Abstract
It is critical to avoid delays in detecting strain manipulations, such as the addition/deletion of a gene or modification of genes for increased virulence or antibiotic resistance, using genome analysis during an epidemic outbreak or a bioterrorist attack. Our objective was to evaluate the efficiency of genome analysis in such an emergency context by using contigs produced by pyrosequencing without time-consuming finishing processes and comparing them to available genomes for the same species. For this purpose, we analyzed a clinical isolate of Francisella tularensis subspecies holarctica (strain URFT1), a potential biological weapon, and compared the data obtained with available genomic sequences of other strains. The technique provided 1,800,530 bp of assembled sequences, resulting in 480 contigs. We found by comparative analysis with other strains that all the gaps but one in the genome sequence were caused by repeats. No new genes were found, but a deletion was detected that included three putative genes and part of a fourth gene. The set of 35 candidate LVS virulence attenuation genes was identified, as well as a DNA gyrase mutation associated with quinolone resistance. Selection for variable sequences in URFT1 allowed the design of a strain-specific, highly effective typing system that was applied to 74 strains and six clinical specimens. The analysis presented herein may be completed within approximately 6 wk, a duration compatible with that required by an urgent context. In the bioterrorism context, it allows the rapid detection of strain manipulation, including intentionally added virulence genes and genes that support antibiotic resistance.
The availability of whole-genome sequences is revolutionizing the fields of bacteriology and infectious disease. By mid-2007, 479 bacterial genomes from 352 distinct species had been sequenced, including representative strains of all notable human pathogens (Fournier et al. 2007). With the increasing capacity of nucleotide sequencing over the last decade, complete bacterial pathogen genome sequencing has allowed, from the description of a unique bacterial genome, the understanding of genome organization and metabolic pathway analysis, and has facilitated the comparison of multiple strains to identify determinants responsible for strain-specific pathogenicity (Beres et al. 2006; Diep et al. 2006). This approach may include hypervirulent or multiresistant strains that eventually may be used as bioweapons or that may cause epidemic outbreaks. From this perspective, the rapid identification of new genes, gene loss, or gene modifications that would explain new virulence or antibiotic resistance, and the identification of strain-specific sequences that would make it possible to trace epidemics, is critical. Such comparisons have been tentatively performed using microarray technology (Broekhuijsen et al. 2003); however, this technique is not well suited to the detection of small sequence variations and is unable to detect added genetic material.
Nearly all bacterial genome sequences have been determined using the Sanger method followed by finishing. This method, though effective, is time-consuming, and time is critical in the event of either an epidemic outbreak or a bioterrorist attack. Recently, pyrosequencing technology for nucleic acids (Ronaghi et al. 1996, 1998) and its use to large scale, which offers a ∼100-fold increase in throughput over current Sanger sequencing technology, was described (Margulies et al. 2005). This technology allowed the sequencing of the small genome of the bacterium Mycoplasma genitalium in only 4 h with 96% coverage and 99.96% accuracy (Margulies et al. 2005). It can thus quickly sequence the complete genome, without the need for cloning, of any emerging or epidemic pathogen. However, pyrosequencing technology does not avoid fastidious and time-consuming finishing procedures to determine the complete genome sequence. Our objective was to determine if the quality of incomplete genomic sequences (i.e., quality of contig sequences without finishing) from pyrosequencing technology is sufficient for performing reliable comparative analysis and determining the gene repertoire of bacterial genomes in an urgent context that requires speedy results, such as an epidemic outbreak or bioterrorist attack.
A locally isolated clinical strain of Francisella tularensis holarctica was used as a paradigm for this evaluation as (1) it is a highly virulent bacterium developed in the past to be used as a bioweapon (Dennis et al. 2001); (2) it was previously tentatively manipulated, as it is easy to transform (Hodges and Penn 2005); (3) four other complete genomes of different F. tularensis subspecies, including one with reduced pathogenicity in humans (LVS), are available for comparison (F. tularensis tularensis SCHU S4, GenBank accession no. AJ749949; F. tularensis holarctica LVS, GenBank accession no. AM233362; F. tularensis holarctica FSC200, GenBank accession no. AASP00000000; F. tularensis holarctica OSU18, GenBank accession no. CP000437); and (4) we have a quinolone-resistant strain with a mutation in the DNA gyrase A gene (strain URFtCIPR). These four previously sequenced genomes are very closely related; the estimated genetic variation between the vaccinal LVS and the virulent OSU18 genomes is 0.11%, and that between LVS and the virulent SCHU S4 is 0.7% (Rohmer et al. 2006).
