Abstract
Mycobacterium marinum, a ubiquitous pathogen of fish and amphibia, is a near relative of Mycobacterium tuberculosis, the etiologic agent of tuberculosis in humans. The genome of the M strain of M. marinum comprises a 6,636,827-bp circular chromosome with 5424 CDS, 10 prophages, and a 23-kb mercury-resistance plasmid. Prominent features are the very large number of genes (57) encoding polyketide synthases (PKSs) and nonribosomal peptide synthases (NRPSs) and the most extensive repertoire yet reported of the mycobacteria-restricted PE and PPE proteins, and related-ESX secretion systems. Some of the NRPS genes comprise a novel family and seem to have been acquired horizontally. M. marinum is used widely as a model organism to study M. tuberculosis pathogenesis, and genome comparisons confirmed the close genetic relationship between these two species, as they share 3000 orthologs with an average amino acid identity of 85%. Comparisons with the more distantly related Mycobacterium avium subspecies paratuberculosis and Mycobacterium smegmatis reveal how an ancestral generalist mycobacterium evolved into M. tuberculosis and M. marinum. M. tuberculosis has undergone genome downsizing and extensive lateral gene transfer to become a specialized pathogen of humans and other primates without retaining an environmental niche. M. marinum has maintained a large genome so as to retain the capacity for environmental survival while becoming a broad host range pathogen that produces disease strikingly similar to M. tuberculosis. The work described herein provides a foundation for using M. marinum to better understand the determinants of pathogenesis of tuberculosis.
In 1926, Joseph D. Aronson isolated a Mycobacterium from tubercles observed predominantly in the spleen and liver of diseased fish that had died in the Philadelphia Aquarium and named it Mycobacterium marinum (Aronson 1927). M. marinum was subsequently shown to also be a human pathogen when it was isolated again much later in a swimming pool-associated outbreak of human granulomatous skin lesions, although in this report the Mycobacterium was mistakenly given a new species name, Mycobacterium balnei (Linell and Norden 1954), a name that is no longer used.
Several phylogenetic studies have shown that M. marinum is most closely related to Mycobacterium ulcerans (>97% nucleotide identity) and then M. tuberculosis (>85% nucleotide identity) (Tønjum et al. 1998; Stinear et al. 2000; Devulder et al. 2005; Gey van Pittius et al. 2006; Yip et al. 2007). Multi-locus sequence typing (MLST) and the recently completed M. ulcerans genome sequence clearly show that M. ulcerans has recently evolved from a M. marinum progenitor by acquisition of a virulence plasmid and reductive evolution (Stinear et al. 2004, 2007; Yip et al. 2007). M. ulcerans causes the devastating human skin disease Buruli ulcer, which is quite distinct in its pathology from other mycobacterial diseases, primarily because of the production from its virulence plasmid of mycolactone, an immunosuppressive polyketide toxin restricted to M. ulcerans (George et al. 1999).
M. marinum infection of humans, called fish tank or aquarium tank granuloma, typically occurs when M. marinum is inoculated through the skin by cuts and scratches following direct contact with an infected fish or contaminated aquatic environments (Petrini 2006). The ensuing granulomatous infection generally limited to the skin and soft tissues extremities is pathologically indistinguishable from M. tuberculosis dermal disease (Travis et al. 1985; MacGregor 1995). Like tuberculosis, fish tank granuloma can resolve spontaneously but frequently requires long-term antibiotic treatment owing to the infecting bacteria developing phenotypic antibiotic resistance in vivo (Aubry et al. 2000, 2002). M. marinum disease can be effectively treated with the anti-tuberculosis agents rifampin, ethambutol, and the quinolones, as well as other agents such as doxycyline and clarithromycin (Aubry et al. 2000, 2002). The systemic granulomatous disease caused by M. marinum in its poikilothermic hosts, for example, frogs and fish, also shares key histological features with human tuberculosis (Pozos and Ramakrishnan 2004; Swaim et al. 2006). For instance, zebrafish develop organized caseating granulomas that resemble active human tuberculosis (Swaim et al. 2006). The two organisms share virulence determinants, and M. tuberculosis virulence genes complement orthologous M. marinum genes with mutations (Cosma et al. 2003, 2006; Gao et al. 2004; Guinn et al. 2004; Volkman et al. 2004; Bhatt et al. 2007a). Because of these findings and the relative ease of its manipulation, M. marinum infection of its natural host species is used as a model to study tuberculosis (Saad et al. 1999; Davis et al. 2002; Cosma et al. 2003, 2004, 2006; Gao et al. 2003, 2004, 2006; Volkman et al. 2004; Swaim et al. 2006; Clay et al. 2007).
M. marinum has both similar and distinct growth and physiological properties as compared to M. tuberculosis. Its optimal growth temperature when grown in Middlebrook 7H9 medium is 35°C, and it grows poorly at 37°C (Clark and Shepard 1963). Its lower optimal growth temperature likely explains its causing systemic disease in poikilotherms and a superficial disease, restricted cooler extremities of the body, in warm-blooded animals (Clark and Shepard 1963). At its optimal growth temperature, it grows relatively rapidly with a generation time of 4–6 h versus >20 h for M. tuberculosis. Like M. tuberculosis, it is a facultative anaerobe and demonstrates a diverse metabolic repertoire in vitro, but in contrast to M. tuberculosis, it is unable to reduce nitrate and produces characteristic bright yellow carotenoid pigments when exposed to light (Tsukamura 1983). These photochromogenic pigments protect it from UV damage in incident sunlight by reducing singlet oxygen species. Indeed, in contrast to M. tuberculosis, which has no environmental reservoirs, M. marinum has been found in various aquatic environments around the world, including swimming pools and drinking water (Dailloux et al. 1980; Slosarek et al. 1994; Toma 1998; Leoni et al. 1999; Bardouniotis et al. 2003; Hall-Stoodley et al. 2006). It can form biofilms (Hall-Stoodley et al. 2006) and replicate within protozoan hosts such as Dictyostelium discoidieum (Solomon et al. 2003; Hagedorn and Soldati 2007) and Acanthamoeba polyphaga (Adekambi et al. 2006) as well as cultured mammalian macrophages (Ramakrishnan and Falkow 1994).
These observations suggest that M. marinum has the characteristics of both a specialist bacterium, adapted to persist within intracellular environments, such as free-living aquatic amoeba and vertebrate hosts, as well as of a generalist, equipped to survive under changing and sometimes extracellular environmental conditions (Stamm and Brown 2004). A dichotomous lifestyle was first proposed by the seminal transmission studies of Clark and Shepard (1963) when they observed that fish, parenterally infected with M. marinum, provided a source of water-borne mycobacteria that could, in turn, infect tadpoles.
In contrast, M. tuberculosis has become highly specialized for intracellular survival in a very restricted range of mammalian hosts, and several recent studies have shown that lateral gene transfer has been a major force in the evolution of the M. tuberculosis complex from an environmental Mycobacterium (Kinsella et al. 2003; Gutierrez et al. 2005; Rosas-Magallanes et al. 2006; Becq et al. 2007). Here, we have determined the genome sequence of M. marinum and then exploited its very close genetic relationship with M. tuberculosis to provide further insight into the multiple changes that shaped M. tuberculosis into a major human pathogen from environmental origins.
Results
General features of the M. marinum genome
Multi-locus sequence typing (MLST) among 26 diverse M. marinum strains from around the world has revealed significant intraspecies sequence homogeneity, with <3% nucleotide variation present between all M. marinum STs (Stinear et al. 2000; Yip et al. 2007). Nine distinct sequence types (ST) were identified, with the “M” strain representing an intermediate ST, designated ST22, and this strain was chosen for sequence analysis.
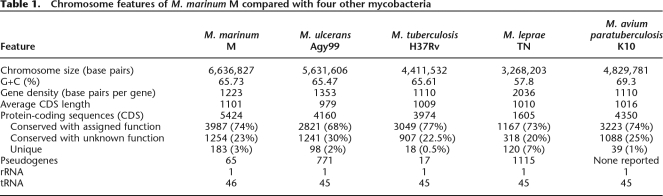
The M. marinum genome is composed of a single circular chromosome of 6,636,827 bp with an average G/C content of 62.5%, 5424 predicted coding sequences (CDS), 65 pseudogenes, 46 tRNA genes, a single rRNA operon, and a 23-kb plasmid named pMM23 (Table 1; Fig. 1A,B). Functional information could be assigned to 74% of all CDS, 23% encode conserved hypothetical proteins, and 3% were unique (Table 1). The chromosome harbors seven distinct, low-copy-number insertion sequences, named ISMyma01–07 (Supplemental Table 1), and 10 putative prophages, named phiMmar01–10 (Supplemental Table 2). GC skew analysis demonstrated a bias in G+C content between the leading and lagging strands (Fig. 1A), indicating a probable origin of replication around dnaA (Lobry 1996). AT skew analysis was also used and revealed some significant sequence biases (Fig. 1C), detected to a lesser extent in M. ulcerans and M. tuberculosis and absent from other mycobacteria. These sequence deviations are caused by the presence of PE-PGRS and nonribosomal peptide synthase (NRPS) genes, with their distinctive codon usage, repeating functional domains and sequence motifs (Supplemental Fig. 1). The plasmid encodes 29 predicted CDS, including replication and partition proteins, along with a 1110-amino acid relaxase/helicase that may function in conjugation or mobilization. The plasmid also contains a functional mercury-resistance operon (M. Schue, L.G. Dover, G.S. Besra, J. Parkhill, and N.L. Brown, unpubl.), flanked by two genes encoding members of the serine-site-specific recombinase family; it is possible that these may confer independent mobility on this element. Analysis by PCR of the distribution of pMM23 among 42 diverse M. marinum isolates representing each of the nine M. marinum MLST genotypes revealed that only the M strain contains this replicon (data not shown).
Table 1.
Chromosome features of M. marinum M compared with four other mycobacteria
Figure 1.
(A) Circular representation of the M. marinum chromosome. The scale is shown in megabases in the outer black circle. Moving inward, the next two circles show forward- and reverse-strand CDS, respectively, with colors representing the functional classification. (Red) Information pathways; (light blue) regulation; (light green) hypothetical protein; (dark green) cell wall and cell processes; (orange) conserved hypothetical protein; (cyan) IS elements; (yellow) intermediate metabolism; (gray) lipid metabolism; (purple) PE/PPE. The location of each copy of a PE or PPE gene is then shown (purple). The following two circles show forward- and reverse-strand genes that are not found in other mycobacteria (colors represent the functional classification), followed by the G+C content and finally the GC skew (G–C)/(G+C) using a 20-kb window. (B) Circular representation of the M. marinum mercury-resistance plasmid pMM23, following the same color scheme as the chromosome map but with a different scale in kilobase pairs as indicated. (C) AT skew analysis generated with Gene-Spaghetti, a method for visualizing DNA composition. The colors reflect AT skew (T – A)/(T + A). (Red) Extreme = 60% T-rich; (blue) extreme = 60% A-rich. The software uses a sliding Gaussian window to estimate local base usage (http://www.vicbioinformatics.com/genespaghetti.shtml). Outlier locus_tags were represented by three types of CDS; (blue) NRP; (gray) PE-PGRS; (red) PPE.
