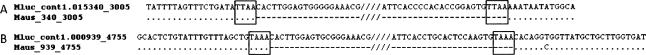Abstract
DNA transposons, or class 2 transposable elements, have successfully propagated in a wide variety of genomes. However, it is widely believed that DNA transposon activity has ceased in mammalian genomes for at least the last 40 million years. We recently reported evidence for the relatively recent activity of hAT and Helitron elements, two distinct groups of DNA transposons, in the lineage of the vespertilionid bat Myotis lucifugus. Here, we describe seven additional families that have also been recently active in the bat lineage. Early vespertilionid genome evolution was dominated by the activity of Helitrons, mariner-like and Tc2-like elements. This was followed by the colonization of Tc1-like elements, and by a more recent explosion of hAT-like elements. Finally, and most recently, piggyBac-like elements have amplified within the Myotis genome and our results indicate that one of these families is probably still expanding in natural populations. Together, these data suggest that there has been tremendous recent activity of various DNA transposons in the bat lineage that far exceeds those previously reported for any mammalian lineage. The diverse and recent populations of DNA transposons in genus Myotis will provide an unprecedented opportunity to study the impact of this class of elements on mammalian genome evolution and to better understand what makes some species more susceptible to invasion by genomic parasites than others.
Transposable elements represent a substantial fraction of many eukaryotic genomes. For example, ∼50% of the human genome is derived from transposable element sequences (Lander et al. 2001). Other genomes, especially those of plants, may consist of substantially higher proportions of transposable element-derived DNA (SanMiguel and Bennetzen 1998; Bennetzen 2000). Transposable elements are typically divided into two classes (Wicker et al. 2007). Class 1 is represented by the retrotransposons (LINEs, SINEs, LTRs, and ERVs). Class 2 includes the “cut-and-paste” DNA transposons, which are characterized by terminal inverted repeats (TIRs) and are mobilized by an element-encoded transposase. Currently, 10 superfamilies of cut-and-paste DNA transposons are recognized in eukaryotes (Feschotte and Pritham 2007).
While class 2 elements are widespread and active in a variety of eukaryotes, they have been thought to be transpositionally inactive in mammalian genomes. This conclusion was based primarily on the initial analyses of the human and mouse genome sequences. While both species harbor a significant number and a diverse assortment of DNA transposons, they show no signs of recent activity (Lander et al. 2001; Waterston et al. 2002). For example, there are more than 300,000 DNA elements recognizable in the human genome, which are grouped into 120 families and belong to five superfamilies. A large subset of these elements (40 families; ∼98,000 copies) were integrated in the last 40–80 million years (Myr), but there remains no evidence for any human DNA transposon families younger than ∼37 Myr (Pace and Feschotte 2007).
In stark contrast to the situation in human and mouse, there is mounting evidence for recent and substantial DNA transposon activity in the vespertilionid bat Myotis lucifugus. In one study, this conclusion was inferred from the presence of minimally diverged nonautonomous hAT transposons, the polymorphic status of some of these transposons in natural Myotis populations, and the discovery of an apparently full-length and potentially functional autonomous hAT transposon (Ray et al. 2007). In a separate report, an extensive and recently amplified population of Helitrons, a distinct subclass of DNA transposons, was discovered in the M. lucifugus genome (Pritham and Feschotte 2007). These surprising observations raised a number of intriguing questions about the activity of DNA transposons and its genomic impact in Myotis and other bats. In particular, we wondered whether the discovery of relatively recent hAT and Helitron activity in Myotis were merely coincidental or whether it reflected some unique biological or genomic features of bats in general or vespertilionids in particular that has allowed DNA transposons to invade and expand in these genomes, while such occurrences appear to be rare or nonexistent in other mammalian genomes. As a prelude to addressing these questions we examined M. lucifugus for additional traces of DNA transposon activity. Consequently, we explored the diversity, abundance, and evolutionary history of DNA transposons in the M. lucifugus genome.
Herein, we characterize seven new families of DNA transposons from the current whole-genome sequence of M. lucifugus. In addition, we provide additional information on the previously described Myotis_hAT transposons (Ray et al. 2007). Together, these elements represent three distinct superfamilies and exhibit clear signs of recent activity in the vespertilionid lineage, that is, within the last 40–50 Myr. We also identified a ninth transposon lineage that likely expanded prior to the chiropteran divergence. One family represents the youngest DNA transposon family so far recorded in any mammalian species, one that is likely still expanding in the genome. The discovery of this unprecedented level of DNA transposon activity in a mammalian genome represents a dramatic shift in our view of mobile element biology in mammals.
Results
hAT superfamily
The consensus sequence for Myotis_hAT1 was previously described (Ray et al. 2007). To summarize, the entire sequence spans 2921 nucleotides with a single ORF consisting of bases 700–2631 that encodes an apparently intact transposase of 643 amino acids (aa). The present analysis confirms the characteristic target-site duplications (TSDs) for Myotis hAT elements, 8 bp with a central TA dinucleotide, and the typical short TIR (terminal inverted repeats) sequence. Myotis_hAT1 and its nonautonomous derivatives are by far the most abundant family of elements analyzed in this study, with >96,000 hits of ≥100 bp in the current WGS data.
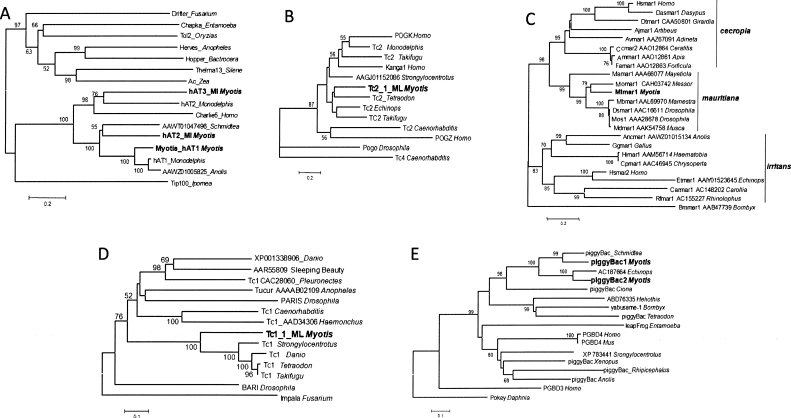
We identified a second family of hAT elements, called hAT2_ML. The TIRs of hAT2_ML are highly similar to Myotis_ hAT1, but the internal sequence is only weakly similar. hAT2_ML is predicted to encode a 428-aa transposase with only minimal sequence similarity and 33% amino acid identity to the Myotis_ hAT1 transposase. Phylogenetic comparison to other hAT transposases confirms that Myotis_hAT1 and hAT2_ML belong to the hAT superfamily and are closely related to hAT1_MD (Repbase Update, volume 12, issue 10), a hAT transposon family recently identified in the opossum Monodelphis domestica (Fig. 1A). RepeatMasker results using the full consensus as a query suggests that hAT2_ML is not nearly as widespread as Myotis_ hAT1 with only 2004 hits of ≥100 bp.
