination of individuals, not just groups of samples. Furthermore, unlike the other approaches, CHARM is highly quantitative, a substantial advantage in application to the study of human disease.
The methylome is defined as the comprehensive picture of DNA methylation across the genome, and it is an important shift in focus from the individual gene level (Feinberg 2001). The rationale for this view is that our focus on methylation in the promoters of known genes is too constrained, that much of methylation is not where one looks. Despite introduction of the word “methylome” into the literature 6 yr ago, DNA methylation has made the least progress of any functional element in its understanding from a genomic perspective (Callinan and Feinberg 2006). This is ironic as DNA methylation is relatively well understood from a gene perspective, i.e., its method of propagation is well known, in comparison to chromatin modification, and DNA methylation has a strong link to the DNA sequence itself, i.e., encoding specifically at CpG dinucleotides, all of these much more so than other types of epigenetic information, such as chromatin modification.
Why has so little progress been made in understanding the methylome? Two major limitations may be responsible. First is a fundamental bias regarding the location of methylation modification in disease and even in studies of variation in tissues, i.e., largely restricted to “CpG islands,” and limitations in the detection methods themselves. Bird introduced the concept of a CpG island in 1987 (Bird et al. 1987), as regions of dense CpG content normally protected from DNA methylation in vertebrates but found frequently to be methylated in cancer (for review, see Esteller 2006). It has been widely believed that CpG island methylation is the most critical target for understanding genomic DNA methylation, although that island-centric view is undergoing rethinking (Jones and Baylin 2007). For example, binding sites for the insulator protein CTCF within differentially methylated regions of imprinted genes appear in short stretches of about 50 nucleotides, with a relatively conserved ∼20-bp core (e.g., Rosa et al. 2005). Thus, it is likely that other minimal units of DNA methylation will be smaller and of different GC content than densely GC-rich regions. Traditional approaches for DNA methylation analysis focused specifically on CpG islands may also miss sites important for topological conformation of DNA within the nucleus and gene regulation. For example, we earlier identified GC-rich regions that did not meet the definitional requirement of CpG island but are normally methylated and over-represented near the ends of chromosomes (Onyango et al. 2000).
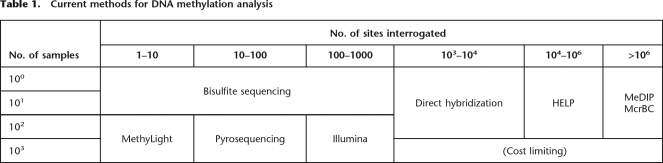
The second reason for the slow pace of understanding the methylome is substantial limitations in current technology affecting sensitivity, specificity, throughput, quantitation, and cost among the currently used detection methods. The most commonly used methods can themselves be divided into three categories (Table 1): (1) Bisulfite DNA sequencing. This involves chemical conversion of cytosine to uracil by sodium bisulfite or metabisulfite, followed by PCR (which incorporates T for U), and then DNA sequencing. While providing single-base resolution, the cost is the highest of all the commonly used methods, tens of thousands of dollars for a megabase of sequence data, itself comprising 40,384 CpG dinucleotides assayed (Eckhardt et al. 2006), and is therefore not currently suitable for whole-genome analysis on multiple samples. (2) A variety of methods that interrogate specific single-CpG dinucleotides or amplicons. These include MethyLight (Eads et al. 2000), COBRA (Xiong and Laird 1997), bisulfite pyrosequencing (Dupont et al. 2004), and the Illumina GoldenGate methylation assay (Bibikova et al. 2006). While sensitive, specific, and relatively inexpensive, none of these methods is suitable for analysis of the whole genome, which includes ∼28 million CpG dinucleotides. (3) Microarray-based methods. These can interrogate much larger numbers of CpG than the other approaches, at extremely low unit cost, since the pricing is similar to other non-methylation-based array methods.
Table 1.
Current methods for DNA methylation analysis
There are four major types of microarray-based methylation analysis. (1) Direct hybridization to CpG island arrays. This was one of the earliest methods; it was used to provide valuable data on tumor-type classification, for example (Gitan et al. 2002), and it still remains a useful discovery tool. However, its earliest developers have migrated away from this approach, since it requires presupposition about the potentially methylated sequences. (2) Methylated DNA immunoprecipitation (MeDIP), in which methylated DNA is fractionated using an antibody and then hybridized, with a differentially labeled total DNA control, to an oligonucleotide array (Weber et al. 2005). (3) Restriction enzyme digestion using methylcytosine-sensitive enzymes, followed by ligation-mediated PCR amplification of the targets. The paradigm of this method is the HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay (Khulan et al. 2006). DNA is digested in parallel with MspI (resistant to DNA methylation), and then the HpaII and MspI products are amplified by ligation-mediated PCR and hybridized using separate fluorochromes to a customized array. As HpaII sites comprise 8% of CpG, that represents a fixed limit of sensitivity of the method. Alternatively, the restriction enzyme-digested DNA can be directly sequenced rather than hybridized to microarrays (Allinen et al. 2004), although one is still limited by the relatively small number of methylcytosine-sensitive restriction sites in the genome. (4) Restriction enzyme digestion of methylated DNA using McrBC, without PCR, and differential hybridization to an array. DNA is digested with McrBC, an enzyme with the unusual and desirable property of cutting methylated DNA promiscuously (recognition sequence RmC(N)55–103RmC), cleaving half of the methylated DNA in the genome and all methylated CpG islands (Sutherland et al. 1992). The enzyme is used on size-selected (1.5–4.0 kb) DNA to fractionate unmethylated (i.e., gel-purified high molecular weight) DNA after digestion, which is comparatively (two-color) hybridized with DNA similarly processed but not cut with the enzyme, on high density arrays. The original method was developed for Arabidopsis (Lippman et al. 2004), where it has value in eliminating the large fraction of methylated repetitive DNA in the plant genome. For mammalian genome application, a selection algorithm has been applied to obtain specific array probes thought to represent the state of a given methylation target (Ordway et al. 2006).
Although all of the microarray approaches are in common use, they have not been directly compared to each other, and our original goals were relatively modest: to directly compare methods using the same DNA samples and the same arrays. However, we found significant limitations generally to hybridization-based methylation analysis that could largely be overcome with novel statistical procedures and array design algorithms. As will be described in the second portion of the paper, a fractionation method-independent approach, termed CHARM (comprehensive high-throughout arrays for relative methylation), can detect DNA genome-wide methylation with ∼100% sensitivity and 90% specificity.