Central metabolism, respiration, and gene regulation
As identified from other mycobacterial genomes, M. marinum has intact glycolysis and pentose phosphate pathways but no Entner-Doudoroff pathway and a bifurcated TCA cycle (Tian et al. 2005). The expanded gene families encoding the key enzymes of the beta-oxidation pathway suggest that, like M. tuberculosis, exogenous fatty acids are important for the growth of M. marinum. FadD (acyl-CoA synthase) catalyzes the formation of acyl-CoA thioesters that are then processively oxidized by FadE, EchA, and FadA (Cole et al. 1998). In M. marinum, 32 fadD paralogs were predicted compared with 23 in M. tuberculosis. There are 46 paralogous fadE genes compared with 34 in M. tuberculosis. There are also multiple copies of other key enzymes in the cycle including 35 EchA and nine FadA proteins (compared with 22 and six, respectively, in M. tuberculosis).
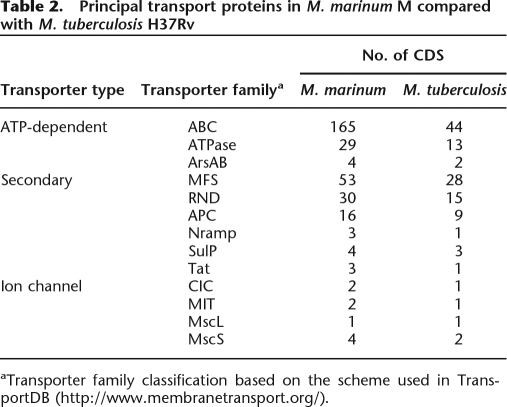
In addition to fatty acids, M. marinum is predicted to use a diverse catabolic repertoire including compounds like pyruvate, glucose, succinate, and acetate as sole carbon sources. This is also supported by 192 CDS predicted to be associated with carbon substrate transport. Overall, M. marinum has at least double the content of every class of transporter compared with M. tuberculosis (Table 2). There is a predominance of ATP-dependent transporters represented by 165 predicted ABC-type, 21 P-type, and eight F-type ATPase family members.
Table 2.
Principal transport proteins in M. marinum M compared with M. tuberculosis H37Rv
aTransporter family classification based on the scheme used in TransportDB (http://www.membranetransport.org/).
Based on gene orthology with M. tuberculosis, complete NADH oxidase, ubiquinol cyctochrome c oxidase, and ATP synthase complexes indicate ATP generation by oxidative phosphorylation during aerobic growth. In M. tuberculosis, anaerobic growth is facilitated by the nitrate and fumarate reductase systems; however, M. marinum lacks the corresponding genes but contains a potential alternative anaerobic respiratory pathway involving nitrite reductase (NrfD) and formate dehydrogenase (MMAR_5194, MMAR_5195), where formate oxidation is coupled to nitrite reduction. These observations are in agreement with the species-defining nitrate reductase negative phenotype of M. marinum. Formate dehydrogenase is a selenoprotein, and its gene is linked to a selenocysteine synthase locus (MMAR_5190). Selenoenzymes are often associated with anaerobic respiratory processes but have not previously been reported in mycobacteria. There are seven putative NarK nitrite transporters, and nitrite is a common component of aquatic ecosystems.
A versatile respiratory potential under aerobic conditions is suggested in M. marinum by more than 590 putative oxidoreductases, dehydrogenases, mono-, and dioxygenases. The cytochrome P450 repertoire is particularly large with 47 P450 monooxygenases, some of which may play a role in specific hydroxylation of certain metabolites, while others will probably be involved in targeted oxidation of different xenobiotics encountered by the bacterium.
Versatility is also reflected in the large number of CDS (330) predicted to be involved in gene regulation, including the response to environmental change, such as those for the 17 sigma factors, 16 complete two-component regulatory systems, and 24 serine–threonine protein kinases (Wehenkel et al. 2007).
Cell wall lipids
Some predictions from the genome regarding the metabolism of key structural lipids of M. marinum have been described previously (Onwueme et al. 2005; Stinear et al. 2007). M. marinum has all the components of the type I and II fatty acid synthase complexes that are central to the production of cell wall mycolates. Similarly, M. marinum has the five type I polyketide synthases (PKS), encoded by the genes ppsA-E, that are required for the production of phenolphthiodiolones, which can be further modified by glycosylation to form the virulence factors and immune modulators, the phenolic glycolipids (PGLs) (Daffe et al. 1992; Reed et al. 2004). Refer to Onwueme et al. (2005) for a detailed analysis of the biosynthesis of PGLs and related molecules in M. marinum.
Recent studies have shown that M. marinum also produces four distinct but highly related species of lipooligosaccharide (LOS), the biosynthesis of which has been linked to the presence of a locus between MMAR_2307 and MMAR_2320 (Burguiere et al. 2005; Ren et al. 2007). LOS production appears important for sliding motility and biofilm formation in M. marinum. Genome comparisons suggest that the equivalent region in M. tuberculosis has become nonfunctional via successive DNA deletions and has continued to decay in Mycobacterium bovis, where the region of difference, RD4, was identified (Brosch et al. 1998; Brosch et al. 2002).
Polyprenyl phosphate (Pol-P) forms the lipid backbone of the sugar-linked intermediates that are required by bacteria during cell wall biosynthesis. Pol-P is formed by sequential condensation of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). In other mycobacteria, including M. tuberculosis, these molecules are synthesized via the non-mevalonate (DXP) pathway. However, M. marinum contains both an intact mevalonate (MVA) and DXP pathway for polyprenoid synthesis, the first example of such in the mycobacteria (Rohdich et al. 2004), although M. ulcerans has a remnant, nonfunctional version of the MVA pathway. Comparative genomics suggests that the 11-kb region spanning the MVA locus was deleted from a progenitor of M. tuberculosis via the activity of IS1558 (Rv2177c).
Secondary metabolism
Little is known of the secondary metabolites produced by M. marinum; however, genome inspection suggests that it has the potential to produce a diverse array of compounds with 34 CDS predicted to encode PKSs. In addition to the pps locus (ppsA–E) discussed above, three other PKSs have M. tuberculosis orthologs of known function, namely, pks12 (MMAR_3025) required for mycoketide production (Matsunaga et al. 2004); pks15/1 (MMAR_1762), encoding a free-standing PKS module required for production of phenolphthiocerol (Constant et al. 2002); and pks13 (MMAR_5364), encoding another free-standing PKS module required for mycolic acid synthesis (Portevin et al. 2004). M. marinum also has the pks10, 7, 8, 9 & 11 locus, encoding a PKS of unknown function, whose predicted module and domain structure are identical to that of M. tuberculosis, where it is essential (Sassetti et al. 2001).
Among the other predicted PKSs, two systems are worthy of comment—MMAR_1180 and the MMAR_3796–MMAR_3800 system. Neither of these has a strict ortholog in other sequenced mycobacterial genomes, although the second module of MMAR_1180 and several of the MMAR_3796–MMAR_3800 modules have strong homology with the mycolactone PKS from M. ulcerans. These PKSs comprise modules that are more closely related to Streptomyces PKSs than other mycobacterial PKSs, which suggests that they might encode small molecule products that are novel among known mycobacteria.
M. marinum also has 14 CDS encoding predicted fatty acyl-AMP ligases (FAAL), six of which are shared with M. tuberculosis (FadD25, 26, 28, 29, 30, 32). FAALs are a family of FadD-like enzymes that have been shown in M. tuberculosis to adenylate long-chain fatty acids to facilitate further chain extension by PKS (Trivedi et al. 2004). They promote biochemical cross-talk between distinct FAS and PKS loci, thereby increasing metabolite diversity (Gokhale et al. 2007).
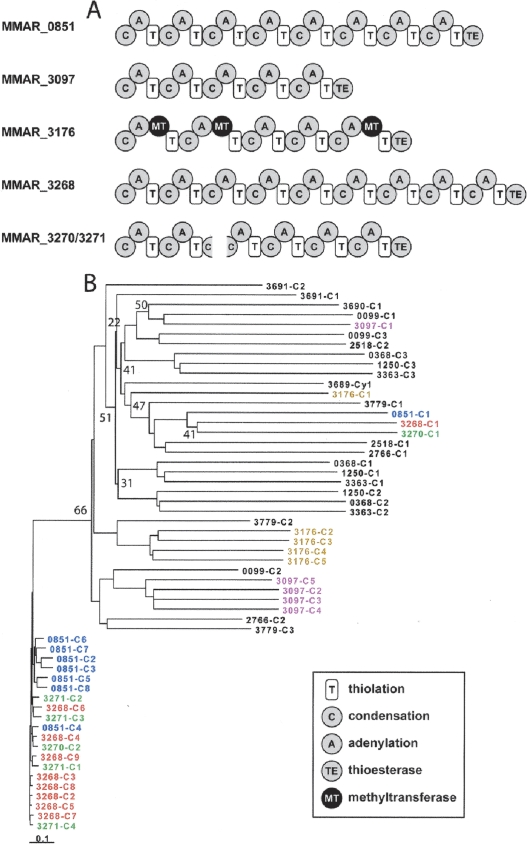
A most prominent feature of the M. marinum genome is its distinctive array of large CDS encoding 23 nonribosomal peptide synthases (NRPSs), 18 of which cannot be ascribed a precise function as they have not been studied and there are no strictly orthologous genes in other mycobacteria. NRPSs are modular enzymes that assemble peptidic small molecules in the fashion of an assembly line: Each module is responsible for inserting a single building block (usually an amino acid) into the growing peptide chain. Thus, the sequence of protein modules in an NRPS determines the composition of the small molecule it produces (Fischbach and Walsh 2006). The characteristics of all the NRPSs identified in M. marinum are summarized in Supplemental Figure 2, whereas Figure 2 depicts the domain arrangement of some novel NRPSs that will be discussed further here, together with their phylogeny.
Figure 2.
(A) Module and domain architecture of selected NRPSs. (B) Inferred phylogeny using the Neighbor-Joining method (Saitou and Nei 1987) of selected M. marinum condensation domains based on aligned predicted amino acid sequences. Different colors highlight condensation domains within an operon. Replicate trees in which the associated domains clustered together on <80% of occasions in a bootstrap test (10,000 replicates) are indicated. The scale is the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set.
The NRPS encoded by MMAR_3268, comprising 9859 amino acids arranged in nine modules (Fig. 2A), is the largest protein in the M. marinum proteome, with the second being another, highly similar, NRPS, MMAR_0851, at 8760 amino acids and eight modules. These NRPSs are, in turn, closely related to two other NRPSs, the products of MMAR_3270 and MMAR_3271. With the exception of the first condensation (C) domains of MMAR_3268, MMAR_0851, and MMAR_3270, all of the other C domains are ∼88% identical at the amino acid level, and the cognate modules appear to have arisen from duplication events, in a similar manner to that seen for the mycolactone PKS in M. ulcerans (Stinear et al. 2004). From the phylogeny of the M. marinum C domains (Fig. 2B), it can be seen that there is a distinct clade (refer to the lower end of Fig. 2B) representing the C domains from these four NRPS. This is consistent with a model in which both a mixture of gene duplication and intragene module duplication have contributed to the evolution of these gene clusters. Supporting this view, the C domains from MMAR_3097 and MMAR_3176 also form their own clades, albeit a bit deeper, suggesting they may have evolved further.