Figure 1.
Results of phylogenetic analyses. (A) hAT transposases, (B) Tc2 transposases, (C) mariner transposases, (D) Tc1 transposases, (E) piggyBac transposases.
We also recovered a third hAT family member, hAT3_ML, from the M. lucifugus genome. The consensus sequence contains a large ORF spanning base pairs 1595–3403 and encoding a putative protein of 602 aa, which nests in phylogenetic analysis within a clade encompassing several mammalian hAT transposases (Fig. 1A). With its 3904-bp consensus, it is the longest of all of the elements described in this study and is second to Myotis_hAT1 in terms of its abundance with >12,000 elements of ≥100 bp identified in the current WGS assembly of M. lucifugus.
Tc1/mariner superfamily
Eukaryotic Tc1/mariner elements are divided into several anciently diverged lineages (Robertson 2002; Feschotte and Pritham 2007). Three distinct Tc1/mariner lineages have been previously identified in mammals and characterized most extensively in human: pogo, Tc2, and D34D mariner. We identified members of all three groups in M. lucifugus as well as the first mammalian member of the Tc1 group. The pogo lineage is represented by Tigger elements in mammals. It is known to include both eutherian-wide and primate-specific families (Robertson 1996; Smit and Riggs 1996; Pace and Feschotte 2007). Thus, the presence of Tigger elements was somewhat expected in M. lucifugus. Indeed, we identified remnants of what appears to be an ancient family of Tigger-like elements called Tigger1_ML. A tentative consensus was reconstructed from the alignment of the seven longest copies. The size (2.8 kb) and TIRs are consistent with other Tigger elements and the consensus is similar to consensus sequences in Repbase as Tigger1a_CAR and Tigger1a_ART, which represent Tigger1-like families specific to carnivores and artiodactyls, respectively. We also identified several highly eroded copies of a related family of Tigger1 transposons in the European hedgehog, Erinaceus europaeus (e.g., accession AANN01830819, position 1599–4234). Given that hedgehog, carnivores, artiodactyls, and chiropterans are all part of the well-supported Laurasiatheria superorder, we hypothesize that Tigger1_ML amplified during the early part of the Laurasiatherian radiation. This is by far the oldest family of DNA transposons that we identified and, as it does not appear to be unique to chiropterans, we have chosen not to focus on the details of this family in the current study.
The Tc2 group is sister to the pogo lineage and was first identified in Caenorhabditis elegans (Ruvolo et al. 1992). There are relatively few Tc2-like elements in human, and their amplification predates the split of eutherian mammals (Pace and Feschotte 2007). In contrast, we identified a distinct and much more recent family of Tc2-like elements in M. lucifugus. The Tc2_1_ML consensus is 1728 bp with 23 bp TIRs similar to known or newly identified Tc2 elements. It contains a single ORF spanning positions 213–1559 and encoding a putative transposase of 448 aa. Phylogenetic analysis confirms that Tc2_1_ML belongs to the Tc2 group, but does not cluster with either of the mammalian Tc2-like transposases identified in human (Kanga), opposum (Tc2_MD2), and tenrec (Tc2_Et), or with the human transposase-derived proteins POGK and POGZ (Fig. 1B). Thus, Tc2_1_ML defines a distinct lineage of Tc2-like elements. Tc2_1_ML is the least abundant of the DNA transposon families described in this study with only 589 hits ≥100 bp.
Two distinct lineages of canonical D34D mariner are known to occur in mammals: the cecropia subfamily, represented by Hsmar1 in anthropoid primates and Mmar1 in mouse, and the irritans subfamily, represented by Hsmar2 in primates (Robertson 2002). We detected an abundant (2036 hits >100 bp) family of transposons in M. lucifugus, Mlmar1, that displays all the features of a canonical D34D mariner. The Mlmar1 consensus is 1287 bp with 30-bp TIRs and a single ORF, spanning position 174–1184 and predicted to encode a 336-aa transposase. Surprisingly, phylogenetic analyses robustly place the predicted Mlmar1 transposase within the mauritiana subfamily of mariner (Fig. 1C), a subfamily typified by the original mos1 element active in Drosophila mauritiana (Hartl et al. 1997; Robertson 2002). We could not detect any members of the irritans or cecropia subfamilies of mariner in M. lucifugus using protein queries representing these lineages, although these two lineages have been identified in a broad range of mammals (Demattei et al. 2000; Robertson 2002; Waterston et al. 2002; Sinzelle et al. 2006), including other bat species (see below).
Tc1-like elements represent another distinct lineage of Tc1/mariner elements that are widespread and common in invertebrates and lower vertebrates (fish, frogs), but no Tc1-like element has previously been identified in mammals (Avancini et al. 1996; Leaver 2001; Sinzelle et al. 2005). Consistent with this observation, TBLASTN searches with representative Tc1-like transposases from fish or nematode return no significant hits in the complete genome of human, mouse, rat, and dog. In contrast, the same query returns hundreds of hits from the M. lucifugus WGS database. A single Tc1-like family, Tc1_1_ML, predominates in the genome. The Tc1_1_ML consensus spans 1222 bp with 29 bp TIR with a 5′-CACTG-3′ terminal motif typical of many Tc1-like elements. The ORF occupies position 150–1190 of the consensus and encodes a putative transposase of 346 residues. Phylogenetic analyses show that the Tc1_1_ML transposase forms a well-supported clade with Tc1-like transposases from fish (Fig. 1D). We detected 3788 Tc1_1_ML fragments of 100 bp or larger in the M. lucifugus WGS database. Many of the Tc1_1_ML elements are inserted in (TA) dinucleotide repeats, an insertion preference also observed for other Tc1 elements (Vigdal et al. 2002). Of note is one subfamily derived from Tc1_1_ML. The consensus of this subfamily has an intact ORF that represents 2/3 (708 bp; 235 aa) of the complete ORF for Tc1_1_ML. Five hundred copies of this subfamily of Tc1 are distributed throughout the M. lucifugus genome.
piggyBac superfamily
piggyBac-like elements have been identified in a wide range of animal species and Entamoeba (Sarkar et al. 2003; Pritham et al. 2005). Among mammals, piggyBac elements have so far only been characterized in the human genome, where they are predominantly represented by two families of nonautonomous elements (MER85 and MER75) and by several stationary “domesticated” transposases (PGBD1-5 genes) (Sarkar et al. 2003). We identified two families of piggyBac-like elements in M. lucifugus. piggyBac1_ML is defined by a 2626-bp consensus with short TIRs (15 bp) that are very similar to other piggyBac transposons. The consensus contains a 1719-bp ORF (position 587–2305) that likely encodes a transposase of 572 residues. Two potentially active elements, with intact ORFs and identifiable TSDs, were located in the available genome data. piggyBac2_ML is similar in length at 2639 bp and with a 583-aa transposase (nt 716–2467). Sequence comparison with other piggyBac transposases and phylogenetic analyses indicate that the two families are relatively closely related and form a strong cluster with the sea squirt, Ciona intestinalis (Fig. 1E). Myotis piggyBac-like elements tend to be flanked by the canonical 5′-TTAA-3′ TSD. piggyBac1_ ML elements number at least 2056 in the M. lucifugus genomes and are likely still mobilizing (see below). However, piggyBac2_ML elements are currently more numerous, with at least 3869 instances.