Results
Overall design
Here we have designed a study to compare three array-based methylation detection technologies, MeDIP as an example of immunoprecipitation-based methods, McrBC fractionation as an example of restriction enzyme fractionation, and HELP as an example of differential methylcytosine sensitive ligation-mediated PCR. As our test samples, two paired cell lines were used: HCT116, a highly methylated colorectal carcinoma line, and a DNA methyltransferase I and 3B double-knockout cell line (DKO), with comparatively low levels of methylation (Rhee et al. 2002). These data were also compared to direct bisulfite methylation analysis using the Illumina GoldenGate methylation assay (Bibikova et al. 2006) on 1466 CpG sites in 466 genes.
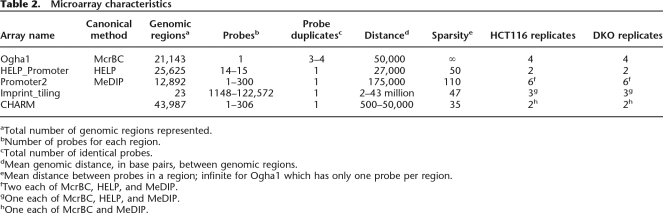
For all three assay types, design-specific arrays have already been designed, and we followed these designs, referred to here as canonical arrays. However, to enable direct comparison on the same arrays, samples were hybridized to NimbleGen’s Promoter 2 array and designed two tiling arrays (see Methods), which are referred to herein as common arrays. Note that in the case of MeDIP, one of the common arrays is the same as the canonical array, i.e., the Promoter 2 array. Because of the flexibility of design, the NimbleGen platform was used in all cases, which has also been used by the originators of these assay systems. In each case, a competitive hybridization approach was performed, in which samples were differentially labeled with Cy3 and Cy5 as described in the experimental protocols, specifically: (1) for MeDIP, methyl-enriched DNA with Cy5 and total DNA with Cy3; (2) for HELP, HpaII amplified with Cy5 and MspI with Cy3; and (3) for McrBC, methyl-depleted with Cy5, and total with Cy3. Note that McrBC dye-swaps were created as recommended by the original publication for mammalian DNA (Ordway et al. 2006). However, we found that the benefit of dye-swaps does not merit their extra cost (see Supplemental Fig. 1), thus the comparisons shown here do not include them. The complete list of comparative experiments and arrays is provided in Table 2.
Table 2.
Microarray characteristics
aTotal number of genomic regions represented.
bNumber of probes for each region.
cTotal number of identical probes.
dMean genomic distance, in base pairs, between genomic regions.
eMean distance between probes in a region; infinite for Ogha1 which has only one probe per region.
fTwo each of McrBC, HELP, and MeDIP.
gOne each of McrBC, HELP, and MeDIP.
hOne each of McrBC and MeDIP.
To decide among various strategies for measuring the same quantity, one looks to optimize sensitivity and specificity. Because specificity can be easily improved at the cost of sensitivity, and vice versa, one needs to assess both independently. We designed our experiments to assess sensitivity and specificity in the practical context of detecting methylated sites. To appropriately assess how experimental variability affects specificity, two technical replicates were performed for each method/sample-type pair (see Table 2). Measurements of methylation should be the same in both replicates, and deviation from equal values serves a measure of precision, which directly affects the specificity for measurements of methylation levels. The assessment of specificity was also facilitated by the use of the DKO samples. These provided many unmethylated sites useful for this assessment: Methods with low specificity will be more likely to call unmethylated sites as methylated. The HCT116 samples permitted a comprehensive assessment of sensitivity as many sites were methylated: Methods with high sensitivity will be more likely to call methylated sites as methylated (true positives). The Illumina GoldenGate assay was used as a reference against which all microarray methods were compared.
Quantification of methylation measurements
For each microarray hybridization, we used the raw feature intensities to form log ratios and denoted these with M, as done in most of the microarray literature (Allison et al. 2006). The M-values were formed so that larger values represented more evidence of methylation, e.g., with MeDIP, the immunoprecipitate intensity was in the numerator and the total DNA intensity in the denominator. Note that each feature on the array was associated with one M-value. Each array was then normalized so that unmethylated regions, on average, produced M-values of 0. Details of the normalization technique are available in the Methods section. The Illumina GoldenGate platform quantifies methylation as a percentage. However, the raw data files report the Cy3 and Cy5 intensities related to the unmethylated and methylated pseudoalleles, thus, M-values were formed in a similar way.
Note that M is a continuous variable, so that methylation could be assessed in a quantitative way, which has not been performed previously for array-based methylation analysis. This is critical for biological analysis, since epigenetic information is often chromosome-specific, e.g., imprinted genes. Furthermore, DNA methylation may have a threshold effect for regulating gene expression, e.g., ∼25% for E-cadherin in a broad range of cell types (Reinhold et al. 2007). Note that transforming M directly into estimates of absolute methylation is not straightforward. However, later in this section we demonstrate that by simply using cut-off values we obtain a strategy with high sensitivity and specificity.
MeDIP is comparatively imprecise
We first assessed the precision of each method, by comparing M-values from replicate arrays, specifically studying the distribution of the differences between replicated M-values: M1i − M2i where i represents a feature, and 1 and 2 represent the two replicate hybridizations. In principle, these values should all be 0, since M1i and M2i were measures of the same quantity. However, as expected, differences were observed due to natural variation in the sample preparation and array hybridization. These differences were studied using the canonical arrays for each method, because each method was likely optimized on their canonical arrays and we wanted to see each method at its optimal condition in addition to the common arrays.
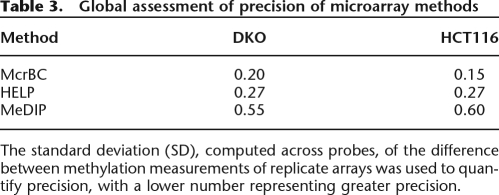
The standard deviation (SD) of these differences, taken across probes, is a useful summary that relates directly to the range of M-values one should expect from samples with no difference in methylation status. For McrBC, the standard deviations (SDs) were 0.20 and 0.15 for the DKO and HCT116 samples, respectively (Table 3). For the HELP method, the SD was 0.27 for both samples. Finally, the MeDIP showed the worst precision with SDs of 0.55 and 0.60 for the DKO and HCT116 samples, respectively (Table 3). These results were for the M-values obtained from the canonical arrays. A graphical assessment of precision is shown in Supplemental Figure 2.
Table 3.
Global assessment of precision of microarray methods
The standard deviation (SD), computed across probes, of the difference between methylation measurements of replicate arrays was used to quantify precision, with a lower number representing greater precision.