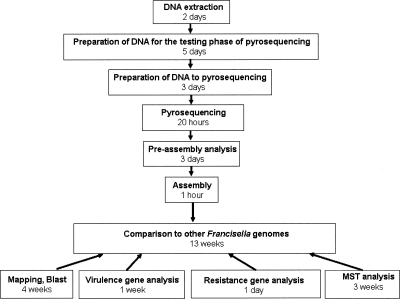
The genome of the URFT1 strain was sequenced using pyrosequencing technology without finishing. This strain was isolated from the lymph node of a patient with oculo-glandular tularemia (Supplemental Fig. S1) and was identified as a member of the holarctica subspecies (Fournier et al. 1998). Bioinformatics and molecular analysis were performed to map contigs to four Francisella genomes in order to identify gene gain and/or loss, gene polymorphism that may influence pathogenicity or antibiotic resistance, and to genotype the strain for its comparison with a collection of Francisella strains using Multi Spacer Typing (MST). Herein, we showed that pyrosequencing technology quickly generated high-quality sequences of the genome of the F. tularensis strain URFT1. We were able to map 143 full-length contigs, covering 92.7% of four other F. tularensis genomes. The gaps obtained between contigs corresponded to repeated genomic regions except one, which corresponded to the loss of three ORFs and part of a fourth. Coverage and quality of sequences allowed the identification and comparison of 35 putative candidate virulence genes responsible for virulence attenuation in LVS (Rohmer et al. 2006) and the DNA gyrase gene in URFT1 contigs. This gene and its mutation, which renders the URFtCIPR strain resistant to quinolone, was detected after its introduction among Rickettsia DNA, showing that pyrosequencing technology allows the detection of single nucleotide variations. Finally, URFT1 contigs contributed to the selection of highly variable spacer sequences, making it possible to distinguish this virulent strain from 80 F. tularensis holarctica strains. This strain-based typing method was even more discriminatory for this purpose than the reference Multiple-Locus Variable-Number Tandem Repeat Analysis (MLVA) method. As the time to obtain such results would be critical in the context of either an epidemic outbreak or a bioterrorist attack, the complete time schedule needed from the extraction of DNA to the end of sequence analysis is also detailed in this study (Fig. 1).
Figure 1.
Major steps and timeline followed for the preparation, pyrosequencing, and analysis of the URFT1 genome. This is calculated on the basis of the work done by one technician or scientist at each step.
Results
Sequencing of the URFT1 genome and comparison to other F. tularensis genomes
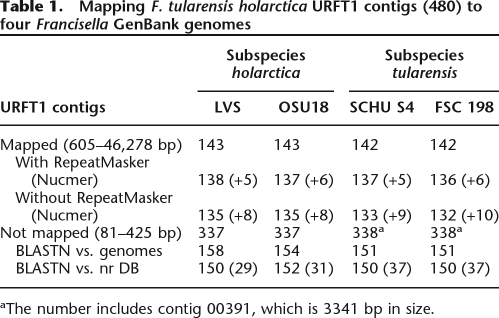
After the completion of two GS20 runs, the pyrosequencing of the virulent F. tularensis URFT1 genome gave a total of 49,836,703 high-quality (HQ) bases and a total of 491,998 HQ reads (HQ/Key Pass = 66.59%–67.23%; Mix + Dots/Key Pass = 15.7%, quality score of Q40). The average length of reads was 101 nucleotides (nt). The average depth of coverage was 26×. There were 471,628 assembled reads with 1,800,530 assembled bases resulting in 480 contigs with sizes ranging from 81 to 46,278 bp, and 4885 singletons were found. Comparative genomic analysis between these contigs and complete virulent as well as avirulent F. tularensis genomes made possible the mapping of 143 and 142 contigs (with sizes ranging from 605 to 46,278 bp) to the LVS and OSU18 genomes as well as to the SCHU S4 and FSC 198 genomes, respectively, using PROJECTOR and MUMmer tools (Table 1; Fig. 2; Supplemental Fig. S2). The mapped contigs covered a size of ∼92.7% of the genome of either LVS (1,895,994 bp) or OSU18 (1,895,727 bp). One contig (number 00398) matched three genes at the start and at the end of the circular LVS genome sequence (see GenBank annotation), suggesting that the URFT1 genome may also be circular. The mapping contigs were also slightly influenced by the bioinformatics tool used. Indeed, we found more mapped contigs with than without the use of the RepeatMasker algorithm (e.g., 138 vs. 135 and 137 vs. 133 for LVS and SCHU S4, respectively). Other contigs (5–10 in parentheses in Table 1) were not mapped using the PROJECTOR tool, but they showed significant matches using the NUCmer algorithm.