In summary, we identified a diverse array of hAT, Tc1/mariner, and piggyBac transposons in the M. lucifugus genome. The Myotis_hAT1 and hAT3_ML elements are by far the most abundant. As with Myotis_hAT1 elements (Ray et al. 2007), all of the newly discovered transposon families are associated with multiple nonautonomous MITE subfamilies. It is beyond the scope of the present study to describe all of these subfamilies in detail, but we note that nonautonomous elements appear to have outnumbered their autonomous partners, as is typically observed for DNA transposon families in humans and most other eukaryotes. (Feschotte et al. 2002; Pace and Feschotte 2007)
Taxonomic distribution of DNA transposons identified in Myotis lucifugus
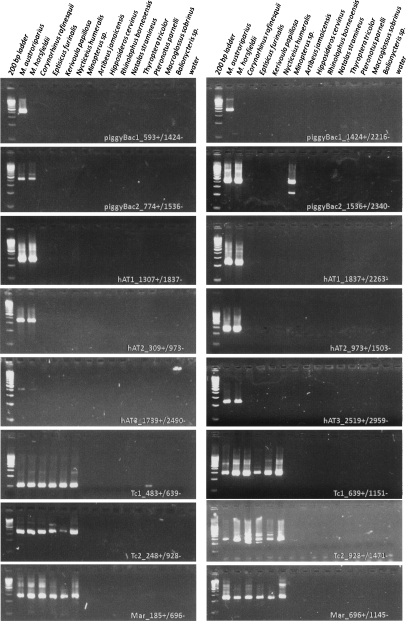
PCR-based analyses of 15 taxa representing nine bat families were performed using two oligonucleotide primer pairs for each transposon family (Fig. 2). Results indicate that the families described have limited taxonomic distributions. Tc1_1_ML, Tc2_1_ML, and Mlmar1 appear to be limited to Vespertilionidae with the possible exception of Tc1-like sequences in Pteronotus parnelli (Family Mormoopidae). All three hAT families are restricted to the genus Myotis. piggyBac2_ML shares a similar distribution to the hAT elements, but positive results were obtained from the lone representative of Miniopteridae, a family that was recently elevated from subfamily status within Vesperdilionidae (Miller-Butterworth et al. 2007). Amplification of the miniopterid and mormoopid representatives suggests the need for additional exploration of these taxa. PCR data using piggyBac1_ML-derived oligonucleotides suggests that these elements are even further restricted. Amplification was obtained in Myotis austroriparius, a fellow North American bat, but not the Asian representative, Myotis horsfieldii. We acknowledge that these results do not confirm that no class 2 transposons are active in other bat lineages. Instead, they lend support to the hypothesis that these particular transposons have a limited taxonomic distribution.
Figure 2.
Electropherogram results from PCR analysis of a taxonomically diverse panel of bat genomes. Oligonucleotide primer pairs are indicated in white.
With the exception of Tigger1_ML, whose amplification likely predates the split of Chiroptera or perhaps even Laurasiatheria, TBLASTN and BLASTN queries of chiropteran taxa with the consensus transposon sequences from M. lucifugus yielded only very few significant hits in other bat species. A handful of hits were obtained with the Mlmar1 predicted transposase in the phyllostomid bats Artibeus jamaicensis and Carollia perspicillata, and in the rhinolophid bat, Rhinolophus ferrumenquinum. Interestingly, the hits correspond to cecropia-like and the previously described irritans-like mariners (Sinzelle et al. 2006) (Fig. 1C). Thus, it appears that distinct types of mariners have colonized different bat lineages. Mlmar1 is most closely related to Mboumar1, a mauritiana-like mariner from the ant Messor bouvieri (see also phylogeny in Fig. 1C). The transposase regions are 75% identical at the nucleotide level and 85% similar at the protein level. This level of sequence similarity is remarkable given the evolutionary distance of their host species, >800 Myr (Gu 1998; Blair and Hedges 2005). We could not detect any traces of other mauritiana-like mariner transposases in any of the ∼30 additional mammalian species nor any other chordate species represented in the NCBI databases.
Age estimations of DNA transposon families
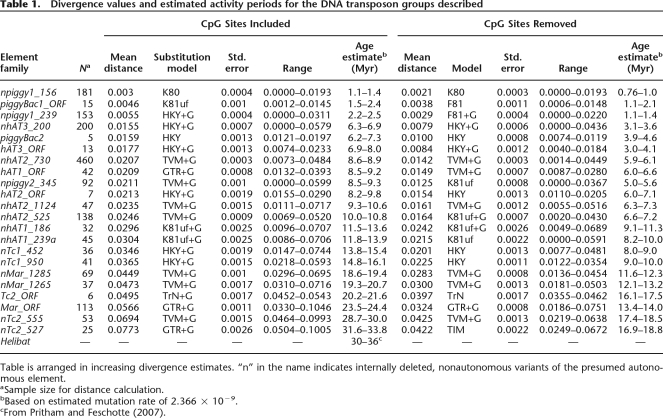
Table 1 and Figure 3 show the estimated ages for each family based on average divergence from the consensus sequence and an estimated neutral mutation rate of ∼2.366 × 10−9 (see Methods). Age analyses of the eight youngest families produce a clear temporal pattern indicative of successive expansions in the genome. The earliest invasions began with Tc2-like and mariner-like elements and were followed by the colonization of Tc1_1_ML, hAT-like, and piggyBac-like families.
Table 1.
Divergence values and estimated activity periods for the DNA transposon groups described
Table is arranged in increasing divergence estimates. “n” in the name indicates internally deleted, nonautonomous variants of the presumed autonomous element.
aSample size for distance calculation.
bBased on estimated mutation rate of 2.366 × 10−9.
cFrom Pritham and Feschotte (2007).
Figure 3.
Divergence estimates and age ranges for each autonomous and nonautonomous transposon family described.