The picture looks similar for the adenylation (A) domains, although their level of conservation is slightly lower than the C domains. This is a very intriguing situation since these NRPSs appear to have very similar A domains in each module, and as a result, the amino acid substrates of these domains cannot be predicted reliably using structure-based and homology-based models. Interestingly, the NRPS Mps, which synthesizes the tetrapeptidyl alcohol backbone of the mycobacterial glycopeptidolipids (GPLs) and was first described in a strain of M. smegmatis (Billman-Jacobe et al. 1999), comprises four modules that—like the M. marinum NRPS modules—seem to have arisen from some form of module duplication. Since it is known that the Mps A domains activate different amino acid substrates (one phenylalanine, one allo-threonine, and two alanine), it is likely that these differences in substrate selectivity resulted from divergent evolution following duplication. The example of Mps has two implications for the M. marinum NRPSs. First, it suggests that despite the high degree of amino acid sequence identity among the A domains in MMAR_3268, MMAR_0851, MMAR_3270, and MMAR_3271, these domains may have evolved divergently to accept different amino acid substrates. Since most known NRPSs are composed of modules with disparate evolutionary origins, the M. marinum NRPSs would be a highly unusual example of NRPS “innovation” by divergent evolution and could have important implications for efforts to custom-design NRPSs that make unnatural products. Second, it raises the possibility that the three-module NRPS MMAR_0099, which has A domains that are related to those of Mps, could synthesize an unusual nonreduced GPL similar to the serine-containing GPL isolated from Mycobacterium xenopi (Besra et al. 1993). In support of this conjecture, MMAR_0099 is clustered with PKS-encoding genes that could synthesize the lipid portion of a noncanonical GPL.
The secondary metabolite repertoire of M. marinum is not limited to fatty acids, polyketides, and nonribosomal peptides. M. marinum, in contrast to M. tuberculosis, is photochromogenic, synthesizing carotenoid pigments upon exposure to light. Mutational analysis identified several genes encoding carotenoid biosynthesis pathway enzymes that are essential for photochromogenicity (Ramakrishnan et al. 1997; Gao et al. 2003). In support of these previous findings, the remainder of this locus appears to be involved in the synthesis of a beta-carotene-like terpene that is predicted to have photochromogenic properties. MMAR_4805 is a trans-isoprenyl diphosphate synthase that would polymerize IPP units to form geranylgeranyl-pyrophosphate, MMAR_4807 is a phytoene synthase (CrtB ortholog) that would catalyze a head-to-head coupling of geranylgeranyl-pyrophosphate to form phytoene, MMAR_4806 is a phytoene dehydrogenase (CrtI ortholog) that would desaturate phytoene to form the conjugated polyene core of the carotenes, and MMAR_4808–4809 encode a two-component lycopene cyclase—homologous to CrtYc/CrtYd from the Brevibacterium linens isorenieratene gene cluster (Krubasik and Sandmann 2000)—that would cyclize the termini to form a beta-carotene or a similar C40 terpene with photochromogenic properties.
All these observations underscore a complex and diverse secondary metabolome within M. marinum and are a point of significant difference compared with M. tuberculosis and other mycobacteria, including M. ulcerans, the latter species having lost many of these regions by deletion (Stinear et al. 2007).
ESX loci and ESAT-6 proteins
The ESX loci are large gene clusters that encode the dedicated ATP-dependent secretory apparatus required to export specific members of the 6-kDa early secreted antigenic target (ESAT-6) protein family, together with their corresponding effector proteins. In M. marinum there are 29 esx genes, compared with 23 in M. tuberculosis (Supplemental Table 3). These are within at least five ESX loci including the prototypical ESX-1 system, which is a major virulence determinant in both M. tuberculosis (Lewis et al. 2003; Brodin et al. 2004; Guinn et al. 2004) and M. marinum (Gao et al. 2004; Volkman et al. 2004). It is noteworthy that a 3.7-kb portion of the ESX-1 region is partially duplicated (ESX-6, MMAR_0184–MMAR_0188), resulting in two copies of esxA and esxB that are almost identical. Although it is not clear at present whether esxA_1 and esxB_1 are functional, this illustrates again how members of the ESX gene family evolve by duplication and subsequent diversification (Tekaia et al. 1999).
ESX-3 (spanning nucleotides 636,000–651,200) is another system shared between M. marinum and M. tuberculosis, whereas ESX-2 may be defective in M. marinum M as indicated by partial deletion of a membrane-bound component, MMAR_5460. ESX-3 is essential for in vitro growth of M. tuberculosis (Sassetti et al. 2001), and its expression is controlled by the zinc uptake regulator Zur/FurB (Maciag et al. 2007), which is also present in M. marinum. Orthologous systems of ESX-3 have been found in all mycobacteria sequenced to date, suggesting a fundamental role for this region in the mycobacterial life cycle. The other shared ESX loci are ESX-4 and ESX-5. While ESX-4 seems to be the most ancestral ESX-system based on its presence in other actinobacteria (Gey Van Pittius et al. 2001), ESX-5 has been shown to be involved in export of the PPE41 protein in M. marinum (Abdallah et al. 2006).
In addition to ESAT-6 proteins, ESX systems also appear to secrete other effector proteins, often encoded by unlinked genes, such as EspA (Rv3616c) in M. tuberculosis. Inactivation of espA, which is paralogous to Rv3864, incapacitates the ESX-1 system, and neither ESAT-6 nor CFP-10 is secreted, thus leading to severe attenuation of virulence (Fortune et al. 2005; MacGurn et al. 2005). In M. marinum, the EspA ortholog (MMAR_4166) belongs to an 18-membered family, which also includes MMAR_5439, the ortholog of Rv3864. Surprisingly, the genes for 15 of these EspA family members occur within a 46-kb region encompassing ESX-1, with 14 of them arranged in tandem upstream of the locus. While it is clear that this is yet another example of gene duplication associated with ESX systems, it is not known whether all of these EspA paralogs are functional or if they are secreted.
In summary, it is striking that the ESX complements of M. marinum and M. tuberculosis are so similar despite their different genome sizes, while M. smegmatis, with a large genome (7.0 Mb), has only three ESX loci and is lacking ESX-1. ESX-1 is required for both intercellular bacteria spread between macrophages as well as granuloma formation—two key requirements for mycobacterial pathogenesis (Cosma et al. 2003; Guinn et al. 2004; Volkman et al. 2004; Swaim et al. 2006). Therefore, the genomic data are consistent with the acquisition of these systems being driven by the selection of an intracellular niche during evolution of these two pathogens. In contrast, M. smegmatis, a nonpathogenic saprophyte, is unable to replicate intracellularly (Wei et al. 2000). Also telling is that M. ulcerans, the only pathogenic Mycobacterium that does not have an intracellular lifestyle within granulomas (its polyketide toxin enables the bacteria to lyse cells and dwell extracellularly), has only 13 esx genes and three intact ESX loci but not ESX-1, which has incurred two deletions (Stinear et al. 2007).
PE/PPE proteins
The PE/PPE proteins are enigmatic mycobacterial proteins, so-called because of their characteristic proline-glutamate (PE), or proline-proline-glutamate (PPE) N-terminal motifs. M. marinum has 281 CDS distributed around its genome encoding 27 PE, 148 PE-PGRS, and 106 PPE proteins (Fig. 1A). Compared with 115 in M. ulcerans and 169 in M. tuberculosis, M. marinum has the most extensive repertoire yet reported of these mycobacteria-specific proteins representing 9.1% of its coding capacity, compared with 7.1% for M. tuberculosis and 3.8% for M. ulcerans. It has been proposed that the PE/PPE genes coevolved with the ESX loci and then underwent a specific and significant expansion in the common progenitor of M. tuberculosis, M. marinum, and M. ulcerans (Gey van Pittius et al. 2006). Such a large collection of these genes in M. marinum may be consistent with its diverse host range as some PE-PGRS proteins from M. marinum are known to be involved in intracellular persistence (Ramakrishnan et al. 2000). Significant amino acid sequence divergence occurs, predominantly within the C-terminal region of these proteins (Garnier et al. 2003; Gey van Pittius et al. 2006), suggesting that they are subject to constant diversifying selection, perhaps to avoid detection or destruction during intracellular life in a variety of hosts.
Lipoproteins
Like the Esx and PE/PPE proteins, lipoproteins play an important role in mycobacterial virulence by interacting with the host immune responses; for example, LpqH-dependent signaling through TOLL-like receptor 2 (Rezwan et al. 2007). Despite their different genome sizes, M. marinum and M. tuberculosis have similar numbers of CDS encoding predicted lipoproteins. M. marinum has 87 predicted lipoproteins, compared with 89 in M. tuberculosis. Furthermore, some lipoprotein sequences vary significantly between each Mycobacterium such that 14 putative lipoproteins in M. tuberculosis, encoded by lppA,B,C,G,O,Q,T,Y, lprL,P, lpqX, lipR,F, and pstS1, have no ortholog in M. marinum. Interestingly too, M. smegmatis has only 24 CDS encoding predicted lipoproteins. Variation in the lipoprotein content may reflect the different host ranges of each Mycobacterium and perhaps indicate the importance of specific lipoproteins for M. marinum and M. tuberculosis (Rezwan et al. 2007).
Evolution of M. tuberculosis and M. marinum from a recent common ancestor
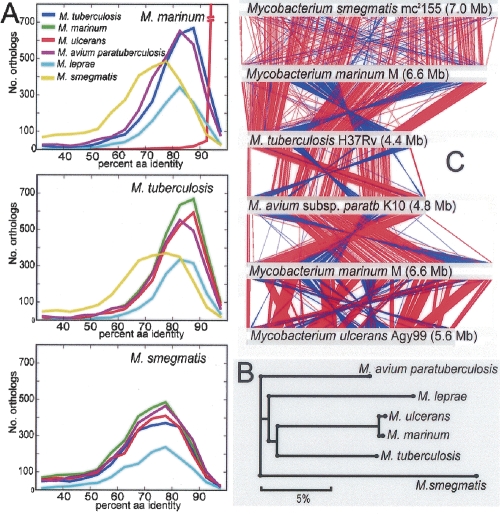
Pairwise reciprocal best-hit FASTA analysis was used to define all orthologs between each of M. marinum M, M. tuberculosis H37Rv, M. ulcerans Agy99, M. avium paratuberculosis K10, Mycobacterium leprae TN, and M. smegmatis mc2155. The results are summarized in ortholog frequency plots (Fig. 3A). These comparisons also revealed a common core set of 1072 orthologs (1755 if M. leprae is excluded). A summary of the average percent amino acid identity for these core CDS is presented in Supplemental Table 4 and was used to infer a phylogeny (Fig. 3B) that demonstrates M. marinum is most closely related to M. ulcerans and then to M. tuberculosis (3000 shared orthologs) and least related to M. smegmatis, a phylogeny in close agreement with those based on 16S rRNA gene sequences (Gey van Pittius et al. 2006).
Figure 3.
(A) Orthology frequency plots based on reciprocal best-hit FASTA analysis (refer to Methods) for each pairwise ortholog comparison among six species of mycobacteria. (B) Phylogenomic analysis of six mycobacterial species based on percent amino acid identity among 1072 core, shared orthologs. All nodes had >99.98% bootstrap support. (C) Diagram depicting linear genomic comparisons generated with the BLASTN, and displayed with the Artemis Comparison Tool (Carver et al. 2005) of (listed from top to bottom) M. smegmatis mc2155, M. marinum, M. tuberculosis H37Rv, M. avium subsp. paratuberculosis K10, M. marinum M, and M. ulcerans Agy99. Regions of DNA:DNA similarity are joined by red lines for direct matches, and blue lines for inverted matches.