Table 1.
Mapping F. tularensis holarctica URFT1 contigs (480) to four Francisella GenBank genomes
aThe number includes contig 00391, which is 3341 bp in size.
Figure 2.
Mapping F. tularensis holarctica URFT1 contigs on LVS genome. Circles are (from inner to outer sides) contigs with sizes ranging from 81 to 425 bp, contigs with sizes of 605–46,278 bp (URFT1 strain), rRNA operons, and CDSs (LVS strain).
On the other hand, a total of 377 short remaining contigs (81–425 bp) were rejected by the bioinformatic tools used for the four Francisella genomes. However, using BLASTN analysis (E-value = 10−4), a first subset of contigs (i.e., 154–158) showed complete or partial matches with Francisella genomes and corresponded mainly to previously mapped contigs (Fig. 2). The second remaining subset of contigs (i.e., 179–187, except 29–37, which matched mainly Francisella sequences) did not match the nr sequence database. These contigs resulted from only two to nine sequencing reads and suggested that they may correspond to sequencing errors. The last contig (number 00391), which was 3341 bp in size and resulted from 914 reads, did not match any F. tularensis tularensis genomes, but matched those of F. tularensis holarctica.
To further compare URFT1 contigs with other Francisella genomes, we identified the LVS genes that are present within the gaps between the 143 mapped contigs. A total of 93 gaps were counted, and the corresponding genes in the LVS genome are listed in Supplemental Table S1. All of these LVS genes were found to be absent or only partially matched the contigs and corresponded mainly to repeated genes, operons, or elements. For example, only one URFT1 rRNA operon was identified among the three Francisella rRNA operons. However, one gap (number 49), identified inside contig 399 (Supplemental Table S1), corresponded to three consecutive genes in the LVS genome (FTL_1081, FTL_1082, and FTL_1083). This region was also found to be present in the three other F. tularensis strains examined after BLASTN analysis. To confirm the presence of this gap in the URFT1 genome, we performed PCR amplification and sequencing using primers designed to sit upstream and downstream of this gap. The sequence of the PCR product showed a size of 557 bp in the URFT1 genome, whereas it corresponded to 2252 predicted bp in the LVS genome (Supplemental Fig. S3). Sequence comparison of this URFT1 product against the LVS genome showed the presence of only two contiguous genes, FTL_1080 and FTL_1084, separated by a 204-bp intergenic spacer. Moreover, we observed that the 5′ end of the gene FTL_1084 was truncated by 184 bp. Thus, we confirmed the deletion of 1695 bp in the URFT1 genome, which included three genes, a fragment of a fourth, and intergenic spacers of the Francisella genomes.
Analysis of LVS attenuation candidate genes in URFT1
The URFT1 contigs obtained from pyrosequencing contained homology with all 35 of the candidate genes putatively responsible for LVS attenuation in the virulent URFT1 strain (Table 2). Of these, 30 URFT1 genes presented differences previously identified between the avirulent LVS strain and the virulent strains OSU18, SCHU S4, and U112. However, five candidate genes, i.e., a pilus assembly protein, a transcriptional regulator protein, desoxyribidipyrimidine photolyase, a hypothetical protein, and a metal ion transporter protein (FTL_0391–FTL_0392, FTL_0062, FTL_0341–FTL_0342, FTL_0997a, and FTL_1697–FTL_1698) were 100% similar between the virulent URFT1 and the avirulent LVS strains, but were different between the virulent URFT1 strain and the virulent OSU18, SCHU S4, and U112 strains.
Table 2.
Loci in LVS for which a nonsynonymous mutation has been detected by comparison to other sequenced strains of F. tularensis as done by Rhomer et al. (2006)
aLocus tag according to LVS genome annotation.
b(P) Present; (A) absent.
cIn the last column, the status of these mutations in the genome of strain URFT1 is indicated.
Detection and analysis of a F. tularensis quinolone resistance gene and its mutation
The gyrase A gene, which contains the mutation associated with quinolone resistance or sensitivity in the URFtCIPR and URFT1 strains, respectively, was identified in URFT1 contigs. In addition, among the assembled contigs obtained in the Rickettsia pyrosequencing run with the 3-kb PCR product that frames the gyrA gene of the URFtCIPR strain, two contigs of 176 bp and 101 bp contained the gyrA sequence and the point mutation associated with quinolone resistance (C to T transition at position 524 of gyrA) (Supplemental Fig. S4).