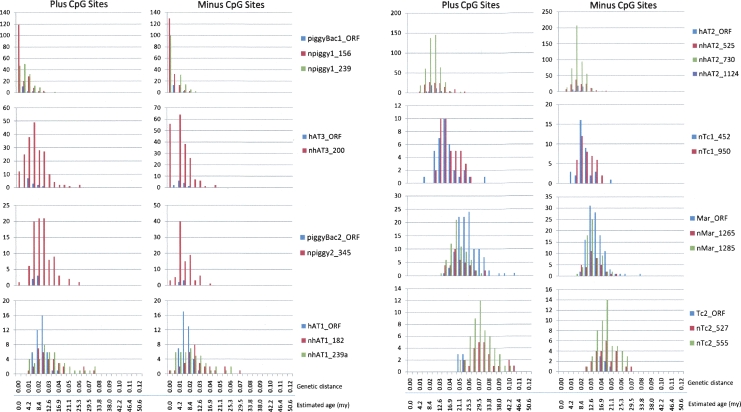
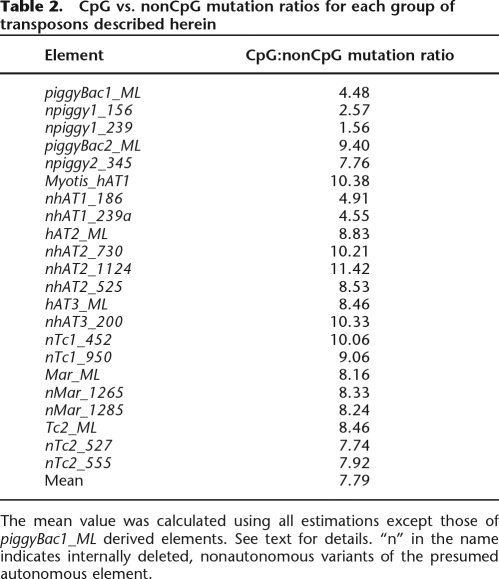
In mammals, CpG dinucleotides within repeats are generally heavily methylated, and thus tend to degrade into TpG or CpA dinucleotides at a substantially higher rate than other base combinations (Coulondre et al. 1978; Razin and Riggs 1980; Xing et al. 2004). Thus, lower divergence estimates were expected and observed when calculated using sequences from which the CpG dinucleotides had been removed. CpG to non-CpG mutations densities were calculated for all families (Table 2). The youngest subfamilies, derived from piggyBac1_ML, produced consistently lower ratios than any other subfamily. This is most likely explained by their very recent deposition in the genome (see below) and bcause they have not resided long enough to accumulate many mutations of any kind. This hypothesis, however, does not explain the relatively low ratios observed for the nhAT1 subfamilies, which we believe were mobilized as much as 10 Myr earlier. Average CpG dinucleotide mutation densities were approximately eight times higher than non-CpG sites. We note that this is higher than the rate observed for primates (∼6×; Xing et al. 2004), bringing to mind the hypothesis that increased methylation, and consequently, high CpG:non-CpG mutation ratios may serve as a genomic defense mechanism against large bursts of transposable element activity (Schmid 1998; Xing et al. 2004).
Table 2.
CpG vs. nonCpG mutation ratios for each group of transposons described herein
The mean value was calculated using all estimations except those of piggyBac1_ML derived elements. See text for details. “n” in the name indicates internally deleted, nonautonomous variants of the presumed autonomous element.
Current activity by piggyBac1_ML
Both genetic distance and CpG analyses above provide evidence for very recent and likely ongoing piggyBac1_ML activity. Indeed, a large fraction of the nonautonomous piggyBac1 elements identified in M. lucifugus are strictly identical (65.7% of the elements in the npiggy_156 subfamily and 30.7% in the npiggy_239 subfamily), which strongly indicates that they have been inserted very recently. As one can observe in Figure 3, most of the DNA transposon families analyzed in this study appear to have reached an amplification peak and then entered a period of reduced activity and extinction. In contrast, the npiggy_156 subfamily appears to be nearing its amplification peak, suggesting that piggyBac1_ML may still be active in the Myotis genome. The identification of two full-length elements with intact ORFs in the current genome sequence data further support this hypothesis. Further evidence for recent piggyBac1_ML activity is in the observation that we were able to experimentally isolate intact preintegration sites for npiggy_156 elements in a related species, M. austroriparius (Fig. 4). Finally, experimental results from a diverse panel of bat species indicate that piggyBac1_ML is present in only a subset of species in the genus (Fig. 2). Thus, we can confidently state that this particular subfamily was mobilized within the last ∼8–12 Myr (Stadelmann et al. 2007).
Figure 4.
Filled and intact preintegration site sequences for individual nonautonomous piggyBac1_ML elements from M. lucifugus and M. austroriparius, respectively. Target site duplications for each insertion are shaded. (A) Locus 340_3005. (B) Locus primer sequences (5′–3′) for the two loci presented are as follows: Mluc_cont1.015340_3005_L—TTGTACCAAAAGGTGCCAAA, Mluc_cont1.015340_3005_R—TTTCCTCATATACCATCCCATTTT; Mluc_cont1.000939_4755_L—CAAAATAAGGGAGAAAGGAAACA, Mluc_cont1.000939_4755_R—GGGCTGAGAAACAAGATCCA. (Mluc) M. lucifugus; (Maus) M. austroriparius.
Nested insertion analysis
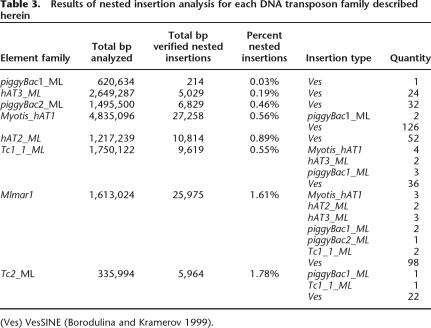
As a second, independent method to estimate relative ages of the elements, we compared the number and identity of nested insertions within individual transposon copies. Assuming that DNA transposon insertions occur fairly randomly throughout the genome, older elements should have higher proportions of nested insertions than younger elements. Furthermore, while it is possible for representatives from older transposon families to become nested within younger families if their periods of activity overlapped, these instances should be relatively rare. RepeatMasker hits for each family of transposons were subjected to a second RepeatMasker analysis using a custom library that included each of the new transposable elements described here and Ves, a SINE found in the suborder Yangochiroptera (Borodulina and Kramerov 1999, 2005). We limited ourselves to these elements because their sequences have been well characterized here or elsewhere (Borodulina and Kramerov 1999; Ray et al. 2007) and their target site duplications are easily identified.
Analysis of nested insertions supports the age hierarchy suggested by divergence estimates. Among the hAT and piggyBac elements examined, only two nested DNA transposon insertions were recovered—two piggyBac1 insertions inside a Myotis_hAT1 element (Table 3)—lending support to the hypothesis that these families are the most recently active DNA transposon families in the M. lucifugus genome. In contrast, the older elements represented by the Tc1, Tc2, and mariner families have been subjected to numerous interruptions by the more recently active elements. Mlmar1 elements have suffered nested insertions from all other families except Tc2_1_ML, which is likely older. It is interesting that the Tc2 elements have only accumulated two nested insertions by their fellow DNA transposons. This could be explained by the low occurrence of these elements (n = 593) and the relatively small size of these insertions (∼560 bp). Mariner elements are present in larger numbers (>2000) and larger average size (778 bp), and thus present approximately five times the number of potential insertion targets during an only slightly shorter time frame. Analyses of Ves SINE nested insertions present a similar picture. The four most recently active elements, piggyBac1_ML, piggyBac2_ML, hAT2_ML, and hAT3_ML have been subjected only to Ves insertions.