Relationships based on shared amino acid identity contrast strikingly with the relatively high conservation of genome size and gene order between M. marinum and the 7.0-Mb genome of M. smegmatis (Fig. 3C), a conservation that probably reflects relatively tight selection constraints on genome architecture for environmental mycobacteria. Genome comparisons between M. marinum, M. tuberculosis H37Rv (4.4 Mb), and M. avium subsp. paratuberculosis K10 (4.8 Mb) show the rearrangements and genome reduction that have occurred within these more closely related species (Fig. 3C).
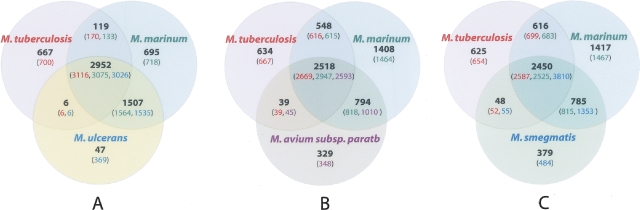
In a complementary approach to identifying orthologs, a BLASTCLUST analysis was applied to M. marinum, M. tuberculosis, M. ulcerans, M. avium subspecies paratuberculosis, and M. smegmatis. The results are summarized in three Venn diagrams (Fig. 4) and show CDS that are both conserved and unique between these species. In the three different three-way comparisons—taking M. tuberculosis and comparing it with either M. marinum and M. ulcerans, M. marinum and M. avium subsp. paratuberculosis, or M. marinum and M. smegmatis—there remained around 633 CDS found only in M. tuberculosis (Fig. 4). Furthermore, these CDS were the same in each three-way comparison, suggesting that M. tuberculosis possesses about 633 specific CDS that have (1) been acquired by lateral gene transfer (LGT) or (2) been subject to endogenous mutagenic processes such as duplication, recombination, or sequence amelioration since diverging from the most recent common ancestor (MRCA) of M. marinum and M. tuberculosis.
Figure 4.
Venn diagrams showing orthologous CDS among four mycobacterial species as determined by BLASTCLUST analysis. Numbers in parentheses include paralogous CDS.
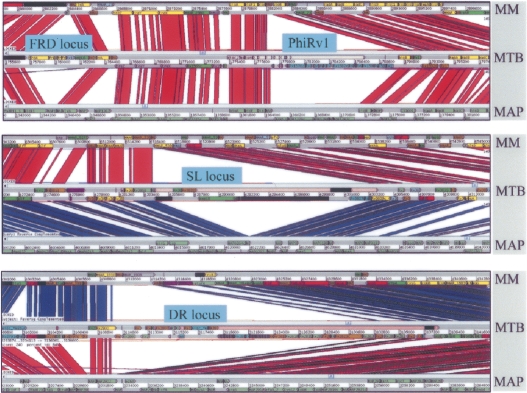
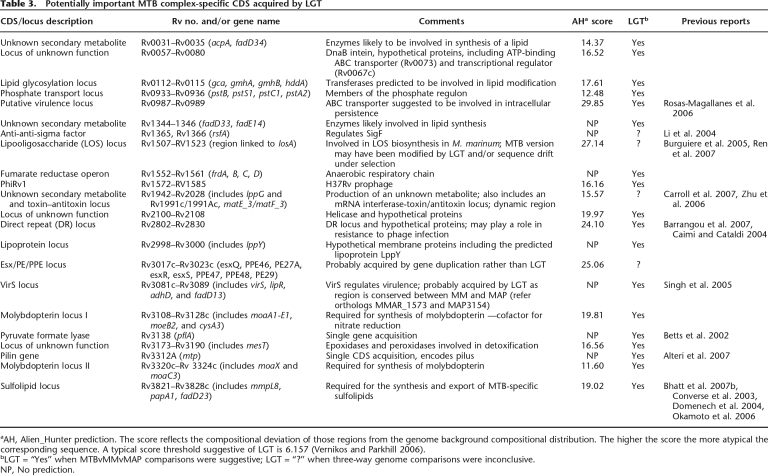
A detailed analysis of the 633 M. tuberculosis-specific CDS identified by BLASTCLUST (Fig. 4B) using the Artemis Comparison Tool in three-way whole-genome BLASTN comparisons, and Alien_Hunter to identify potential foreign DNA (Fig. 5), shows that these CDS span ∼600 kb and comprise 140 distinct regions of difference between M. marinum and M. tuberculosis, of which 340 kb, spanning at least 80 regions and containing 360 CDS, appears to have been gained by LGT. Some of the “M. tuberculosis-specific” regions identified by this analysis code for known virulence factors or have been proposed to play a role in pathogenesis (Table 3; Supplemental Table 5). These include CDS that produce secondary metabolites such as the sulfolipids, which have been reported to suppress host immune responses (Domenech et al. 2004; Okamoto et al. 2006), virS encoding a putative regulator of virulence functions (Singh et al. 2005), other regions such as a potential lipid glycosylation locus (Rv0112–Rv0115), and a putative adhesin/transporter locus (Rv0986–Rv0987) (Rosas-Magallanes et al. 2006), as well as the recently reported pilin gene (Rv3312a) (Alteri et al. 2007).
Figure 5.
Four examples of LGT in M. tuberculosis H37Rv as predicted by Alien_Hunter (sections highlighted in pink) and whole-genome comparisons with M. marinum and M. avium subsp. paratuberculosis, displayed using the Artemis Comparison Tool. Regions of DNA:DNA similarity are joined by red lines, while inverted regions are shown as blue lines (reversed and complemented here for clarity).
Table 3.
Potentially important MTB complex-specific CDS acquired by LGT
aAH, Alien_Hunter prediction. The score reflects the compositional deviation of those regions from the genome background compositional distribution. The higher the score the more atypical the corresponding sequence. A typical score threshold suggestive of LGT is 6.157 (Vernikos and Parkhill 2006).
bLGT = “Yes” when MTBvMMvMAP comparisons were suggestive; LGT = “?” when three-way genome comparisons were inconclusive.
NP, No prediction.
The fumarate reductase operon, required for growth during anaerobiosis, also appears to have been gained by LGT; coupled with the acquisition of loci required for synthesis of the cofactor molybdopterin, essential for nitrate respiration (MacGurn and Cox 2007), these latter adaptations are consistent with a bacterium adapted to survive under specific reduced oxygen conditions, such as those within granulomas. The direct repeat (DR) region, another genomic segment confined to members of the M. tuberculosis complex, which serves as the basis of spoligotyping (van Embden et al. 2000), also appears to have been gained by LGT, confirming previous observations (Caimi and Cataldi 2004). This region is an example of a CRISPR locus, recently shown to be involved in bacteriophage resistance (Barrangou et al. 2007), and often variable in presence and content between different bacteria.
The same BLASTCLUST analysis also revealed 695 CDS present in M. marinum, but not in M. ulcerans or M. tuberculosis (Fig. 4A), that are distributed over at least 55 distinct chromosomal segments, spanning a total of 750 kb. Many of these regions overlap some of the 157 previously reported M. ulcerans regions of difference (MURDs) that are segments of the M. marinum M genome not present in M. ulcerans Agy99 (Stinear et al. 2007). A summary of these CDS and their predicted products is contained in Supplemental Table 6.
Classification by function shows that among the 55 regions, 11 appear to be involved in various metabolic processes, 16 encode predicted membrane proteins, and 10 correspond to prophages. There is a large locus, spanning MMAR_3382–MMAR_3414, which includes pmiA, encoding a protein involved in synthesis of cell wall components, that inhibits phagosome maturation (Robinson et al. 2007). Similarly, a 20-kb region between MMAR_2955 and MMAR_2988 (MURD73) harbors mimD, a gene that appears to be required for survival within macrophages (Mehta et al. 2006).
A three-way comparison with M. tuberculosis and M. smegmatis (Fig. 4C) revealed 785 CDS shared between M. marinum and M. smegmatis that are predicted to encode cell wall-associated proteins (148) or enzymes involved in intermediary metabolism such as hydroxylases, oxidases, glyoxylases, decarboxylases, and reductases (212); lipid metabolism (70); regulatory proteins (102); and conserved hypothetical proteins (207).
Discussion
The close relationship between M. marinum and M. tuberculosis uncovered by this study provides deep insight into the evolution of pathogenesis in both these species. Their recent common origin is reflected in their shared and substantial common genetic core. There is a M. marinum ortholog for 80% of all CDS in M. tuberculosis with an average shared amino acid identity of 85%. However, somewhat paradoxically, whole-genome comparisons reveal a higher level of colinearity, synteny, and conservation of chromosome size between M. marinum and the distant saprophyte, M. smegmatis (76% average amino acid identity), than with M. tuberculosis. The most parsimonious explanation for these observations is that M. marinum, as a generalist, has preserved more of the characteristics of the hypothetical MRCA, while M. tuberculosis has undergone a process of reductive evolution and adaptation, presumably for survival within mammalian hosts. In addition, the much lower content of IS elements in M. marinum and M. smegmatis has also contributed to maintaining synteny, as these elements are known to drive genome rearrangements (Parkhill et al. 2003), particularly in pathogens like M. ulcerans that have undergone reductive evolution (Stinear et al. 2007).
Two striking features of the M. marinum genome are the extensive range of genes encoding NRPSs and the huge repertoire of PE, PPE, and ESX systems. As seen previously in comparison of mycobacterial genomes (Garnier et al. 2003; Stinear et al. 2007), there is great variation in the size, sequence, and structure of loci encoding members of the PE, PPE, and ESAT-6 protein families both between M. marinum and M. ulcerans, but also with respect to tubercle bacilli. Of particular interest is the duplication of part of the ESX-1 locus, a major virulence determinant of M. marinum and M. tuberculosis, and the massive expansion of the espA paralogs upstream of the ESX-1 region of the genome. The latter is without precedent and suggests that under certain conditions selected EspA members may play different physiological roles, perhaps in response to a change in host. Proteomic analysis of the M. marinum secretome could provide evidence for this and help identify which ESX-system is required for secretion of specific EspA proteins.
A major conclusion of our study is the scale of DNA acquisition by LGT in M. tuberculosis, and this is further evidence of adaptation. Indeed, several authors have proposed that M. tuberculosis evolved from an environmental progenitor by LGT (Kinsella et al. 2003; Rosas-Magallanes et al. 2006). Our three-way comparisons between M. marinum, M. tuberculosis, and M. avium subsp. paratuberculosis revealed at least 80 distinct regions that have probably been acquired by LGT in M. tuberculosis. These span more than 360 CDS and account for 8% of the genome, a level twice that reported in a recent study using di-, tri-, and tetranucleotide signatures of M. tuberculosis (Becq et al. 2007). Whether acquired by LGT or other means, some of these M. tuberculosis-specific regions contain known virulence genes or code for adaptation factors (Table 3). In the same vein, M. marinum has also gained genes by LGT with notable examples being some of the NRPS genes, such as MMAR_0851 and its paralogs, that may have been acquired from aquatic Streptomyces spp.