McrBC and HELP can discriminate DKO from HCT116
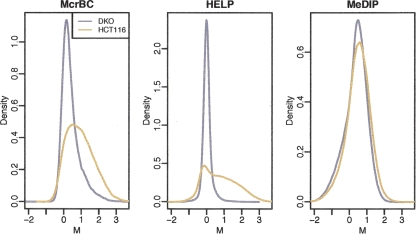
A global assessment of sensitivity was performed by comparing the distribution of the M-values from the HCT116 and DKO samples, i.e., a highly methylated and a highly unmethylated reference sample, respectively. Thus the expected M-values for DKO sample should mostly be centered at 0, and HCT116 should be shifted to a substantial number of positive values. Figure 1 demonstrates that the MeDIP method can barely distinguish between the two cell lines of differing methylation on a global scale, although at individual loci differences are clearly seen (discussed below). The McrBC and HELP arrays perform better at globally distinguishing the DKO from the HCT116 sample, with HELP to a somewhat greater degree.
Figure 1.
Density estimates (smoothed histograms) of the M-values comparing DKO (gray) and HCT116 (brown) samples. Note that the display of the overall M distribution masks differences at individual sites.
Site-specific comparison of methods
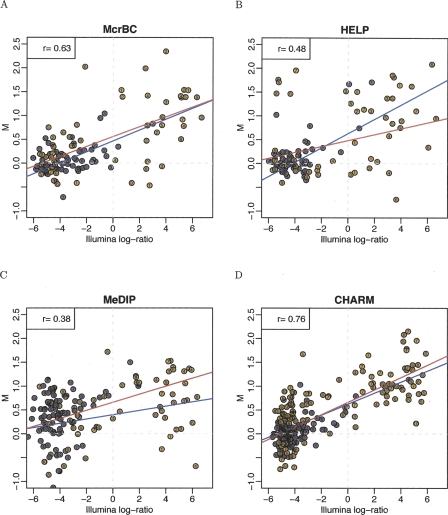
The ability to distinguish sample methylation globally is not nearly as important as the ability to detect methylation at high genomic resolution. We therefore compared the performance of each method at the individual CpG level using the Illumina platform as reference standard, based on studies from us and others (Bibikova et al. 2006; Ladd-Acosta 2007), as well as data from this study shown in Supplemental Figure 3. For each method/array combination, each CpG assayed by the Illumina platform was matched to one M-value obtained from the microarray data. We now describe how this mapping was obtained for each method. For the McrBC method, we predicted the start and end of the resulting genomic segments after cutting at every ACG or GCG. These are referred to as the McrBC segments. For each probe on the Illumina platform representing an ACG or GCG we assigned two segments: those ending and starting on that CG. Next, for each probe on the microarray, we determined which McrBC segment contained it. Finally, the median M-value for all the microarray probes mapped to each Illumina probe was assigned as the microarray M-value. The McrBC canonical arrays used three to four replicate probes for 21,143 locations, as recommended by those authors (Ordway et al. 2006). Thus, at least four probes were used with each Illumina probe. A similar approach was used for the HELP method except the cleavage occurs at CCGG sites (Khulan et al. 2006). The HELP canonical arrays used 14–15 tiled probes in each of the HELP segments. The canonical arrays for MeDIP were the Promoter 2 arrays, which represent 12,892 promoter regions. We matched every CpG inside these promoter regions to the closest probe also in that region. We were then able to map CpGs represented by an Illumina probe, and included in one of the promoter regions, to one microarray probe M-value. Figure 2 demonstrates this comparison, using 587, 57, 51, and 1188 Illumina CpGs corresponding to specific CpGs on the MeDIP, McrBC, HELP, and CHARM arrays, respectively. The set of CpG covered by all platforms was too small to provide meaningful results, thus we based our comparisons on the different sets mapped by the different array types.
Figure 2.
Comparison of method-specific methylation measurements to reference data. For the HCT116 (brown) and DKO (gray) samples, M-values from high-throughput methods are plotted against M-values from the Illumina reference platform. To illustrate the CpG observed-to-expected ratio, a 500-bp window was formed around each probe; this ratio (multiplied by 10) is displayed inside each point. A regression line was calculated and is displayed for probes with ratios <0.6 (blue line) and >0.6 (red line).
Sensitivity of HELP and MeDIP depends greatly on the CpG content
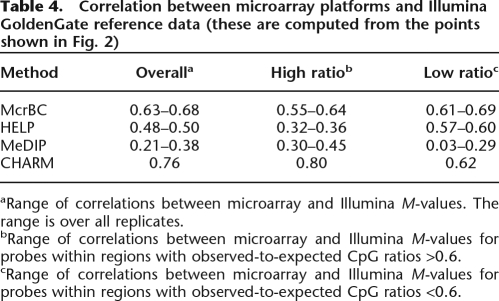
Figure 2 plots M-values from each of the microarray platforms against the corresponding M-values obtained from the Illumina platform. Values from the HCT116 and DKO samples were combined. For clarity, in Figure 2, data are shown from one HCT116 and one DKO array for each method. Results for all other arrays, i.e., the replicates, are similar and are shown in Supplemental Figure 4. Figure 2 stratifies points by CpG density. The observed-to-expected ratio for 500-bp regions was computed around each microarray probe shown in Figure 2 (ratios are denoted with color and with a small number inside each point). In this window we defined the expected number of CpGs as the proportion of Cs multiplied by the proportion of Gs. The observed-to-expected ratio is simply the proportion of CpGs divided by the expected proportion of CpGs. Notice that the traditional definition of a CpG island requires this ratio to be >0.6. The probes were stratified into two groups: low CpG density (ratio ≤ 0.6) and high CpG density (ratio > 0.6). A regression line was fitted to each group (shown as red and blue lines for the low- and high-density groups, respectively). The correlation between Illumina M-values and microarray M-values is shown in Table 4. While McrBC showed similar sensitivity for both high- and low-density groups, HELP showed better sensitivity for the lower CpG density group than for the higher CpG density group.
Table 4.
Correlation between microarray platforms and Illumina GoldenGate reference data (these are computed from the points shown in Fig. 2)
aRange of correlations between microarray and Illumina M-values. The range is over all replicates.
bRange of correlations between microarray and Illumina M-values for probes within regions with observed-to-expected CpG ratios >0.6.
cRange of correlations between microarray and Illumina M-values for probes within regions with observed-to-expected CpG ratios <0.6.