Discrimination of Francisella subspecies and strains based on MST (Multi Spacer Typing) comparative genomics
The comparison of URFT1 intergenic regions (spacers) with those of the avirulent LVS and virulent SCHU S4 strains of F. tularensis strains allowed the identification of 245 variable spacers with a size ≥250 bp present in the three F. tularensis genomes. Among these, eight highly variable spacers were selected (S1–S8, Table 3). These intergenic spacers enabled discrimination between 76 strains of F. tularensis holarctica (74 Swedish clinical isolates, LVS and URFT1) as well as the single strain of F. tularensis tularensis (SCHU S4) into 16 (S1), six (S2), three (S3), two (S4), two (S5), two (S6), three (S7), and two (S8) genotypes. When combining the most polymorphic spacers, S1–S4, Francisella strains were distinguished into 27 MST genotypes (Table 4). The spacers S1–S4 were also amplified in five out of six clinical samples using the designed primers. This allowed the detection of an additional genotype with spacer S1 in one clinical sample, leading to the identification of 27 MST genotypes within F. tularensis holartica. The SCHU S4 strain, i.e., a member of F. tularensis tularensis, alone showed MST genotype 28 and was distinguished from all F. tularensis holartica materials. The MLVA method also enabled the categorization of the 76 strains of F. tularensis holarctica (URFT1, LVS, and 74 Swedish clinical isolates), the five our of six clinical samples, and the single strain of F. tularensis tularensis (SCHU S4) into only 20 genotypes after a combination of the six VNTR loci (Fig. 3). Each intergenic spacer discriminated the SCHU S4 strain from the 76 strains of F. tularensis holarctica, whereas only three VNTR loci (Ft-M3, Ft-M21, and Ft-M22) can do that.
Table 3.
Summary information of the eight intergenic spacers and the primers used for amplifying and sequencing these spacers
aIntergenic spacer designations consist of the name of the 5′-ORF-the name of the 3′-ORF.
bThe position of the spacers on the genome, the spacer sizes, and the PCR product sizes were deduced from F. tularensis SCHU S4 strain (GenBank accession no. NC006570), but may vary with different strains.
cPart of a complete intergenic spacer.
Table 4.
Results of MST performed on DNA from strains URFT1, LVS, 74 Swedish clinical isolates, and five clinical samples
GenBank accession numbers for spacer types are as follows: S1: 1, EF175177; 2, EF175178; 3, EF175179; 4, EF175180; 5, EF175181; 6, EF175182; 7, EF175183; 8, EF175184; 9, EF175185; 10, EF175186; 11, EF175187; 12, EF175188; 13, EF175189; 14, EF175190; 15, EF175191; 16, EF175192; 17, EF175193; 18, EF175194; S2: 1, EF175195; 2, EF175196; 3, EF175197; 4, EF175198; 5, EF175199; 6, EF175200; 7, EF175201; S3: 1, EF175202; 2, EF175203; 3, EF175204; 4, EF175205; S4: 1, EF175206; 2, EF175207; 3, EF175208.
Figure 3.
Neighbor-joining phylogenetic tree obtained from four concatenated intergenic spacer sequences (S1, S2, S3, and S4) of 82 F. tularensis strains. Strains belonging to each MST genotype are indicated within parentheses. On the right, genotypes delineated by using MLVA are indicated. Genotypes that do not group the same strains when using both methods are indicated (red). (Arrows) Repartition of strains according to the method used.
Phylogenetic trees obtained from the MST genotypes using the Neighbor-Joining (Fig. 3) and Maximum Parsimony (data not shown) methods on S1–S4 concatenated spacer sequences showed similar clustering. However, the phylogenetic trees represented by MLVA genotypes exhibited some differences compared with the MST trees, especially for SCHU S4, which was not separated from the holarctica strains. However, the separation of strains into clusters was nearly identical for both methods (Fig. 3).