Table 3.
Results of nested insertion analysis for each DNA transposon family described herein
(Ves) VesSINE (Borodulina and Kramerov 1999).
Nested self-insertions are theoretically possible and were expected when the analysis was performed. Indeed, there were some possible instances observed. However, in each of these cases the TSDs were not clearly identifiable. Because TSD identification was one a priori criterion for assessing the presence of any nested insertion (see Methods), we were forced to exclude these from Table 3.
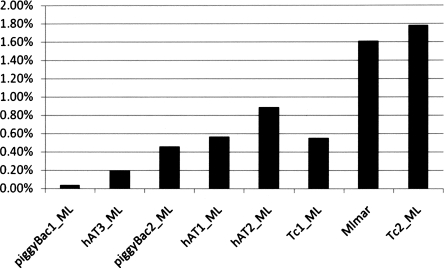
Finally, one would expect older elements to have accumulated more nested insertions than more recently mobilized elements. To test this hypothesis, we examined the percent nested insertion content for all instances of each family of elements recovered from the M. lucifugus genome. Figure 5 illustrates a clear pattern of younger elements (i.e., piggyBac-like and hAT-like) having lower relative nested insertion content when compared with older elements from the Tc1/mariner superfamily. The one exception to this pattern is Tc1_1_ML, which exhibits a relatively low percentage of nested insertions. It is unclear as to why this family exists as an outlier.
Figure 5.
Proportion of insertions nested within instances of each DNA transposon family described. Families are arranged along the X-axis in order of age based on genetic divergence estimates in Table 3. Precise values for can be found in Table 3.
Discussion
DNA transposons are widespread and a diversity of elements have been recently active in the genomes of many eukaryotes, including lower vertebrates (Aparicio et al. 2002; Koga et al. 2006). In contrast, initial analyses of the transposable element landscape in the complete human, mouse, rat, and dog genomes have led to the common belief that mammalian genomes are devoid of recently active DNA transposons (Lander et al. 2001; Waterston et al. 2002; Gibbs et al. 2004; Lindblad-Toh et al. 2005). Supporting this idea, a detailed investigation of the evolutionary history of human DNA transposons revealed that many transposons were intensively active during early primate evolution (∼80–40 million years ago [Mya]), but this activity ceased approximately ∼40 Mya (Pace and Feschotte 2007). Analyses of the rodent and carnivore lineages reveal a similar trend, with no DNA transposon family identified in either mouse or dog that is significantly younger than 50 Myr (Waterston et al. 2002; J. Pace and C. Feschotte, unpubl.).
We have identified seven new families of DNA transposons in the M. lucifugus genome which, in stark contrast to observations made for other mammalian genomes, have apparently been active within the last 40 Myr. Several lines of evidence support this conclusion. First, the elements appear to be almost exclusively limited to Vespertilionidae, and in some cases to selected taxa within the family. Second, the level of sequence divergence among copies (and to their respective consensus sequences) suggests that these families were active from ∼36 Mya to the present. Analysis of nested insertions suggests a hierarchical pattern of insertion consistent with sequence divergence estimates. Third, two potentially full-length and intact piggyBac1_ML transposons could be identified in the available genome sequence data, supporting the hypothesis that the expansion of these elements and their nonautonomous relatives is ongoing. Finally, recent activity of piggyBac1_ML was verified by recovering “empty” orthologous sites in a closely related Myotis species and by the limited taxonomic distribution of piggyBac1_ML to North American Myotis species. Together with the recent study reporting the massive amplification of HeliBats 30–36 Mya in the lineage of M. lucifugus (Pritham and Feschotte 2007), our data demonstrate that the genomes of vespertilionid bats have been subjected to multiple waves of amplification of diverse DNA transposons, from ∼40 Mya to the present. One lineage of piggyBac, piggyBac1_ML, is likely still active. To our knowledge, this is the first documented evidence of the recent activity of a diverse population of DNA transposons in any mammal.
At least 3.5% of the available genome data is made up of sequence derived from the transposons described herein. It should be noted, however, that this number is very likely an underestimate. The previous study of hAT1 elements in M. lucifugus revealed an abundance of nonautonomous nhAT families, which have similar TIRs, but little internal sequence similarity with each other or with larger autonomous elements (Ray et al. 2007). Because of this lack of similarity, RepeatMasker fails to identify a majority of the previously identified nhAT elements when the consensus for Myotis_hAT1 is used as the part of the database. Indeed, masking the genome using the Myotis_hAT1 sequence produces only ∼21,000 hits greater than 100 bp. Creating a database that contains the full-length consensus and the nhAT elements described by Ray et al. (2007) as a single repeat database increases the number of hits to >96,000. We believe that the other transposon families described herein will follow a similar pattern, and therefore, our estimate of the amount of DNA derived from DNA transposons is almost certainly too low. Add the impact of HeliBats, which accounts for at least an additional 3% of the M. lucifugus genome (Pritham and Feschotte 2007), and the as yet uncharacterized influence of retrotransposons suggests an extremely dynamic genomic landscape over the past 40 Myr of vespertilionid bat evolution.
Myotis is one of the most species-rich of all mammalian genera, and repeated waves of transposon activity suggest a mechanism for generating the genetic variability necessary to produce its tremendous species diversity. Interestingly, a recent analysis of Myotis phylogeny based on nuclear and mitochondrial DNA sequences becomes more intriguing when one considers the data presented here. Stadelmann et al. (2007) found that a burst of Myotis diversification occurred ∼12–13 Mya. These dates correspond well to the estimated time during which the most active DNA transposon families were expanding in the Myotis genome (Fig. 2).
The young age and narrow taxonomic distribution of all (but one) of the transposon families identified in M. lucifugus raises the puzzling question of their evolutionary origin. Based on the data presented here and elsewhere (Pace and Feschotte 2007; Ray et al. 2007), we currently favor the hypothesis that most of these families hail from repeated episodes of horizontal introduction of founder autonomous transposons from yet unknown source(s) occurring at different evolutionary time points in the lineage of M. lucifugus. The only alternative hypothesis would be that each family arose from active transposons that were vertically inherited during chordate evolution. We consider this hypothesis less likely, because the lack of closely related elements in any of the 30+ other mammalian species (including three nonvespertilionid bats) for which complete or large amounts of genome sequence data are currently available would entail that these elements have persisted in the lineage of vespertilionid bats, while being systematically lost in all other lineages. Under a scenario dominated by horizontal introduction, it appears that mariner, Tc1, and Tc2-like elements colonized the ancestral vespertilionid genome prior to diversification of the family. Later invasions of the hAT-like and piggyBac-like elements may have occurred prior to diversification of Myotis and, in the case of piggyBac1_ML, New World Myotis. Such a scenario of repeated horizontal transfer is consistent with the well-known propensity and the apparent necessity of class 2 elements to undergo horizontal transfers (Hartl et al. 1997; Robertson 2002). A more thorough analysis of the distribution of these elements in bats and other vertebrates will shed additional light onto their origin.