Severe bias in current methods related to segment characteristics
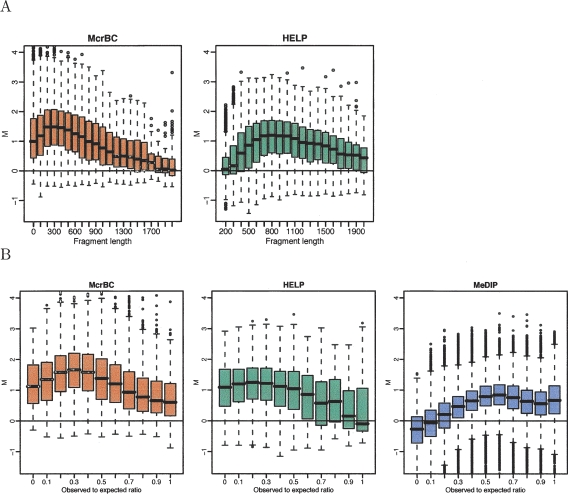
For HCT116 samples, we stratified the M-values obtained from the McrBC and HELP canonical arrays by segment size to produce Figure 3A. Because in this sample one expects many methylated CpGs, many large M-values are expected independent of the segment size. However, the strata related to large and small fragments had substantially fewer large M-values than the middle-sized segments. Notice in particular that the HELP method had no sensitivity for CpGs associated with segments smaller than 300 bp. The McrBC method had no sensitivity for CpGs associated with segments larger than 1500 bp. Best results were observed for segments of sizes 200–600 and 700–1200 bp for McrBC and HELP, respectively. The segment sizes for MeDIP are unpredictable, thus, this method was not included in this figure.
Figure 3.
DNA fragment-length–related biases. (A) M-values for the HCT116 sample are stratified by the DNA fragment size predicted by the McrBC (left panel) and HELP (right panel) enzyme digestions. (B) For all three methods, a 500-bp window was formed around each probe, the observed-to-expected ratio of CpG was calculated, and box-plots of the M-values are displayed by these ratios. Only probes related to fragments of sizes between 50 and 600 bp for McrBC, and between 600 and 1200 bp for HELP, are included.
We also assessed the effect of CpG density with this stratification approach. As in Figure 2, we formed a 500-bp segment around the location of each probe and calculated the observed-to-expected ratio. These were then stratified by their observed-to-expected ratio (Fig. 3B). As first noticed in Figure 2, the HELP method has low sensitivity for high CpG density and the MeDIP method had low sensitivity for low CpG densities.
General limitations in single-CpG accuracy substantially improved by genome-weighted smoothing
Figure 2 also demonstrates that, at the individual CpG level, the agreement between microarray and Illumina reference measurements leaves much room for improvement. Notice that even for the best performing microarray-based method, McrBC, the variability seen in the microarray M-values suggests that none of the methods will be useful in practice if one uses individual probe level data or individual segment data. In particular, notice that a substantial number of the M-values for the CpGs called methylated by the reference standard (Illumina, right of the dashed vertical line) are in the same range as most of the M-values called unmethylated by the reference standard (Illumina, left of the dashed vertical line), i.e., between −0.75 and 0.75 on the Y-axis.
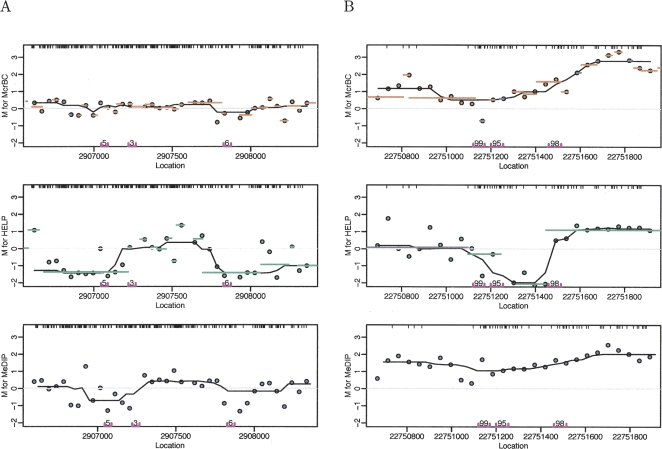
The fact that the methylation status of neighboring CpGs tends to be highly correlated (Eckhardt et al. 2006) motivated our introduction of a novel strategy for methylation analysis of genome-weighted smoothing: averaging probes within small contiguous genomic regions taking into account the biases illustrated in Figure 3. A novel aspect of our approach is that we combine information derived from the genome sequence with microarray data. By characterizing each of the segments induced by laboratory protocols, one can quantify the utility of the associated microarray data. This information is then used to adapt the averaging used in the smoothing step by assigning weights. Details on our novel smoothing strategy are provided in the Methods section. The canonical arrays designed for the McrBC and HELP methods use multiple array features to probe a selected subset of the McrBC and HELP segments described above. These segments in the canonical designs are not contiguous, thus smoothing is not possible with data from these arrays. Therefore, to enable genome-weighted smoothing, we hybridized the samples using each of the methods, not only to their canonical arrays but also to the common arrays defined in Table 2, namely, the Promoter 2 and Imprinting arrays. Figure 4, A and B, shows the resulting M-values for a highly unmethylated region and a highly methylated region, respectively (actual methylation status was determined by the Illumina reference method).
Figure 4.
M values plotted against contiguous locations on the genome for all three methods. The points are the observed M-values. The M-values for probes in the same predicted segments for McrBC and HELP were averaged and are represented in the figure with orange and green lines, respectively. The data were smoothed using running medians with a window size of 7 and showed the results with black curves. CpG locations are shown as black tick marks at the top of the plots. (A) Segment showing lack of methylation determined by the Illumina platform. (B) Segment with high methylation as determined by the Illumina platform. The Illumina probes and measured methylation percentages are shown on the bottom of the plot.
Figure 4 demonstrates the advantage of genome-weighted smoothing. In this figure, M-values are plotted against location on the genome. The points are the M-values observed for each probe. The averaged M-values for probes in the same McrBC and HELP segments are shown with orange and green lines for McrBC and HELP, respectively. The results obtained using genome-weighted smoothing (described above) are shown with black curves. Note that for the McrBC and MeDIP methods, the range of the probe-level and segment M-values associated with unmethylated (Fig. 4A) and methylated (Fig. 4B) regions overlap; the results from smoothing do not. For example, for McrBC the segment M-values range from −0.75 to 0.5 and from −0.75 to 3 for the unmethylated and methylated regions, respectively. The values obtained from smoothing range from −0.2 to 0.25 and from 0.6 to 2.5 for the unmethylated and methylated regions, respectively. The averaging performed in the smoothing procedure greatly reduces noise, and the fact that the averaging is local, i.e., performed in small regions, permits us to preserve the ability to discriminate. Supplemental Figure 5 shows examples of various other regions illustrating the value of this approach.