These genome comparison data indicate that M. marinum and M. tuberculosis have diverged from a common generalist environmental Mycobacterium with M. marinum retaining genes required for its dual lifestyle and M. tuberculosis undergoing reductive evolution compatible with adaptation to its niche. It seems that, throughout the evolution of mycobacterial species, the process of genome reduction is a recurring theme in the adaptation of an environmental bacterium toward an intracellular, parasitic lifestyle. However, it is intriguing that M. marinum as the closest relative of M. tuberculosis has a genome that is 50% bigger than that of M. tuberculosis, while the members of the M. avium complex (∼4.8 Mb) are characterized by genome sizes that are closer to M. tuberculosis. This means that either significant genome downsizing has occurred independently in various phylogenetic lineages of slow-growing mycobacteria or that the contribution of LGT to the M. marinum gene pool must be considerable, as suggested by NRPS gene acquisition (Fig. 1B). However, the relatively large genome size of M. ulcerans (∼5.2 Mb after compensating for amplification of IS elements and acquisition of pMUM001, the mycolactone plasmid), a recent descendant of M. marinum that has adapted to particular environmental niche and entered reductive evolution, is in favor of the hypothesis where members of the M. tuberculosis and M. avium complexes downsized their genomes independently. A more comprehensive phylogenomic reconstruction of the genus Mycobacterium will become possible with the publication of the M. smegmatis genome and the genomes of several other recently completed environmental mycobacteria such as Mycobacterium vanbaalenii. Further insight into this process will also be provided by analyses of the genome sequences of Mycobacterium prototuberculosis and Mycobacterium canettii, which precede M. tuberculosis in the current evolutionary scheme (Brosch et al. 2002; Gutierrez et al. 2005).
Methods
M. marinum M strain
M. marinum M (ATCC BAA-535) is a human patient isolate from Moffett Hospital, University of California, San Francisco in 1992. Multi-locus sequence typing shows that this strain belongs to sequence type 22 (Yip et al. 2007).
Genome sequencing
Genomic DNA was sheared by sonication, size fractionated, and used to generate several shotgun libraries. Clones were sequenced from these libraries as follows: 21,773 reads from an m13mp18 library with an insert size of 1.4–2 kb; 66,388 paired reads from a pUC18 library with an insert size of 3–3.3 kb; and 55,654 paired reads from a pMAQ1b library with an insert size of 5.5–6 kb. This shotgun generated approximately 9.3-fold coverage of the genome. To scaffold these sequences, 6120 paired reads were generated from several pBACe3.6 libraries with insert sizes ranging from 15 to 48 kb, generating around 11-fold genome coverage in BAC clones. A further 27,357 directed sequences were generated during the gap-closure and finishing phase. DNA was sequenced using ABI BigDye terminator reactions and run on AB3730 capillary sequencers. Sequence assembly was performed by phrap and finished to standard criteria using GAP4 as previously described (Cole et al. 1998). PCR was used to check the distribution of the pMM23 plasmid among 41 different isolates of M. marinum, spanning the nine reported MLST genotypes by amplification of the plasmid genes merB (merB-F 5′-CTCGACGCCCTG GATGTT-3′ and merB-R 5′-AAACGCGGTCCGAATAGAG-3′; 432-bp fragment) and parA (parA-F 5′-CGTTTCGGCAGATAAGCAGT-3′ and parA-R 5′-GTGGTCAGCTCCCCCAAT-3′; 421-bp fragment).
Genome annotation
Annotation and database construction were performed with Wasabi, an in-house, Web-interfaced MySQL database for management of genome annotation, Artemis, and GenoList. Complete databases are available (http://www.sanger.ac.uk/Projects/M_marinum/ and http://genolist.pasteur.fr/MarinoList/). CDS were predicted using a combination of Glimmer (Delcher et al. 1999), EasyGene (Anders Krogh, pers. comm.), Orpheus (Frishman et al. 1998), and GeneMark (Besemer et al. 2001); followed by manual curation as previously described (Parkhill et al. 2003); assigned the locus_tag prefix MMAR_XXXX; and compared to public sequence databases using the BLAST suite (Altschul et al. 1990). IS were identified with reference to IS-Finder database (http://www-is.biotoul.fr/); cytochrome P450s were identified using the cytochrome P450 database (http://drnelson.utmem.edu/CytochromeP450.html) and transport proteins with reference to the transporter database (http://www.membranetransport.org/).
Whole-genome comparisons and lateral gene transfer
Whole-genome DNA:DNA comparisons of M. marinum M were performed using BLASTN analysis (Expect threshold = 10) against M. tuberculosis H37Rv, M. ulcerans Agy99, M. avium subsp. paratuberculosis K10, M. leprae TN, and M. smegmatis mc2155. The results of these comparisons were visualized using the Artemis Comparison Tool (ACT) (Carver et al. 2005). In addition, to detect CDS that may have arisen from lateral gene transfer events, two new software tools were used. Gene Spaghetti is GNU public license software for visualizing base composition and codon usage in a genome. The software uses a sliding Gaussian window to estimate local base usage. Here, a window with a standard deviation of 200 was used. Further details and access to the software are available at http://www.vicbioinformatics.com/genespaghetti.shtml. Alien_Hunter uses a combination of all nucleotide motifs of between 1 bp and 8 bp to identify regions of the genome that differ in nucleotide composition from the backbone, and then a change-point Hidden Markov Model to refine the boundaries of the anomalous DNA (Vernikos and Parkhill 2006).
Ortholog analysis
Orthologs were defined in two ways. Reciprocal FASTA analysis was performed on all CDS from M. marinum, M. tuberculosis, M. ulcerans, M. avium subsp. paratuberculosis, M. smegmatis, and M. leprae, with reciprocal top hits requiring >80% amino acid identity over 30% or more of the length of both sequences. This FASTA analysis was also used to define a core set of mycobacterial orthologs among the six species. A pairwise comparison was then undertaken of the average percentage amino acid identity from the core set of orthologs. A phylogeny of the six species was inferred from the resulting distance matrix (each distance being calculated as 100% minus the average percent identity) using the Neighbor-Joining method of tree construction (Saitou and Nei 1987) with the “Neighbor” program in the Phylip package. The robustness of the resulting tree topology was tested by bootstrap analysis. Ten thousand bootstrap samples were constructed by selecting ortholog groups at random with replacement from the original 1072 ortholog groups identified by the FASTA analysis above. The resulting trees were compared using Phylip’s “Consense” program.
In a second complementary approach, CDS from M. marinum, M. tuberculosis, M. ulcerans, and M. avium subsp. paratuberculosis were clustered into orthologous groups using the BLASTCLUST tool from the NCBI BLAST software suite version 2.2.14, with default settings in protein mode. This enforced a maximum of 10% length variation within a cluster and a minimum bit score to length ratio of 1.75. There was high concordance obtained by both clustering techniques. All BLASTCLUST clusters were checked manually and verified using BLASTN and TBLASTX (Altschul et al. 1990).
Acknowledgments
We thank the Sanger Institute core sequencing and informatics groups for support, L. Jones and F. Tekaia for database support, Jessica L. Porter for technical assistance, Derrick Fouts for use of the unpublished M. smegmatis genome, and A. Colorni for critical review of the manuscript. We gratefully acknowledge the financial support of the Wellcome Trust, Fondation Raoul Follereau (STC), the National Health and Medical Research Council of Australia (TPS), and grant R01 AI036396 from the National Institutes of Health (LR).
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. CP000854 (chromosome) and CP000895 (pMM23 plasmid).]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075069.107.
References
- Abdallah A.M., Verboom T., Hannes F., Safi M., Strong M., Eisenberg D., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Verboom T., Hannes F., Safi M., Strong M., Eisenberg D., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Hannes F., Safi M., Strong M., Eisenberg D., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Safi M., Strong M., Eisenberg D., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Strong M., Eisenberg D., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Eisenberg D., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Musters R.J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Luirink J., Appelmelk B.J., Luirink J., Luirink J., et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- Adekambi T., Ben Salah S., Khlif M., Raoult D., Drancourt M., Ben Salah S., Khlif M., Raoult D., Drancourt M., Khlif M., Raoult D., Drancourt M., Raoult D., Drancourt M., Drancourt M. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 2006;72:5974–5981. doi: 10.1128/AEM.03075-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri C.J., Xicohténcatl-Cortes J., Hess S., Caballero-Olín G., Girón J.A., Friedman R.L., Xicohténcatl-Cortes J., Hess S., Caballero-Olín G., Girón J.A., Friedman R.L., Hess S., Caballero-Olín G., Girón J.A., Friedman R.L., Caballero-Olín G., Girón J.A., Friedman R.L., Girón J.A., Friedman R.L., Friedman R.L. Mycobacterium tuberculosis produces pili during human infection. Proc. Natl. Acad. Sci. 2007;104:5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aronson J.D. Spontaneous tuberculosis in salt water fish. J. Infect. Dis. 1927;39:315–320. [Google Scholar]
- Aubry A., Jarlier V., Escolano S., Truffot-Pernot C., Cambau E., Jarlier V., Escolano S., Truffot-Pernot C., Cambau E., Escolano S., Truffot-Pernot C., Cambau E., Truffot-Pernot C., Cambau E., Cambau E. Antibiotic susceptibility pattern of Mycobacterium marinum. Antimicrob. Agents Chemother. 2000;44:3133–3136. doi: 10.1128/aac.44.11.3133-3136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry A., Chosidow O., Caumes E., Robert J., Cambau E., Chosidow O., Caumes E., Robert J., Cambau E., Caumes E., Robert J., Cambau E., Robert J., Cambau E., Cambau E. Sixty-three cases of Mycobacterium marinum infection: Clinical features, treatment, and antibiotic susceptibility of causative isolates. Arch. Intern. Med. 2002;162:1746–1752. doi: 10.1001/archinte.162.15.1746. [DOI] [PubMed] [Google Scholar]