These data also raised the puzzling question as to why bats (and Vespertilionidae in particular) exhibit such a high level of recent transposon activity, while these types of elements have seemingly gone extinct in other mammalian lineages examined. While we are currently unable to answer this question, we note that bats exhibit a variety of life history traits unique among mammals (flight, high population sizes and densities, torpor) that provide potential avenues for future research. One of the most intriguing of these characteristics is the fact that bats are notorious viral reservoirs (Calisher et al. 2006). Like viruses, DNA transposons are examples of genomic parasites, and the ability of bats to safely harbor large loads of a variety of viruses may suggest similar genetic tolerance with regard to DNA transposons. On the other hand, viruses remain the best candidates as potential vectors for the horizontal introduction of DNA transposons and other TEs (Miller and Miller 1982; Fraser et al. 1983; Jehle et al. 1998; Piskurek and Okada 2007). Thus, the propensity of bats to tolerate massive and diverse viral infections may have facilitated the recurrent horizontal introduction of DNA transposons and/or their evolutionary persistence in vespertilionid bats.
In conclusion, we have provided evidence that the previously identified trend toward DNA transposon extinction in mammals is not universal and that a wide diversity of DNA transposons have been active throughout the diversification of the vespertillionid bats, that is, within the last 40 Myr. Furthermore, we propose that the genus Myotis is an excellent candidate for studying the impact and influence of mobile elements, especially class 2 elements, on the evolution of genomic, species, and ecological diversity in mammals. Concomitantly, studies of bats may represent a unique opportunity to investigate life history characteristics that make some organisms more susceptible to transposon activity than others.
Methods
Transposon identification and consensus reconstruction and classification
We carried out initial computational searches from January through April 2007 against the M. lucifugus WGS data deposited at NCBI (http://www.ncbi.nlm.nih.gov/, GenBank accession no. AAPE00000000; K. Lindblad-Toh, pers. comm.). At that time, the WGS data set represented ∼73% of the unassembled M. lucifugus genome with contig sizes averaging ∼2.4 kb (Pritham and Feschotte 2007). The WGS data was queried using TBLASTN to detect the presence of coding sequences related to all known DNA transposon superfamilies (list of queries available upon request). When significant hits were obtained (generally E-value << 10−5), complete sequence entries spanning the top 10–50 subject hits (depending on the overall abundance and sequence heterogeneity of the hits) were retrieved from the database and aligned using a local installation of MUSCLE (Edgar 2004) or CLUSTAL (Thompson et al. 1994). Sequences were trimmed to eliminate unalignable flanks and leave only TSDs, noncoding regions, and the presumed coding regions of the elements. Complete alignments used to determine the full-length consensus sequence for each family are presented as Supplemental Alignments 1–8. Consensus sequences were derived from each multiple alignment based on a majority rule. For each consensus, coding sequences were predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) and, if necessary, refined manually at ambiguous positions in the consensus using multiple alignments of protein sequences from individual copies. Unless previous nomenclature had been established, families and subfamilies were disclosed according to the standard principles of TE classification (Wicker et al. 2007).
hAT3_ML was discovered independently using methods described in Arensburger et al. (2005). After its initial characterization, it was subjected to the same methods as all other elements described herein. All transposon consensus sequences used for this manuscript have been deposited in Repbase (http://www.girinst.org/repbase/index.html).
Age estimation of M. lucifugus transposons
We estimated ages for each transposon family by extracting full- or near full-length ORF's based on the coordinates of the repeats drawn from the output files of a locally implemented version of RepeatMasker 3.1.6. Additionally, we hypothesized that nonautonomous subfamilies were likely mobilized by autonomous elements sharing the same TIRs, and thus were deposited during the same time period as the autonomous elements. We therefore collected representatives from a limited number of nonautonomous derivatives for all families. All extracted elements were aligned with their respective consensus sequence using MUSCLE (Edgar 2004). The most appropriate model for estimating nucleotide divergence from each consensus was determined using ModelTest v3.7 (Posada and Crandall 1998) and genetic distances were calculated in PAUP* v4.0b10 (Swofford 2002).
Specific neutral mutation rate estimates for bats are not currently available. Thus, we utilized published sequences from representative vespertilionid (Nycticeinops schlieffeni, Myotis tricolor, Scotophilus dinganii, Eptesicus hottentotus, Cistugo lesueuri, and Cistugo seabrai) and miniopterid (Miniopterus natalensis, M. inflatus, M. fraterculus, M. australis, and M. macrocneme) taxa to obtain an estimate. Specifically, we obtained sequence data generated by Eick et al. (2005) for four introns (AJ865401-5, AJ865438-43, AJ865646-50, AJ865683-88, AJ866297-301, AJ866329-34, AJ866349-53, AJ866383-87). We constructed a concatenated alignment using MUSCLE (Edgar 2004) and determined the most appropriate evolutionary model for the data using Modeltest v3.7 (Posada and Crandall 1998), TIM+G (Γ = 1.2597). We then estimated the average genetic distance between families Miniopteridae and Vespertilionidae as defined by Eick et al. (2005) using PAUP* v4.0b10 (Swofford 2002). By incorporating the estimated divergence time between these two families, ∼45 Mya (Eick et al. 2005; Miller-Butterworth et al. 2007), we arrived at an estimated mutation rate of 2.366 × 10−9, which was then used to determine likely periods of activity. Representative alignments for several nonautonomous subfamilies are presented in Supplemental Figure S1. All alignments are available upon request.
We estimated ages for each subfamily of elements using sequences as recovered from the genome and sequences from which the CpG dinucleotides had been removed (see Table 1). We also estimated the relative ratio of CpG to non-CpG mutations for each family (Table 2). This was accomplished using a modified Perl script originally designed to estimate CpG dinucleotide mutation rates in primate Alu elements (Xing et al. 2004) and modified for this work.
Nested insertion analysis
We extracted all RepeatMasker hits for each family of transposons and subjected each to a second RepeatMasker analysis using a custom library that included each of the new transposable elements described here and Ves. We validated each nested insertion manually by checking for the presence of the expected target-site duplications.