The HELP method sometimes produced contradictory results at the same loci that were not apparent in the canonical design but were easy to see in the common array design (Fig. 4; Supplemental Fig. 5). This likely explained the lack of agreement with the reference method (Fig. 2). Because the HELP segments are small for the region shown in Figure 4, this result was expected, as Figure 3 demonstrates that the HELP method is not sensitive for small fragments. Supplemental Figure 5 shows several other examples.
CHARM, comprehensive high-throughput arrays for relative methylation
Based on the data described above, and in particular the importance of genome-weighted smoothing and array design, we have developed a novel platform for array-based DNA methylation analysis. The new method is independent of platform, and it combines the design of a novel array design and statistical procedures that perform genome-weighted averaging from neighboring genomic locations. More details are provided below.
The first component of our method is a new tiling array specifically designed to maximize the number of assayed CpGs. For the reasons stated above, we did not want to restrict our attention to CpG islands. Instead, the number of CpGs assayed, for which we could reliably detect methylation status, were maximized. For example, because we rely on smoothing, isolated CpGs were not assayed. A careful analysis of different numbers of probes included in the smoothing demonstrated that at least 15 probe intensities were needed to obtain useful results (data not shown). The procedure for creating the array was as follows:
We identified all the CpGs in the genome. Any region of 300 bp with no CpGs was discarded.
We removed probes with multiple matches, including fuzzy matches as defined by NimbleGen (http://www.nimblegen.com/products/chip/index.html), to the genome.
Any region having a gap of ≥300 bp between consecutive probes was divided into two new regions.
We discarded any region with fewer than 15 probes.
We tiled regions as possible, using 50-mers 35 bp apart. One can also prioritize for economy to limit to a single array by calculating the ratio of CpGs per probes in the region and by assigning higher priority to those with a higher ratio.
This array design would improve the detection strategy for any of the methods because it facilitates the smoothing strategy and assays many more CpGs. Probes associated with problematic segments (e.g., very small segments in the HELP assay) could be removed in the analysis stage. However, we selected McrBC for the application of this approach because of its superior sensitivity and specificity described earlier. Going forward, samples were also hybridized using the CHARM design as well as the MeDIP assay as well. We did not continue to use the HELP assay mainly because of its limited number of detectable sites (HpaII dependence).
To detect methylated regions in the CHARM method, the M-values were normalized, as described in the Methods section, and processed using genome-weighted smoothing, as described above. Figure 2D shows the smoothed M-values obtained from CHARM plotted against the reference M-values. Comparing Figure 2D with Figure 2, A–C, demonstrates how CHARM greatly improved the results obtained with the other methods.
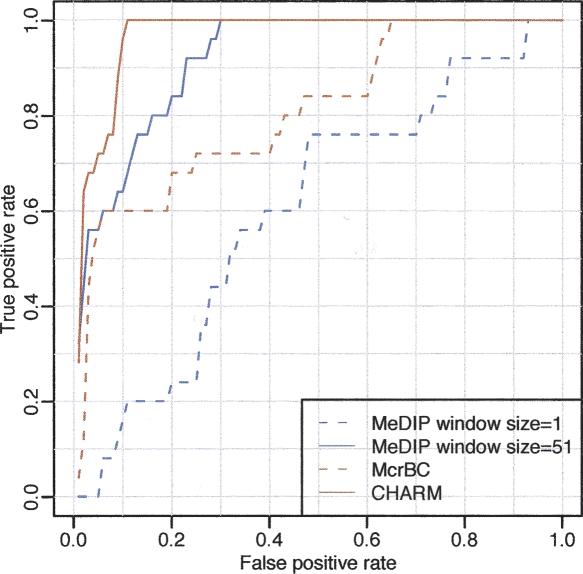
Although it is potentially useful to treat methylation state as a continuous variable (Rakyan et al. 2004), the state of individual CpGs is strongly bimodal. Therefore, besides comparing quantitative results among methods, it is also important to also determine the ability to discriminate highly methylated from highly unmethylated sequences, a common question in molecular biology, e.g., in generating lists of candidate genes subject to epigenetic regulation or alteration in disease. This binary classification also enabled us to construct receiver operating characteristic (ROC) curves (Fig. 5). ROC curves plot the sensitivity vs. (1 − specificity) for a binary classifier system (methylated or not) as discrimination thresholds (values of M) are varied. For this purpose, a genomic region was defined as “methylated” if all probes from the Illumina platform in the region were >90%. Similarly, unmethylated regions were defined as those with all probes <10%; 100 Illumina probe sets fulfilled these criteria. If the smoothed M-value within any of these regions was above a predetermined threshold, the region was considered methylated. Various thresholds were considered, and each defines a point in the ROC curve. The results greatly improved with CHARM. Notice that for a specificity of 90%, the McrBC sensitivity improved from 60% without CHARM to 100% with CHARM.
Figure 5.
ROC curve demonstrating the advantage of genome-weighted smoothing. We considered all gene regions represented on the Illumina platform. For the purpose of ROC calculation, highly methylated and unmethylated regions were compared. If all probes in the region showed on the reference Illumina platform a methylation percentage >90%, the region was considered a true positive. If all probes in the region reported a percentage <10% they were considered a true negative. To define a positive from the microarray data using a window size of 1, a cut-off for the M-values was chosen. If any probe intensity within the region was above that cut-off, it was defined as positive. A running median with a window size of 51 was then analyzed and defined a positive in the same way, except that the smoothed results instead of the individual probe intensities were used. Results are shown for both McrBC and MeDIP. For a given threshold, the true-positive rate is defined as the percentage of true-positive regions for which the microarray data surpasses that threshold. The false-positive rate is defined in the same way but for the true-negative regions.
Finally, we note that the CHARM method, unlike MeDIP, HELP, or nonsmoothed McrBC, is highly quantitative, meaning that there was a linear relationship between methylation measured on the array and the reference methylation platform (Illumina), as shown clearly in Figure 2. The correlation coefficient comparing these two values was substantially better for CHARM compared to the other methods (Table 4), as was the ROC curve (Fig. 5).