- Bardouniotis E., Ceri H., Olson M.E., Ceri H., Olson M.E., Olson M.E. Biofilm formation and biocide susceptibility testing of Mycobacterium fortuitum and Mycobacterium marinum. Curr. Microbiol. 2003;46:28–32. doi: 10.1007/s00284-002-3796-4. [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P., Boyaval P., Moineau S., Romero D.A., Horvath P., Moineau S., Romero D.A., Horvath P., Romero D.A., Horvath P., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Becq J., Gutierrez M., Rosas-Magallanes V., Rauzier J., Gicquel B., Neyrolles O., Deschavanne P., Gutierrez M., Rosas-Magallanes V., Rauzier J., Gicquel B., Neyrolles O., Deschavanne P., Rosas-Magallanes V., Rauzier J., Gicquel B., Neyrolles O., Deschavanne P., Rauzier J., Gicquel B., Neyrolles O., Deschavanne P., Gicquel B., Neyrolles O., Deschavanne P., Neyrolles O., Deschavanne P., Deschavanne P. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol. Biol. Evol. 2007;24:1867–1871. doi: 10.1093/molbev/msm111. [DOI] [PubMed] [Google Scholar]
- Besemer J., Lomsadze A., Borodovsky M., Lomsadze A., Borodovsky M., Borodovsky M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besra G.S., McNeil M.R., Rivoire B., Khoo K.H., Morris H.R., Dell A., Brennan P.J., McNeil M.R., Rivoire B., Khoo K.H., Morris H.R., Dell A., Brennan P.J., Rivoire B., Khoo K.H., Morris H.R., Dell A., Brennan P.J., Khoo K.H., Morris H.R., Dell A., Brennan P.J., Morris H.R., Dell A., Brennan P.J., Dell A., Brennan P.J., Brennan P.J. Further structural definition of a new family of glycopeptidolipids from Mycobacterium xenopi. Biochemistry. 1993;32:347–355. doi: 10.1021/bi00052a043. [DOI] [PubMed] [Google Scholar]
- Betts J.C., Lukey P.T., Robb L.C., McAdam R.A., Duncan K., Lukey P.T., Robb L.C., McAdam R.A., Duncan K., Robb L.C., McAdam R.A., Duncan K., McAdam R.A., Duncan K., Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Bhatt A., Fujiwara N., Bhatt K., Gurcha S.S., Kremer L., Chen B., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Fujiwara N., Bhatt K., Gurcha S.S., Kremer L., Chen B., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Bhatt K., Gurcha S.S., Kremer L., Chen B., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Gurcha S.S., Kremer L., Chen B., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Kremer L., Chen B., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Chen B., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Chan J., Porcelli S.A., Kobayashi K., Besra G.S., Porcelli S.A., Kobayashi K., Besra G.S., Kobayashi K., Besra G.S., Besra G.S., et al. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. 2007a;104:5157–5162. doi: 10.1073/pnas.0608654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt K., Gurcha S.S., Bhatt A., Besra G.S., Jacobs W.R., Gurcha S.S., Bhatt A., Besra G.S., Jacobs W.R., Bhatt A., Besra G.S., Jacobs W.R., Besra G.S., Jacobs W.R., Jacobs W.R. Two polyketide-synthase-associated acyltransferases are required for sulfolipid biosynthesis in Mycobacterium tuberculosis. Microbiology. 2007b;153:513–520. doi: 10.1099/mic.0.2006/003103-0. [DOI] [PubMed] [Google Scholar]
- Billman-Jacobe H., McConville M.J., Haites R.E., Kovacevic S., Coppel R.L., McConville M.J., Haites R.E., Kovacevic S., Coppel R.L., Haites R.E., Kovacevic S., Coppel R.L., Kovacevic S., Coppel R.L., Coppel R.L. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 1999;33:1244–1253. doi: 10.1046/j.1365-2958.1999.01572.x. [DOI] [PubMed] [Google Scholar]
- Brodin P., Rosenkrands I., Andersen P., Cole S.T., Brosch R., Rosenkrands I., Andersen P., Cole S.T., Brosch R., Andersen P., Cole S.T., Brosch R., Cole S.T., Brosch R., Brosch R. ESAT-6 proteins: Protective antigens and virulence factors? Trends Microbiol. 2004;12:500–508. doi: 10.1016/j.tim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Brosch R., Gordon S.V., Billault A., Garnier T., Eiglmeier K., Soravito C., Barrell B.G., Cole S.T., Gordon S.V., Billault A., Garnier T., Eiglmeier K., Soravito C., Barrell B.G., Cole S.T., Billault A., Garnier T., Eiglmeier K., Soravito C., Barrell B.G., Cole S.T., Garnier T., Eiglmeier K., Soravito C., Barrell B.G., Cole S.T., Eiglmeier K., Soravito C., Barrell B.G., Cole S.T., Soravito C., Barrell B.G., Cole S.T., Barrell B.G., Cole S.T., Cole S.T. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R., Gordon S.V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Gordon S.V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Garnier T., Gutierrez C., Hewinson G., Kremer K., Gutierrez C., Hewinson G., Kremer K., Hewinson G., Kremer K., Kremer K., et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguiere A., Hitchen P.G., Dover L.G., Kremer L., Ridell M., Alexander D.C., Liu J., Morris H.R., Minnikin D.E., Dell A., Hitchen P.G., Dover L.G., Kremer L., Ridell M., Alexander D.C., Liu J., Morris H.R., Minnikin D.E., Dell A., Dover L.G., Kremer L., Ridell M., Alexander D.C., Liu J., Morris H.R., Minnikin D.E., Dell A., Kremer L., Ridell M., Alexander D.C., Liu J., Morris H.R., Minnikin D.E., Dell A., Ridell M., Alexander D.C., Liu J., Morris H.R., Minnikin D.E., Dell A., Alexander D.C., Liu J., Morris H.R., Minnikin D.E., Dell A., Liu J., Morris H.R., Minnikin D.E., Dell A., Morris H.R., Minnikin D.E., Dell A., Minnikin D.E., Dell A., Dell A., et al. LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 2005;280:42124–42133. doi: 10.1074/jbc.M507500200. [DOI] [PubMed] [Google Scholar]
- Caimi K., Cataldi A., Cataldi A. A fragment of 21 ORFs around the direct repeat region of Mycobacterium tuberculosis is absent from the other sequenced mycobacterial genomes: Implications for the evolution of the DR region. Comp. Funct. Genomics. 2004;5:116–122. doi: 10.1002/cfg.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll P., Brown A.C., Hartridge A.R., Parish T., Brown A.C., Hartridge A.R., Parish T., Hartridge A.R., Parish T., Parish T. Expression of Mycobacterium tuberculosis Rv1991c using an arabinose-inducible promoter demonstrates its role as a toxin. FEMS Microbiol. Lett. 2007;274:73–82. doi: 10.1111/j.1574-6968.2007.00842.x. [DOI] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Clark H.F., Shepard C.C., Shepard C.C. Effect of environmental temperatures on infection with Mycobacterium marinum (balnei) of mice and a number of poikilothermic species. J. Bacteriol. 1963;86:1057–1069. doi: 10.1128/jb.86.5.1057-1069.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H., Davis J.M., Beery D., Huttenlocher A., Lyons S.E., Ramakrishnan L., Davis J.M., Beery D., Huttenlocher A., Lyons S.E., Ramakrishnan L., Beery D., Huttenlocher A., Lyons S.E., Ramakrishnan L., Huttenlocher A., Lyons S.E., Ramakrishnan L., Lyons S.E., Ramakrishnan L., Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. 2007;2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., Eiglmeier K., Gas S., Barry C.E., Gas S., Barry C.E., Barry C.E., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Constant P., Perez E., Malaga W., Laneelle M.A., Saurel O., Daffe M., Guilhot C., Perez E., Malaga W., Laneelle M.A., Saurel O., Daffe M., Guilhot C., Malaga W., Laneelle M.A., Saurel O., Daffe M., Guilhot C., Laneelle M.A., Saurel O., Daffe M., Guilhot C., Saurel O., Daffe M., Guilhot C., Daffe M., Guilhot C., Guilhot C. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methly esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 2002;277:38148–38158. doi: 10.1074/jbc.M206538200. [DOI] [PubMed] [Google Scholar]
- Converse S.E., Mougous J.D., Leavell M.D., Leary J.A., Bertozzi C.R., Cox J.S., Mougous J.D., Leavell M.D., Leary J.A., Bertozzi C.R., Cox J.S., Leavell M.D., Leary J.A., Bertozzi C.R., Cox J.S., Leary J.A., Bertozzi C.R., Cox J.S., Bertozzi C.R., Cox J.S., Cox J.S. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. 2003;100:6121–6126. doi: 10.1073/pnas.1030024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma C.L., Sherman D.R., Ramakrishnan L., Sherman D.R., Ramakrishnan L., Ramakrishnan L. The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 2003;57:641–676. doi: 10.1146/annurev.micro.57.030502.091033. [DOI] [PubMed] [Google Scholar]
- Cosma C.L., Humbert O., Ramakrishnan L., Humbert O., Ramakrishnan L., Ramakrishnan L. Superinfecting mycobacteria home to established tuberculous granulomas. Nat. Immunol. 2004;5:828–835. doi: 10.1038/ni1091. [DOI] [PubMed] [Google Scholar]
- Cosma C.L., Klein K., Kim R., Beery D., Ramakrishnan L., Klein K., Kim R., Beery D., Ramakrishnan L., Kim R., Beery D., Ramakrishnan L., Beery D., Ramakrishnan L., Ramakrishnan L. Mycobacterium marinum Erp is a virulence determinant required for cell wall integrity and intracellular survival. Infect. Immun. 2006;74:3125–3133. doi: 10.1128/IAI.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffe M., Varnerot A., Levy-Frebault V.V., Varnerot A., Levy-Frebault V.V., Levy-Frebault V.V. The phenolic mycoside of Mycobacterium ulcerans: Structure and taxonomic implications. J. Gen. Microbiol. 1992;138:131–137. doi: 10.1099/00221287-138-1-131. [DOI] [PubMed] [Google Scholar]
- Dailloux M., Hartemann P., Beurey J., Hartemann P., Beurey J., Beurey J. Study on the relationship between isolation of mycobacteria and classical microbiological and chemical indicators of water quality in swimming pools. Zentralbl. Bakteriol. Mikrobiol. Hyg. [B] 1980;171:473–486. [PubMed] [Google Scholar]
- Davis J.M., Clay H., Lewis J.L., Ghori N., Herbomel P., Ramakrishnan L., Clay H., Lewis J.L., Ghori N., Herbomel P., Ramakrishnan L., Lewis J.L., Ghori N., Herbomel P., Ramakrishnan L., Ghori N., Herbomel P., Ramakrishnan L., Herbomel P., Ramakrishnan L., Ramakrishnan L. Real-time visualization of mycobacterium–macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L., Harmon D., Kasif S., White O., Salzberg S.L., Kasif S., White O., Salzberg S.L., White O., Salzberg S.L., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devulder G., de Perouse Montclos M., Flandrois J.P., de Perouse Montclos M., Flandrois J.P., Flandrois J.P. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 2005;55:293–302. doi: 10.1099/ijs.0.63222-0. [DOI] [PubMed] [Google Scholar]