Identification of intact preintegration sites in M. austroriparius
To test the hypothesis that piggyBac1_ML has been recently active in Myotis, we designed oligonucleotide primers to survey the presence/absence of 15 individual insertions of a nonautonomous piggyBac1_ML subfamily (npiggy_156) in M. austroriparius, which diverged from M. lucifugus ∼8–12 Mya (Stadelmann et al. 2007). Primers were designed in the genomic regions flanking the elements using sequence data from M. lucifugus and tested on a panel of 10 individuals from natural populations of M. austroriparius. Details on sample collection, DNA extraction, amplification, cloning, and sequencing are as previously described (Ray et al. 2007). Sequences from the two intact preintegration site amplicons shown in Figure 3 have been deposited in GenBank under accession nos. EU177095 and EU177096.
Taxonomic distribution of M. lucifugus transposons
To investigate the presence of related transposon families in other chiropteran or mammalian lineages, we used BLASTN or TBLASTN with nucleotide or amino acid sequence queries corresponding to each family of transposon in separate searches of custom databases representing the following taxa: Chiroptera (taxid:9397, excluding Myotis lucifugus), Xenarthra (taxid:9348), Afrotheria (taxid:311790), Laurasiatheria (taxid:314145), Euarchontoglires (taxid:314146), Mammalia (taxid:40674). We also queried the trace data from the sequencing efforts for the flying fox Pteropus vampyrus (7,782,409 sequences averaging 970 bp).
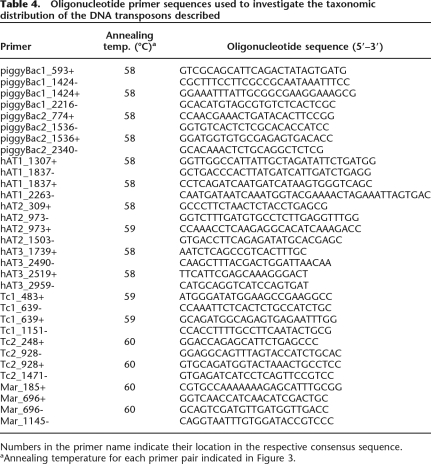
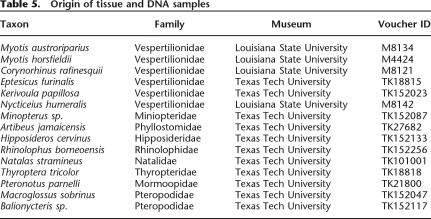
Oligonucleotide primers complementary to portions of the coding region of each autonomous element (Table 4) were used to survey a taxonomically diverse panel of bat genomic DNAs (Table 5) via PCR. Reaction conditions were as described previously (Ray et al. 2007) and annealing temperatures for each primer pair are found in Table 4.
Table 4.
Oligonucleotide primer sequences used to investigate the taxonomic distribution of the DNA transposons described
Numbers in the primer name indicate their location in the respective consensus sequence.
aAnnealing temperature for each primer pair indicated in Figure 3.
Table 5.
Origin of tissue and DNA samples
Phylogenetic analyses
Amino acid sequences for known transposases from each superfamily were obtained from database searches and aligned using MUSCLE (Edgar 2004). Phylogenetic analyses were conducted using MEGA 4 (Kumar et al. 2004). Neighbor-joining trees were constructed using the equal input model with 5000 bootstrap replicates.
Acknowledgments
This work was supported by the Eberly College of Arts and Sciences at West Virginia University (D.A.R.), by start-up funds from UT Arlington (E.J.P. and C.F.) and grant R01GM77582-01 from the National Institute of Health (C.F.). N.L.C. is an investigator of the Howard Hughes Medical Institute. We thank D. Hedges for modifying his original Perl script for the CpG analyses, S. DiFazio for contributing Perl scripts to aid in processing RepeatMasker output, and R. Baker (Texas Tech University) and R. Stevens (Louisiana State University) for contributing tissue and DNA samples for this study. We thank the Broad Institute Genome Sequencing Platform and Whole genome Assembly group, K. Lindblad-Toh, and F. diPalma for making the data available.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.071886.107.
References
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Arensburger P., Kim Y.J., Orsetti J., Aluvihare C., O'Brochta D.A., Atkinson P.W., Kim Y.J., Orsetti J., Aluvihare C., O'Brochta D.A., Atkinson P.W., Orsetti J., Aluvihare C., O'Brochta D.A., Atkinson P.W., Aluvihare C., O'Brochta D.A., Atkinson P.W., O'Brochta D.A., Atkinson P.W., Atkinson P.W. An active transposable element, Herves, from the African malaria mosquito Anopheles gambiae. Genetics. 2005;169:697–708. doi: 10.1534/genetics.104.036145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avancini R.M., Walden K.K., Robertson H.M., Walden K.K., Robertson H.M., Robertson H.M. The genomes of most animals have multiple members of the Tc1 family of transposable elements. Genetica. 1996;98:131–140. doi: 10.1007/BF00121361. [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- Blair J.E., Hedges S.B., Hedges S.B. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Borodulina O.R., Kramerov D.A., Kramerov D.A. Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett. 1999;457:409–413. doi: 10.1016/s0014-5793(99)01059-5. [DOI] [PubMed] [Google Scholar]
- Borodulina O.R., Kramerov D.A., Kramerov D.A. PCR-based approach to SINE isolation: Simple and complex SINEs. Gene. 2005;349:197–205. doi: 10.1016/j.gene.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T., Childs J.E., Field H.E., Holmes K.V., Schountz T., Field H.E., Holmes K.V., Schountz T., Holmes K.V., Schountz T., Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J.H., Farabaugh P.J., Gilbert W., Miller J.H., Farabaugh P.J., Gilbert W., Farabaugh P.J., Gilbert W., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Demattei M.V., Auge-Gouillou C., Pollet N., Hamelin M.H., Meunier-Rotival M., Bigot Y., Auge-Gouillou C., Pollet N., Hamelin M.H., Meunier-Rotival M., Bigot Y., Pollet N., Hamelin M.H., Meunier-Rotival M., Bigot Y., Hamelin M.H., Meunier-Rotival M., Bigot Y., Meunier-Rotival M., Bigot Y., Bigot Y. Features of the mammal mar1 transposons in the human, sheep, cow, and mouse genomes and implications for their evolution. Mamm. Genome. 2000;11:1111–1116. doi: 10.1007/s003350010204. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick G.N., Jacobs D.S., Matthee C.A., Jacobs D.S., Matthee C.A., Matthee C.A. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (chiroptera) Mol. Biol. Evol. 2005;22:1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- Feschotte C., Pritham E.J., Pritham E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Zhang X., Wessler S.R., Zhang X., Wessler S.R., Wessler S.R. Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craig N.L., et al., editors. Mobile DNA II. ASM Press; Washington, DC: 2002. pp. 1147–1158. [Google Scholar]
- Fraser M.J., Smith G.E., Summers M.D., Smith G.E., Summers M.D., Summers M.D. Acquisition of host cell DNA sequences by baculoviruses: Relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J. Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scott G., Steffen D., Worley K.C., Burch P.E., Steffen D., Worley K.C., Burch P.E., Worley K.C., Burch P.E., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Gu X. Early metazoan divergence was about 830 million years ago. J. Mol. Evol. 1998;47:369–371. doi: 10.1007/pl00013150. [DOI] [PubMed] [Google Scholar]