Discussion
In summary, there are two major results of this work. First, we have shown that there are substantial limitations to all three commonly used approaches for array-based DNA methylation analysis. In the case of MeDIP, the assay is of relatively worse specificity, and the method is not sensitive, particularly outside of CpG islands. HELP, while accurately distinguishing markedly different cell types globally, does not cover many CpG dinucleotides because of the dependence on HpaII restriction sites and often shows lack of agreement with the reference method. Of the three approaches, McrBC performed the best, but as seen in the ROC curves, the sensitivity was only 60% at 90% specificity as previously practiced. Second, since neighboring CpGs have been shown to be closely correlated, we developed a novel genome-weighted smoothing algorithm to measure methylation from raw microarray data. Combining this novel approach with the most robust method for fractionating methylated DNA (McrBC), we designed custom arrays ideally suited for methylation detection, as defined in the Results section. This approach is termed “comprehensive high-throughput arrays for relative methylation” (CHARM). CHARM offers the possibility of relatively inexpensive genome-wide analysis with high precision and accuracy. On the NimbleGen HD2 arrays, 2.1 million features can be studied in this way. The approach was data-driven, in that it used an independent assessment of 1466 CpG sites. Furthermore, the genome coverage on the array is genome sequence-driven, rather than based on arbitrary assumptions about the likely location of methylated sites (e.g., promoters) that might miss substantial numbers of regulatory sequences. Even with this unbiased, non-promoter-driven selection strategy, 87% of the Illumina-selected methylation cancer panel 1 genes are present on the HD2 array.
What were the likely inherent limitations of MeDIP and HELP shown by these experiments? The results obtained with the MeDIP method barely distinguished the HCT116 and DKO samples. A likely reason is that the immunoprecipitation (IP) step is not specific, i.e., unmethylated CpGs pass the filter of IP. This is consistent with the observation that detection was biased toward very high CpG content. Furthermore, note that the IP sample will be enriched with CpGs regardless of the number of segments that pass the filter. This is likely to result in cross-hybridization problems, e.g., probes with more CpGs might result in higher intensities only because of cross-hybridization with the high CpG content sample. In expression arrays it has been shown that background noise, such as cross-hybridization, greatly reduces sensitivity in cases were nominal amounts of target are low (Irizarry et al. 2006). If the same phenomenon is true in methylation arrays, then one would expect low sensitivity in regions with a small number of CpGs (low amount of target) as seen in the MeDIP arrays.
While HELP outperformed other methods in distinguishing the highly methylated HCT116 from the relatively unmethylated DKO globally, at the single-CpG level, the HELP method performed barely better than MeDIP. A possible explanation for this apparent contradiction is that the HELP method depends upon differences in ability of a fragment to be amplified, but the PCR step does not always amplify as expected. For example, in dense CpG regions, the smaller pieces, which are expected to amplify, might be too small for the PCR to work properly. Evidence that this phenomenon is occurring is the fact that the microarray data for HELP sometimes appears flipped in plots, such as in Figure 4: fragments were methylation-amplified opposite from the expected. It is important to note that the canonical design for HELP carefully selects regions where this phenomenon is unlikely to occur. But as mentioned, this greatly limits the coverage of the method. More sophisticated post-processing algorithms have been and likely will be further developed to correct for measurement discrepancies (Khulan et al. 2006). However, even with minimal post-processing as done here, one can obtain very good concordance with reference measurements, as seen in Figure 2, and more importantly good sensitivity and specificity at detecting methylation sites as seen in the ROC curves in Figure 5 and Supplemental Figure 6. CHARM also circumvents a major problem for genomic methylation analysis, namely, the importance of detecting changes in regions of relatively low CpG content. As shown here, MeDIP cannot detect methylation in these regions. The limitations for HELP in these regions is that it is more likely to miss these regions and that one cannot smooth the data to obtain high precision, since by definition one does not observe data from adjacent segments.
McrBC fractionation was originally applied to analysis of the plant genome (Martienssen et al. 2005) and subsequently used as a discovery tool for methylated CpG islands (Ordway et al. 2006), but it has not come into common use. The original whole-genome array design represented a few thousand segments with one probe each. In microarrays and other technologies that use hybridization, a large component of the variability seen across measures from different probes is due to sequence effects (Wu and Irizarry 2005). Averaging replicate probes does not reduce this variability as they have the same sequence. Therefore the precision is relatively low in data provided by a single probe replicated four times, as performed by Ordway et al. (2006). McrBC fractionation also suffers to some degree within CpG islands in the ability to discriminate highly methylated from highly unmethylated sequences, although it still outperforms MeDIP which works only within CpG islands (Supplemental Fig. 6).
Future work on CHARM includes the development of preprocessing algorithms that correct for sequence and segment effects. The resulting methods should improve the performance of CHARM within CpG islands. Finally, we note that while CHARM offers state-of-the-art, cost-effective methylation analysis, ∼1/20 penny per measurement, second-generation sequencing will reduce the relative utility of arrays generally, particularly when the goal of a $1000 genome is met. Currently, complete genome coverage by second-generation sequencing, e.g., after bisulfite treatment, would cost hundreds of times this amount per sample. An alternative is fractionation of the DNA followed by sequencing, reducing the complexity of the target genome, as has been done for specific chromatin marks (Mikkelsen et al. 2007). These approaches hold considerable promise, but they also raise the interesting question of what to capture. The data shown here indicate that great care must be used in the capture strategy, and the CHARM assay is an important step toward that. In addition, the capture strategy also uses hybridization, so one must still deal with hybridization-related biases until complete whole-genome sequencing becomes inexpensive.
Methods
Cell culture and genomic DNA isolation
HCT116 cells (American Type Culture Collection) and DNMT1/DNMT3B (DKO) cells (Rhee et al. 2002) were cultured in McCoy’s 5A modified medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. Genomic DNA was isolated from HCT116 and DKO cell lines and was prepared using the MasterPure DNA purification kit (EpiCentre) as specified by the manufacturer.
McrBC assay sample preparation
Genomic DNA (10 μg) was prepared, and McrBC digestion and gel fractionation were performed exactly as published (Ordway et al. 2006). As shown in Table 2, HCT116 and DKO samples prepared using McrBC were analyzed on the Ogha1 array (canonical), Promoter 2 array (common), Imprinting array (common), and the novel CHARM array.
HELP assay sample preparation
HCT116 and DKO samples were prepared as previously described by Khulan et al. (2006), in the laboratory of John Greally, to avoid any issues of technical handling (Albert Einstein College of Medicine, New York), using a total of 20 μg per sample (Khulan et al. 2006). The LM-PCR products were labeled with Cy3- or Cy5-conjugated oligonucleotide and random primers as previously described (Selzer et al. 2005). As shown in Table 2, HCT116 and DKO samples prepared using HpaII and MspI were analyzed on the HELP promoter array (canonical), Promoter 2 array (common), and Imprinting array (common).
MeDIP assay sample preparation
MeDIP assay was conducted according to published methods (Weber et al. 2005). As shown in Table 2, HCT116 and DKO samples prepared using MeDIP were analyzed on the Promoter 2 array (canonical), Imprinting array (common), and CHARM (common) array. As a positive control, MeDIP was validated using real-time PCR of Sat2 (Jiang et al. 2004).