- Domenech P., Reed M.B., Dowd C.S., Manca C., Kaplan G., Barry C.E., Reed M.B., Dowd C.S., Manca C., Kaplan G., Barry C.E., Dowd C.S., Manca C., Kaplan G., Barry C.E., Manca C., Kaplan G., Barry C.E., Kaplan G., Barry C.E., Barry C.E. The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:21257–21265. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- Fischbach M.A., Walsh C.T., Walsh C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Fortune S.M., Jaeger A., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Jaeger A., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Sherman D.R., Bloom B.R., Rubin E.J., Bloom B.R., Rubin E.J., Rubin E.J. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D., Mironov A., Mewes H.W., Gelfand M., Mironov A., Mewes H.W., Gelfand M., Mewes H.W., Gelfand M., Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.Y., Groger R., Cox J.S., Beverley S.M., Lawson E.H., Brown E.J., Groger R., Cox J.S., Beverley S.M., Lawson E.H., Brown E.J., Cox J.S., Beverley S.M., Lawson E.H., Brown E.J., Beverley S.M., Lawson E.H., Brown E.J., Lawson E.H., Brown E.J., Brown E.J. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 2003;71:922–929. doi: 10.1128/IAI.71.2.922-929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.Y., Guo S., McLaughlin B., Morisaki H., Engel J.N., Brown E.J., Guo S., McLaughlin B., Morisaki H., Engel J.N., Brown E.J., McLaughlin B., Morisaki H., Engel J.N., Brown E.J., Morisaki H., Engel J.N., Brown E.J., Engel J.N., Brown E.J., Brown E.J. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Gao L.Y., Pak M., Kish R., Kajihara K., Brown E.J., Pak M., Kish R., Kajihara K., Brown E.J., Kish R., Kajihara K., Brown E.J., Kajihara K., Brown E.J., Brown E.J. A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect. Immun. 2006;74:1757–1767. doi: 10.1128/IAI.74.3.1757-1767.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T., Eiglmeier K., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Eiglmeier K., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Camus J.C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Duthoy S., Grondin S., Lacroix C., Monsempe C., Grondin S., Lacroix C., Monsempe C., Lacroix C., Monsempe C., Monsempe C., et al. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George K.M., Chatterjee D., Gunawardana G., Welty D., Hayman J., Lee R., Small P.L., Chatterjee D., Gunawardana G., Welty D., Hayman J., Lee R., Small P.L., Gunawardana G., Welty D., Hayman J., Lee R., Small P.L., Welty D., Hayman J., Lee R., Small P.L., Hayman J., Lee R., Small P.L., Lee R., Small P.L., Small P.L. Mycolactone: A polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- Van Gey Pittius N.C., Gamieldien J., Hide W., Brown G.D., Siezen R.J., Beyers A.D., Gamieldien J., Hide W., Brown G.D., Siezen R.J., Beyers A.D., Hide W., Brown G.D., Siezen R.J., Beyers A.D., Brown G.D., Siezen R.J., Beyers A.D., Siezen R.J., Beyers A.D., Beyers A.D. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2:RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gey Pittius N.C., Sampson S.L., Lee H., Kim Y., van Helden P.D., Warren R.M., Sampson S.L., Lee H., Kim Y., van Helden P.D., Warren R.M., Lee H., Kim Y., van Helden P.D., Warren R.M., Kim Y., van Helden P.D., Warren R.M., van Helden P.D., Warren R.M., Warren R.M. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale R.S., Saxena P., Chopra T., Mohanty D., Saxena P., Chopra T., Mohanty D., Chopra T., Mohanty D., Mohanty D. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat. Prod. Rep. 2007;24:267–277. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- Guinn K.M., Hickey M.J., Mathur S.K., Zakel K.L., Grotzke J.E., Lewinsohn D.M., Smith S., Sherman D.R., Hickey M.J., Mathur S.K., Zakel K.L., Grotzke J.E., Lewinsohn D.M., Smith S., Sherman D.R., Mathur S.K., Zakel K.L., Grotzke J.E., Lewinsohn D.M., Smith S., Sherman D.R., Zakel K.L., Grotzke J.E., Lewinsohn D.M., Smith S., Sherman D.R., Grotzke J.E., Lewinsohn D.M., Smith S., Sherman D.R., Lewinsohn D.M., Smith S., Sherman D.R., Smith S., Sherman D.R., Sherman D.R. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.C., Brisse S., Brosch R., Fabre M., Omais B., Marmiesse M., Supply P., Vincent V., Brisse S., Brosch R., Fabre M., Omais B., Marmiesse M., Supply P., Vincent V., Brosch R., Fabre M., Omais B., Marmiesse M., Supply P., Vincent V., Fabre M., Omais B., Marmiesse M., Supply P., Vincent V., Omais B., Marmiesse M., Supply P., Vincent V., Marmiesse M., Supply P., Vincent V., Supply P., Vincent V., Vincent V. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn M., Soldati T., Soldati T. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell. Microbiol. 2007;9:2984. doi: 10.1111/j.1462-5822.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Brun O.S., Polshyna G., Barker L.P., Brun O.S., Polshyna G., Barker L.P., Polshyna G., Barker L.P., Barker L.P. Mycobacterium marinum biofilm formation reveals cording morphology. FEMS Microbiol. Lett. 2006;257:43–49. doi: 10.1111/j.1574-6968.2006.00143.x. [DOI] [PubMed] [Google Scholar]
- Kinsella R.J., Fitzpatrick D.A., Creevey C.J., McInerney J.O., Fitzpatrick D.A., Creevey C.J., McInerney J.O., Creevey C.J., McInerney J.O., McInerney J.O. Fatty acid biosynthesis in Mycobacterium tuberculosis: Lateral gene transfer, adaptive evolution, and gene duplication. Proc. Natl. Acad. Sci. 2003;100:10320–10325. doi: 10.1073/pnas.1737230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krubasik P., Sandmann G., Sandmann G. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 2000;263:423–432. doi: 10.1007/s004380051186. [DOI] [PubMed] [Google Scholar]
- Leoni E., Legnani P., Mucci M.T., Pirani R., Legnani P., Mucci M.T., Pirani R., Mucci M.T., Pirani R., Pirani R. Prevalence of mycobacteria in a swimming pool environment. J. Appl. Microbiol. 1999;87:683–688. doi: 10.1046/j.1365-2672.1999.00909.x. [DOI] [PubMed] [Google Scholar]
- Lewis K.N., Liao R., Guinn K.M., Hickey M.J., Smith S., Behr M.A., Sherman D.R., Liao R., Guinn K.M., Hickey M.J., Smith S., Behr M.A., Sherman D.R., Guinn K.M., Hickey M.J., Smith S., Behr M.A., Sherman D.R., Hickey M.J., Smith S., Behr M.A., Sherman D.R., Smith S., Behr M.A., Sherman D.R., Behr M.A., Sherman D.R., Sherman D.R. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.S., Waddell S.J., Monahan I.M., Mangan J.A., Martin S.L., Everett M.J., Butcher P.D., Waddell S.J., Monahan I.M., Mangan J.A., Martin S.L., Everett M.J., Butcher P.D., Monahan I.M., Mangan J.A., Martin S.L., Everett M.J., Butcher P.D., Mangan J.A., Martin S.L., Everett M.J., Butcher P.D., Martin S.L., Everett M.J., Butcher P.D., Everett M.J., Butcher P.D., Butcher P.D. Increased transcription of a potential sigma factor regulatory gene Rv1364c in Mycobacterium bovis BCG while residing in macrophages indicates use of alternative promoters. FEMS Microbiol. Lett. 2004;233:333–339. doi: 10.1016/j.femsle.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Linell F., Norden A., Norden A. Mycobacterium balnei, a new acid-fast bacillus occurring in swimming pools and capable of producing skin lesions in humans. Acta Tuberc. Scand. Suppl. 1954;33:1–84. [PubMed] [Google Scholar]
- Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- MacGregor R.R. Cutaneous tuberculosis. Clin. Dermatol. 1995;13:245–255. doi: 10.1016/0738-081x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- MacGurn J.A., Cox J.S., Cox J.S. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 2007;75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn J.A., Raghavan S., Stanley S.A., Cox J.S., Raghavan S., Stanley S.A., Cox J.S., Stanley S.A., Cox J.S., Cox J.S. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol. Microbiol. 2005;57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- Maciag A., Dainese E., Rodriguez G.M., Milano A., Provvedi R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R., Dainese E., Rodriguez G.M., Milano A., Provvedi R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R., Rodriguez G.M., Milano A., Provvedi R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R., Milano A., Provvedi R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R., Provvedi R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R., Smith I., Palu G., Riccardi G., Manganelli R., Palu G., Riccardi G., Manganelli R., Riccardi G., Manganelli R., Manganelli R. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga I., Bhatt A., Young D.C., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Bhatt A., Young D.C., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Young D.C., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Porcelli S.A., Costello C.E., Jacobs W.R., Costello C.E., Jacobs W.R., Jacobs W.R., et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J. Exp. Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P.K., Pandey A.K., Subbian S., El-Etr S.H., Cirillo S.L., Samrakandi M.M., Cirillo J.D., Pandey A.K., Subbian S., El-Etr S.H., Cirillo S.L., Samrakandi M.M., Cirillo J.D., Subbian S., El-Etr S.H., Cirillo S.L., Samrakandi M.M., Cirillo J.D., El-Etr S.H., Cirillo S.L., Samrakandi M.M., Cirillo J.D., Cirillo S.L., Samrakandi M.M., Cirillo J.D., Samrakandi M.M., Cirillo J.D., Cirillo J.D. Identification of Mycobacterium marinum macrophage infection mutants. Microb. Pathog. 2006;40:139–151. doi: 10.1016/j.micpath.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Okamoto Y., Fujita Y., Naka T., Hirai M., Tomiyasu I., Yano I., Fujita Y., Naka T., Hirai M., Tomiyasu I., Yano I., Naka T., Hirai M., Tomiyasu I., Yano I., Hirai M., Tomiyasu I., Yano I., Tomiyasu I., Yano I., Yano I. Mycobacterial sulfolipid shows a virulence by inhibiting cord factor induced granuloma formation and TNF-alpha release. Microb. Pathog. 2006;40:245–253. doi: 10.1016/j.micpath.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Onwueme K.C., Vos C.J., Zurita J., Ferreras J.A., Quadri L.E., Vos C.J., Zurita J., Ferreras J.A., Quadri L.E., Zurita J., Ferreras J.A., Quadri L.E., Ferreras J.A., Quadri L.E., Quadri L.E. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid Res. 2005;44:259–302. doi: 10.1016/j.plipres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Churcher C.M., Bentley S.D., Mungall K.L., Bentley S.D., Mungall K.L., Mungall K.L., et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Petrini B. Mycobacterium marinum: Ubiquitous agent of waterborne granulomatous skin infections. Eur. J. Clin. Microbiol. Infect. Dis. 2006;25:609–613. doi: 10.1007/s10096-006-0201-4. [DOI] [PubMed] [Google Scholar]
- Portevin D., De Sousa-D’Auria C., Houssin C., Grimaldi C., Chami M., Daffe M., Guilhot C., De Sousa-D’Auria C., Houssin C., Grimaldi C., Chami M., Daffe M., Guilhot C., Houssin C., Grimaldi C., Chami M., Daffe M., Guilhot C., Grimaldi C., Chami M., Daffe M., Guilhot C., Chami M., Daffe M., Guilhot C., Daffe M., Guilhot C., Guilhot C. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. 2004;101:314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozos T.C., Ramakrishnan L., Ramakrishnan L. New models for the study of Mycobacterium–host interactions. Curr. Opin. Immunol. 2004;16:499–505. doi: 10.1016/j.coi.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan L., Falkow S., Falkow S. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 1994;62:3222–3229. doi: 10.1128/iai.62.8.3222-3229.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan L., Tran H.T., Federspiel N.A., Falkow S., Tran H.T., Federspiel N.A., Falkow S., Federspiel N.A., Falkow S., Falkow S. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: Isolation, characterization, and gene disruption via homologous recombination. J. Bacteriol. 1997;179:5862–5868. doi: 10.1128/jb.179.18.5862-5868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan L., Federspiel N.A., Falkow S., Federspiel N.A., Falkow S., Falkow S. Granuloma-specific expression of mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- Reed M.B., Domenech P., Manca C., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., Domenech P., Manca C., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., Manca C., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., Kreiswirth B.N., Kaplan G., Barry C.E., Kaplan G., Barry C.E., Barry C.E. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- Ren H., Dover L.G., Islam S.T., Alexander D.C., Chen J.M., Besra G.S., Liu J., Dover L.G., Islam S.T., Alexander D.C., Chen J.M., Besra G.S., Liu J., Islam S.T., Alexander D.C., Chen J.M., Besra G.S., Liu J., Alexander D.C., Chen J.M., Besra G.S., Liu J., Chen J.M., Besra G.S., Liu J., Besra G.S., Liu J., Liu J. Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 2007;63:1345–1359. doi: 10.1111/j.1365-2958.2007.05603.x. [DOI] [PubMed] [Google Scholar]