- Hartl D.L., Lohe A.R., Lozovskaya E.R., Lohe A.R., Lozovskaya E.R., Lozovskaya E.R. Modern thoughts on an ancyent marinere: Function, evolution, regulation. Annu. Rev. Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- Jehle J.A., Nickel A., Vlak J.M., Backhaus H., Nickel A., Vlak J.M., Backhaus H., Vlak J.M., Backhaus H., Backhaus H. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J. Mol. Evol. 1998;46:215–224. doi: 10.1007/pl00006296. [DOI] [PubMed] [Google Scholar]
- Koga A., Iida A., Hori H., Shimada A., Shima A., Iida A., Hori H., Shimada A., Shima A., Hori H., Shimada A., Shima A., Shimada A., Shima A., Shima A. Vertebrate DNA transposon as a natural mutator: The medaka fish Tol2 element contributes to genetic variation without recognizable traces. Mol. Biol. Evol. 2006;23:1414–1419. doi: 10.1093/molbev/msl003. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M., Tamura K., Nei M., Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leaver M.J. A family of Tc1-like transposons from the genomes of fishes and frogs: Evidence for horizontal transmission. Gene. 2001;271:203–214. doi: 10.1016/s0378-1119(01)00530-3. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Miller D.W., Miller L.K., Miller L.K. A virus mutant with an insertion of a copia-like transposable element. Nature. 1982;299:562–564. doi: 10.1038/299562a0. [DOI] [PubMed] [Google Scholar]
- Miller-Butterworth C.M., Murphy W.J., O'Brien S.J., Jacobs D.S., Springer M.S., Teeling E.C., Murphy W.J., O'Brien S.J., Jacobs D.S., Springer M.S., Teeling E.C., O'Brien S.J., Jacobs D.S., Springer M.S., Teeling E.C., Jacobs D.S., Springer M.S., Teeling E.C., Springer M.S., Teeling E.C., Teeling E.C. A family matter: Conclusive resolution of the taxonomic position of the long-fingered bats, miniopterus. Mol. Biol. Evol. 2007;24:1553–1561. doi: 10.1093/molbev/msm076. [DOI] [PubMed] [Google Scholar]
- Pace J.K., Feschotte C., Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek O., Okada N., Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pritham E.J., Feschotte C., Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl. Acad. Sci. 2007;17:422–432. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham E.J., Feschotte C., Wessler S.R., Feschotte C., Wessler S.R., Wessler S.R. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol. Biol. Evol. 2005;22:1751–1763. doi: 10.1093/molbev/msi169. [DOI] [PubMed] [Google Scholar]
- Ray D.A., Pagan H.J.T., Thompson M.L., Stevens R.D., Pagan H.J.T., Thompson M.L., Stevens R.D., Thompson M.L., Stevens R.D., Stevens R.D. Bats with hATs: Evidence for recent DNA transposon activity in genus myotis. Mol. Biol. Evol. 2007;24:632–639. doi: 10.1093/molbev/msl192. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A.D., Riggs A.D. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Robertson H.M. Members of the pogo superfamily of DNA-mediated transposons in the human genome. Mol. Gen. Genet. 1996;252:761–766. doi: 10.1007/BF02173985. [DOI] [PubMed] [Google Scholar]
- Robertson H.M. Evolution of DNA transposons in eukaryotes. In: Craig N.L., et al., editors. Mobile DNA II. ASM Press; Washington, DC: 2002. pp. 1093–1110. [Google Scholar]
- Ruvolo V., Hill J.E., Levitt A., Hill J.E., Levitt A., Levitt A. The Tc2 transposon of Caenorhabditis elegans has the structure of a self-regulated element. DNA Cell Biol. 1992;11:111–122. doi: 10.1089/dna.1992.11.111. [DOI] [PubMed] [Google Scholar]
- SanMiguel P., Bennetzen J.L., Bennetzen J.L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. (Lond.) 1998;82:37–44. [Google Scholar]
- Sarkar A., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Fraser M.J., Robertson H.M., Collins F.H., Robertson H.M., Collins F.H., Collins F.H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Genet. Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- Schmid C.W. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzelle L., Pollet N., Bigot Y., Mazabraud A., Pollet N., Bigot Y., Mazabraud A., Bigot Y., Mazabraud A., Mazabraud A. Characterization of multiple lineages of Tc1-like elements within the genome of the amphibian Xenopus tropicalis. Gene. 2005;349:187–196. doi: 10.1016/j.gene.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Sinzelle L., Chesneau A., Bigot Y., Mazabraud A., Pollet N., Chesneau A., Bigot Y., Mazabraud A., Pollet N., Bigot Y., Mazabraud A., Pollet N., Mazabraud A., Pollet N., Pollet N. The mariner transposons belonging to the irritans subfamily were maintained in chordate genomes by vertical transmission. J. Mol. Evol. 2006;62:53–65. doi: 10.1007/s00239-005-0013-7. [DOI] [PubMed] [Google Scholar]
- Smit A.F., Riggs A.D., Riggs A.D. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmann B., Lin L.K., Kunz T.H., Ruedi M., Lin L.K., Kunz T.H., Ruedi M., Kunz T.H., Ruedi M., Ruedi M. Molecular phylogeny of New World Myotis (Chiroptera, Vespertilionidae) inferred from mitochondrial and nuclear DNA genes. Mol. Phylogenet. Evol. 2007;43:32–48. doi: 10.1016/j.ympev.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. 2002. PAUP*: Phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Sunderland, MA. [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigdal T.J., Kaufman C.D., Izsvak Z., Voytas D.F., Ivics Z., Kaufman C.D., Izsvak Z., Voytas D.F., Ivics Z., Izsvak Z., Voytas D.F., Ivics Z., Voytas D.F., Ivics Z., Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wicker T., Sabot F., Hua-Van A., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., Sabot F., Hua-Van A., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., Hua-Van A., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., Flavell A., Leroy P., Morgante M., Panaud O., Leroy P., Morgante M., Panaud O., Morgante M., Panaud O., Panaud O., et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Xing J., Hedges D.J., Han K., Wang H., Cordaux R., Batzer M.A., Hedges D.J., Han K., Wang H., Cordaux R., Batzer M.A., Han K., Wang H., Cordaux R., Batzer M.A., Wang H., Cordaux R., Batzer M.A., Cordaux R., Batzer M.A., Batzer M.A. Alu element mutation spectra: Molecular clocks and the effect of DNA methylation. J. Mol. Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]