Illumina GoldenGate assay sample preparation
Bisulfite conversion of 500 ng of genomic DNA was achieved through use of the EZ DNA Methylation-Gold kit (Zymo Research). All HCT116 and DKO samples were processed as described previously on the Illumina GoldenGate methylation cancer panel I (Bibikova et al. 2006). A β-value of 0–1.0 was reported signifying percent methylation, from 0% to 100%, respectively, for each CpG site. β-values were calculated by subtracting background using negative controls on the array and taking the ratio of the methylated signal intensity against the sum of both methylated and unmethylated signals.
Quantitative methylation analysis using pyrosequencing
One microgram of genomic DNA was bisulfite treated using the EpiTect kit (Qiagen) according to the manufacturer’s recommendations. Bisulfite treatment of genomic DNA results in unmethylated cytosine nucleotides being changed to thymidine while methylated cytosines remain unchanged. This difference is then detected as a C/T nucleotide polymorphism at each CpG site.
CpG-unbiased primers were designed to PCR amplify 38, 16, and 14 CpG sites, respectively, in three genes, HLA-F, KCNK4, and HLTF (previously known as SMARCA3), showing conflicting methylation across MeDIP, McrBC, and Illumina assays (Supplemental Table 1). Nested PCR was performed under standard conditions. Amplicons were analyzed on a PSQ HS 96 pyrosequencer as specified by the manufacturer (Biotage) and CpG sites quantified, from 0% to 100% methylation, using Pyro Q-CpG software.
Microarray design
Ogha1 is the canonical array for the McrBC method: 21,143 McrBC segments are represented by one probe each. Three to four replicate are used for each probe. The locations of these segments were chosen by the designer based on transcriptional start sites and CpG islands as described in their paper (Ordway et al. 2006). The HELP promoter array is the canonical array for the HELP method. The Promoter 2 array is one of NimbleGen’s off-the-shelf products, with 12,892 promoter regions. The Imprint_ tiling array represents 23 regions chosen by our group to study imprinted genes. Region sizes ranged from 133,475 to 13,096,022 bp, and probes of size 50 bp were tiled at 47 bp from each other with an occasional large jump to avoid repeat elements. Table 2 provides a summary of these arrays.
Data analysis
Normalization
As described above, the basic measurement used to quantify methylation is the log-ratio of the intensities observed in the treated and control channels. Existing statistical methods have used the unadjusted log-ratios, assumed linear dye log-scale effect and removed these effects by simply subtracting the median, and fitting and removing the effect within an ANOVA model (Kerr and Churchill 2001). These methods fail to correct for the strong nonlinear effects seen in M versus A plots. In expression arrays, Loess normalization has been widely and successfully used to solve this problem (Yang et al. 2002). The basic idea in this procedure is to assume that for most probes genes are not expressed and M = 0 and that A-dependent deviations are a smooth function. The bias is estimated and removed using Loess regression. This approach has been successful in expression arrays. However, because in methylation experiments we expect many sites to be methylated, one can no longer make this assumption. If Loess normalization were used here, and we define M = 0 as the average value of unmethlyated sites, then one would incorrectly force M = 0 for many of the probes associated with methylation.
We therefore developed a method that does not require the assumption that M = 0 for most probes. This method used genome sequence information and our knowledge of the fragment selection method to select what we call pseudo-housekeeping probes for which one can in fact assume M = 0. We then apply the Loess normalization procedure developed for expression arrays to the pseudo-housekeeping genes, obtain the correction curve, and use this curve to correct M-values for all probes. An additional advantage of this approach is that it provides a flexible way to adapt existing techniques, developed for expression arrays, to methylation data. Details of this normalization procedure are described in Bolstad et al. (2003).
Genome-weighted smoothing
To obtain a smoothed M-value at any given genomic location, we average all the M-values that were within a prespecified distance from the location in question. A weighted average was used with the weights determined from the results presented in Figure 3. Notice that this part of the procedure is method-dependent. The interval providing the values that are averaged is referred to as the “smoothing window” and its length is referred to as the “window size.” Many smoothing algorithms exist, each one averaging in a different way, e.g., assigning distance-dependent weights. For the results presented in this paper, the running median algorithm (Tukey 1977; Hardle and Steiger 1995) was used, with a window size of 51 probes (∼1500 bp), due to its simplicity and robustness to outliers.
Acknowledgments
This work was supported by NIH grant HG003233. We thank John Greally and Masako Suzuki for performing the HELP ligation and amplification steps to ensure that we did not perform this procedure incorrectly.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7301508.
References
- Allinen M., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Beroukhim R., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Cai L., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Brennan C., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Lahti-Domenici J., Huang H., Porter D., Hu M., Chin L., Richardson A., Huang H., Porter D., Hu M., Chin L., Richardson A., Porter D., Hu M., Chin L., Richardson A., Hu M., Chin L., Richardson A., Chin L., Richardson A., Richardson A., et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Allison D.B., Cui X., Page G.P., Sabripour M., Cui X., Page G.P., Sabripour M., Page G.P., Sabripour M., Sabripour M. Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Bibikova M., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Doucet D., Thomas N.J., Wang Y., Vollmer E., Thomas N.J., Wang Y., Vollmer E., Wang Y., Vollmer E., Vollmer E., et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.P., Taggart M.H., Nicholls R.D., Higgs D.R., Taggart M.H., Nicholls R.D., Higgs D.R., Nicholls R.D., Higgs D.R., Higgs D.R. Non-methylated CpG-rich islands at the human alpha-globin locus: Implications for evolution of the alpha-globin pseudogene. EMBO J. 1987;6:999–1004. doi: 10.1002/j.1460-2075.1987.tb04851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P., Irizarry R.A., Astrand M., Speed T.P., Astrand M., Speed T.P., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Callinan P.A., Feinberg A.P., Feinberg A.P. The emerging science of epigenomics. Hum. Mol. Genet. 2006;15:R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- Dupont J.M., Tost J., Jammes H., Gut I.G., Tost J., Jammes H., Gut I.G., Jammes H., Gut I.G., Gut I.G. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W., Danenberg K.D., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W., Blake C., Shibata D., Danenberg P.V., Laird P.W., Shibata D., Danenberg P.V., Laird P.W., Danenberg P.V., Laird P.W., Laird P.W. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:e32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Burton J., Cox T.V., Davies R., Down T.A., Cox T.V., Davies R., Down T.A., Davies R., Down T.A., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br. J. Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P. Methylation meets genomics. Nat. Genet. 2001;27:9–10. doi: 10.1038/83825. [DOI] [PubMed] [Google Scholar]