- Rezwan M., Grau T., Tschumi A., Sander P., Grau T., Tschumi A., Sander P., Tschumi A., Sander P., Sander P. Lipoprotein synthesis in mycobacteria. Microbiology. 2007;153:652–658. doi: 10.1099/mic.0.2006/000216-0. [DOI] [PubMed] [Google Scholar]
- Robinson N., Wolke M., Ernestus K., Plum G., Wolke M., Ernestus K., Plum G., Ernestus K., Plum G., Plum G. A mycobacterial gene involved in synthesis of an outer cell envelope lipid is a key factor in prevention of phagosome maturation. Infect. Immun. 2007;75:581–591. doi: 10.1128/IAI.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F., Bacher A., Eisenreich W., Bacher A., Eisenreich W., Eisenreich W. Perspectives in anti-infective drug design. The late steps in the biosynthesis of the universal terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. Bioorg. Chem. 2004;32:292–308. doi: 10.1016/j.bioorg.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Rosas-Magallanes V., Deschavanne P., Quintana-Murci L., Brosch R., Gicquel B., Neyrolles O., Deschavanne P., Quintana-Murci L., Brosch R., Gicquel B., Neyrolles O., Quintana-Murci L., Brosch R., Gicquel B., Neyrolles O., Brosch R., Gicquel B., Neyrolles O., Gicquel B., Neyrolles O., Neyrolles O. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol. Biol. Evol. 2006;23:1129–1135. doi: 10.1093/molbev/msj120. [DOI] [PubMed] [Google Scholar]
- Saad M.H., Fonseca L.D., Ferrazoli L., Fandinho F., Palaci M., Grinsztejn B., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Fonseca L.D., Ferrazoli L., Fandinho F., Palaci M., Grinsztejn B., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Ferrazoli L., Fandinho F., Palaci M., Grinsztejn B., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Fandinho F., Palaci M., Grinsztejn B., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Palaci M., Grinsztejn B., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Grinsztejn B., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Kritski A., Werneck A., Poltoratskaia N., Johnson W.D., Werneck A., Poltoratskaia N., Johnson W.D., Poltoratskaia N., Johnson W.D., Johnson W.D., et al. IS1245 genotypic analysis of Mycobacterium avium isolates from patients in Brazil. Int. J. Infect. Dis. 1999;3:192–196. doi: 10.1016/s1201-9712(99)90023-1. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sassetti C.M., Boyd D.H., Rubin E.J., Boyd D.H., Rubin E.J., Rubin E.J. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Singh A., Tyagi A.K., Singh A., Tyagi A.K., Tyagi A.K. Deciphering the genes involved in pathogenesis of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 2005;85:325–335. doi: 10.1016/j.tube.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Slosarek M., Kubin M., Pokorny J., Kubin M., Pokorny J., Pokorny J. Water as a possible factor of transmission in mycobacterial infections. Cent. Eur. J. Public Health. 1994;2:103–105. [PubMed] [Google Scholar]
- Solomon J.M., Leung G.S., Isberg R.R., Leung G.S., Isberg R.R., Isberg R.R. Intracellular replication of Mycobacterium marinum within Dictyostelium discoideum: Efficient replication in the absence of host coronin. Infect. Immun. 2003;71:3578–3586. doi: 10.1128/IAI.71.6.3578-3586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm L.M., Brown E.J., Brown E.J. Mycobacterium marinum: The generalization and specialization of a pathogenic mycobacterium. Microbes Infect. 2004;6:1418–1428. doi: 10.1016/j.micinf.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Stinear T.P., Jenkin G.A., Johnson P.D.R., Davies J.K., Jenkin G.A., Johnson P.D.R., Davies J.K., Johnson P.D.R., Davies J.K., Davies J.K. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 2000;182:6322–6330. doi: 10.1128/jb.182.22.6322-6330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T.P., Mve-Obiang A., Small P.L., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Mve-Obiang A., Small P.L., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Small P.L., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Johnson P.D., Davies J.K., Lee R.E., Davies J.K., Lee R.E., Lee R.E., et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T.P., Seemann T., Pidot S., Frigui W., Reysset G., Garnier T., Meurice G., Simon D., Bouchier C., Ma L., Seemann T., Pidot S., Frigui W., Reysset G., Garnier T., Meurice G., Simon D., Bouchier C., Ma L., Pidot S., Frigui W., Reysset G., Garnier T., Meurice G., Simon D., Bouchier C., Ma L., Frigui W., Reysset G., Garnier T., Meurice G., Simon D., Bouchier C., Ma L., Reysset G., Garnier T., Meurice G., Simon D., Bouchier C., Ma L., Garnier T., Meurice G., Simon D., Bouchier C., Ma L., Meurice G., Simon D., Bouchier C., Ma L., Simon D., Bouchier C., Ma L., Bouchier C., Ma L., Ma L., et al. Reductive evolution and niche-adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;7:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim L.E., Connolly L.E., Volkman H.E., Humbert O., Born D.E., Ramakrishnan L., Connolly L.E., Volkman H.E., Humbert O., Born D.E., Ramakrishnan L., Volkman H.E., Humbert O., Born D.E., Ramakrishnan L., Humbert O., Born D.E., Ramakrishnan L., Born D.E., Ramakrishnan L., Ramakrishnan L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect. Immun. 2006;74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F., Gordon S.V., Garnier T., Brosch R., Barrell B.G., Cole S.T., Gordon S.V., Garnier T., Brosch R., Barrell B.G., Cole S.T., Garnier T., Brosch R., Barrell B.G., Cole S.T., Brosch R., Barrell B.G., Cole S.T., Barrell B.G., Cole S.T., Cole S.T. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- Tian J., Bryk R., Itoh M., Suematsu M., Nathan C., Bryk R., Itoh M., Suematsu M., Nathan C., Itoh M., Suematsu M., Nathan C., Suematsu M., Nathan C., Nathan C. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: Identification of alpha-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. 2005;102:10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma F. [Nontuberculous mycobacteria in the hospital environment] Bacteriol. Virusol. Parazitol. Epidemiol. 1998;43:229–235. [PubMed] [Google Scholar]
- Tønjum T., Welty D.B., Jantzen E., Small P.L., Welty D.B., Jantzen E., Small P.L., Jantzen E., Small P.L., Small P.L. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: Mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA–DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 1998;36:918–925. doi: 10.1128/jcm.36.4.918-925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis W.D., Travis L.B., Roberts G.D., Su D.W., Weiland L.W., Travis L.B., Roberts G.D., Su D.W., Weiland L.W., Roberts G.D., Su D.W., Weiland L.W., Su D.W., Weiland L.W., Weiland L.W. The histopathologic spectrum in Mycobacterium marinum infection. Arch. Pathol. Lab. Med. 1985;109:1109–1113. [PubMed] [Google Scholar]
- Trivedi O.A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R.S., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R.S., Sridharan V., Tickoo R., Mohanty D., Gokhale R.S., Tickoo R., Mohanty D., Gokhale R.S., Mohanty D., Gokhale R.S., Gokhale R.S. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- Tsukamura M. Numerical classification of 280 strains of slowly growing mycobacteria. Proposal of Mycobacterium tuberculosis series, Mycobacterium avium series, and Mycobacterium nonchromogenicum series. Microbiol. Immunol. 1983;27:315–334. doi: 10.1111/j.1348-0421.1983.tb00591.x. [DOI] [PubMed] [Google Scholar]
- van Embden J.D., van Gorkom T., Kremer K., Jansen R., van Der Zeijst B.A., Schouls L.M., van Gorkom T., Kremer K., Jansen R., van Der Zeijst B.A., Schouls L.M., Kremer K., Jansen R., van Der Zeijst B.A., Schouls L.M., Jansen R., van Der Zeijst B.A., Schouls L.M., van Der Zeijst B.A., Schouls L.M., Schouls L.M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G.S., Parkhill J., Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: Revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Volkman H.E., Clay H., Beery D., Chang J.C., Sherman D.R., Ramakrishnan L., Clay H., Beery D., Chang J.C., Sherman D.R., Ramakrishnan L., Beery D., Chang J.C., Sherman D.R., Ramakrishnan L., Chang J.C., Sherman D.R., Ramakrishnan L., Sherman D.R., Ramakrishnan L., Ramakrishnan L. Tuberculous granuloma formation is enhanced by a Mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehenkel A., Bellinzoni M., Grana M., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cervenansky C., Bellinzoni M., Grana M., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cervenansky C., Grana M., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cervenansky C., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cervenansky C., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cervenansky C., Fernandez P., Andre-Leroux G., England P., Takiff H., Cervenansky C., Andre-Leroux G., England P., Takiff H., Cervenansky C., England P., Takiff H., Cervenansky C., Takiff H., Cervenansky C., Cervenansky C., et al. Mycobacterial Ser/Thr protein kinases and phosphatases: Physiological roles and therapeutic potential. Biochim. Biophys. Acta. 2007;1784:193–202. doi: 10.1016/j.bbapap.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wei J., Dahl J.L., Moulder J.W., Roberts E.A., O’Gaora P., Young D.B., Friedman R.L., Dahl J.L., Moulder J.W., Roberts E.A., O’Gaora P., Young D.B., Friedman R.L., Moulder J.W., Roberts E.A., O’Gaora P., Young D.B., Friedman R.L., Roberts E.A., O’Gaora P., Young D.B., Friedman R.L., O’Gaora P., Young D.B., Friedman R.L., Young D.B., Friedman R.L., Friedman R.L. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 2000;182:377–384. doi: 10.1128/jb.182.2.377-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M.J., Porter J.L., Fyfe J.A., Lavender C.J., Portaels F., Rhodes M., Kator H., Colorni A., Jenkin G.A., Stinear T., Porter J.L., Fyfe J.A., Lavender C.J., Portaels F., Rhodes M., Kator H., Colorni A., Jenkin G.A., Stinear T., Fyfe J.A., Lavender C.J., Portaels F., Rhodes M., Kator H., Colorni A., Jenkin G.A., Stinear T., Lavender C.J., Portaels F., Rhodes M., Kator H., Colorni A., Jenkin G.A., Stinear T., Portaels F., Rhodes M., Kator H., Colorni A., Jenkin G.A., Stinear T., Rhodes M., Kator H., Colorni A., Jenkin G.A., Stinear T., Kator H., Colorni A., Jenkin G.A., Stinear T., Colorni A., Jenkin G.A., Stinear T., Jenkin G.A., Stinear T., Stinear T. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 2007;189:2021–2029. doi: 10.1128/JB.01442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Zhang Y., Teh J.S., Zhang J., Connell N., Rubin H., Inouye M., Zhang Y., Teh J.S., Zhang J., Connell N., Rubin H., Inouye M., Teh J.S., Zhang J., Connell N., Rubin H., Inouye M., Zhang J., Connell N., Rubin H., Inouye M., Connell N., Rubin H., Inouye M., Rubin H., Inouye M., Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]