- Gitan R.S., Shi H., Chen C.M., Yan P.S., Huang T.H., Shi H., Chen C.M., Yan P.S., Huang T.H., Chen C.M., Yan P.S., Huang T.H., Yan P.S., Huang T.H., Huang T.H. Methylation-specific oligonucleotide microarray: A new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardle W., Steiger W., Steiger W. Algorithm AS 296: Optimal median smoothing. Appl. Stat. 1995;44:258–264. [Google Scholar]
- Irizarry R.A., Wu Z., Jaffee H.A., Wu Z., Jaffee H.A., Jaffee H.A. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;22:789–794. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- Jiang G., Yang F., Sanchez C., Ehrlich M., Yang F., Sanchez C., Ehrlich M., Sanchez C., Ehrlich M., Ehrlich M. Histone modification in constitutive heterochromatin versus unexpressed euchromatin in human cells. J. Cell. Biochem. 2004;93:286–300. doi: 10.1002/jcb.20146. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr M.K., Churchill G.A., Churchill G.A. Experimental design for gene expression microarrays. Biostatistics. 2001;2:183–201. doi: 10.1093/biostatistics/2.2.183. [DOI] [PubMed] [Google Scholar]
- Khulan B., Thompson R.F., Ye K., Fazzari M.J., Suzuki M., Stasiek E., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Thompson R.F., Ye K., Fazzari M.J., Suzuki M., Stasiek E., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Ye K., Fazzari M.J., Suzuki M., Stasiek E., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Fazzari M.J., Suzuki M., Stasiek E., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Suzuki M., Stasiek E., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Stasiek E., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Figueroa M.E., Glass J.L., Chen Q., Montagna C., Glass J.L., Chen Q., Montagna C., Chen Q., Montagna C., Montagna C., et al. Comparative isoschizomer profiling of cytosine methylation: The HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C.P.J., Sabunciyan S., Yolken R., Webster M., Dinkins T., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P., Sabunciyan S., Yolken R., Webster M., Dinkins T., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P., Yolken R., Webster M., Dinkins T., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P., Webster M., Dinkins T., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P., Dinkins T., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P., Fan J.-B., Potash J.B., Feinberg A.P., Potash J.B., Feinberg A.P., Feinberg A.P. DNA methylation signatures within the human brain. Am. J. Hum. Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Lavine K., Mittal V., May B., Kasschau K.D., Mittal V., May B., Kasschau K.D., May B., Kasschau K.D., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Martienssen R.A., Doerge R.W., Colot V., Doerge R.W., Colot V., Colot V. Epigenomic mapping in Arabidopsis using tiling microarrays. Chromosome Res. 2005;13:299–308. doi: 10.1007/s10577-005-1507-2. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Alvarez P., Brockman W., Kim T.K., Koche R.P., Brockman W., Kim T.K., Koche R.P., Kim T.K., Koche R.P., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango P., Miller W., Lehoczky J., Leung C.T., Birren B., Wheelan S., Dewar K., Feinberg A.P., Miller W., Lehoczky J., Leung C.T., Birren B., Wheelan S., Dewar K., Feinberg A.P., Lehoczky J., Leung C.T., Birren B., Wheelan S., Dewar K., Feinberg A.P., Leung C.T., Birren B., Wheelan S., Dewar K., Feinberg A.P., Birren B., Wheelan S., Dewar K., Feinberg A.P., Wheelan S., Dewar K., Feinberg A.P., Dewar K., Feinberg A.P., Feinberg A.P. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 2000;10:1697–1710. doi: 10.1101/gr.161800. [DOI] [PubMed] [Google Scholar]
- Ordway J.M., Bedell J.A., Citek R.W., Nunberg A., Garrido A., Kendall R., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Bedell J.A., Citek R.W., Nunberg A., Garrido A., Kendall R., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Citek R.W., Nunberg A., Garrido A., Kendall R., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Nunberg A., Garrido A., Kendall R., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Garrido A., Kendall R., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Kendall R., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Stevens J.R., Cao D., Doerge R.W., Korshunova Y., Cao D., Doerge R.W., Korshunova Y., Doerge R.W., Korshunova Y., Korshunova Y., et al. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27:2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- Rakyan V.K., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Andrews T.D., Howe K.L., Otto T., Olek A., Howe K.L., Otto T., Olek A., Otto T., Olek A., Olek A., et al. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold W.C., Reimers M.A., Maunakea A.K., Kim S., Lababidi S., Scherf U., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Reimers M.A., Maunakea A.K., Kim S., Lababidi S., Scherf U., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Maunakea A.K., Kim S., Lababidi S., Scherf U., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Kim S., Lababidi S., Scherf U., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Lababidi S., Scherf U., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Scherf U., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Shankavaram U.T., Ziegler M.S., Stewart C., Kouros-Mehr H., Ziegler M.S., Stewart C., Kouros-Mehr H., Stewart C., Kouros-Mehr H., Kouros-Mehr H., et al. Detailed DNA methylation profiles of the E-cadherin promoter in the NCI-60 cancer cells. Mol. Cancer Ther. 2007;6:391–403. doi: 10.1158/1535-7163.MCT-06-0609. [DOI] [PubMed] [Google Scholar]
- Rhee I., Bachman K.E., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Bachman K.E., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., Feinberg A.P., Lengauer C., Kinzler K.W., Lengauer C., Kinzler K.W., Kinzler K.W., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Rosa A.L., Wu Y.Q., Kwabi-Addo B., Coveler K.J., Reid Sutton V., Shaffer L.G., Wu Y.Q., Kwabi-Addo B., Coveler K.J., Reid Sutton V., Shaffer L.G., Kwabi-Addo B., Coveler K.J., Reid Sutton V., Shaffer L.G., Coveler K.J., Reid Sutton V., Shaffer L.G., Reid Sutton V., Shaffer L.G., Shaffer L.G. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res. 2005;13:809–818. doi: 10.1007/s10577-005-1015-4. [DOI] [PubMed] [Google Scholar]
- Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Nair P., Brothman A.R., Stallings R.L., Brothman A.R., Stallings R.L., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Sutherland E., Coe L., Raleigh E.A., Coe L., Raleigh E.A., Raleigh E.A. McrBC: A multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- Tukey J. Exploratory data analysis. Addison-Wesley; Reading, MA: 1977. [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Haase M., Lam W.L., Schubeler D., Lam W.L., Schubeler D., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Irizarry R.A. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- Xiong Z., Laird P.W., Laird P.W. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Lin D.M., Peng V., Ngai J., Speed T.P., Peng V., Ngai J., Speed T.P., Ngai J., Speed T.P., Speed T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]