Discussion
In clinical microbiology, most of the major human bacterial pathogen genomes (either complete or unfinished), including the F. tularensis species, have already been sequenced. In both clinical and bioterrorism contexts, the identification of genes added, either intentionally or naturally, and of genes associated with virulence or antibiotic resistance should be rapidly performed. These genes can be compared with microbial genomes available in general and specialized databases, which suggests that finishing the assembly of the microbial genome is not a priority for such emergency situations. The recent development of the pyrosequencing technology has both great promise and pitfalls. The short lengths of the sequences obtained (∼100 bp) makes the presence of repeated regions above this size an obstacle to the molecular closing of the genome. In the case of isolates of the same species that have already been sequenced, and when only gene content is useful, we speculate that repeated regions may not be a constraint for the detection of important genes, avoiding time-consuming closing. Our study evaluated the recent pyrosequencing technology in the rapid sequencing of a virulent strain URFT1 of F. tularensis holarctica and in the quality as well as the coverage of genome contigs by a comparative analysis with known Francisella genomes. The genome was sequenced without cloning and was thus analyzed without trying to close the area between contigs as is usually done for genome comparison (Chen et al. 2007). Pyrosequencing using GS20 without closing the molecule for comparison was previously tested on a Helicobacter pylori isolate, a bacterium responsible for chronic gastric infection, and was suggested as a new approach to studying genetic changes in this species over time or between infected humans according to their evolution (Oh et al. 2006).
We found that contigs of the virulent strain URFT1, obtained by pyrosequcing, covered ∼92.7% of the avirulent LVS and the virulent SCHU S4 genomes. All major contigs (143–142) gave significant matches and were mapped to the avirulent LVS genome, as well as to the virulent OSU18, SCHU S4, and FSC198 genomes. Another URFT1 contig contained genes corresponding to the beginning and the end of the LVS genome, indicating that the URFT1 genome is circular, as are other Francisella genomes. However, several gaps were observed between URFT1 contigs. This result did not influence the detection of the important genes within the Francisella genomes. Indeed, as expected from the previously sequenced F. tularensis genome (Larsson et al. 2005), most of the gaps corresponded to repeated regions, such as rRNA and transposases, whereas a single gap corresponded to three nonrepetitive genes, a fragment of a fourth, and intergenic spacers of the LVS genome that were demonstrated to be absent in the URTF1 genome (Supplemental Table S1; Supplemental Fig. S3). These results indicated that all Francisella genes were identified in our study and shows that not completing the Francisella genome did not influence the detection of important genes within, such as candidate virulence attenuation genes or resistance genes, as discussed below. Moreover, the URFT1 strain showed a single contig that distinguished F. tularensis holarctica genomes (strains LVS and OSU18) from the genomes of F. tularensis tularensis (strains SCHU S4 and FSC198). This biomarker could be used for epidemiological tracing of F. tularensis holarctica strains.
Another aim was to determine if pyrosequencing gives genomic sequences of comparable quality to those produced by Sanger sequencing, in order to enable detection of single nucleotide polymorphisms. To address this issue, we focused our analysis on a set of 35 genes previously identified by Rhomer et al. as possibly being responsible for the reduced virulence of the LVS strain (Rohmer et al. 2006). In a multiple sequence comparison of the 35 LVS gene sequences between avirulent LVS and other virulent F. tularensis genomes including URFT1, the URFT1 sequences of 30 out of 35 genes was 100% similar to that of virulent strains. However, for five genes, we observed exactly the same sequences for both avirulent LVS and virulent URFT1. Among these five genes, polymorphisms in four (FTL_0062, FTL_0341, FTL_0997a, and FTL_0698) were stated as being unlikely to be responsible for causing loss of virulence by comparison with other F. tularensis genomes. They were predicted neither to affect the normal function of genes nor to cause a gain of function in genes that are not functional in virulent strains (Rohmer et al. 2006). By adding sequence data from an additional virulent strain, our results confirm that the modification of these four genes likely does not account for the virulence attenuation in LVS. The fifth candidate identified as being identical between LVS and URFT1 was locus FTL_0391–FTL_0392, which encodes a type IV pilus assembly protein. This was among the most likely candidate for virulence attenuation, as the 0.5-kb deletion was predicted to affect the function of a gene that is functional in all virulent strains. Moreover, this locus was previously demonstrated in a murine model to be associated with virulence attenuation for infection via a peripheral route (Forslund et al. 2006). Finding the same deletion in the URFT1 virulent strain is thus surprising. However, this deletion has been occasionally observed in virulent strains, and in those cases, the hypothesis is that the deletion in this gene occurs during in vitro culture where there is no selection pressure to maintain this gene (Svensson et al. 2005). As URFT1 was subcultured more than five times before it was produced for genome sequencing, it is possible that such an in vitro deletion occurred. In conclusion, the finding that the unique polymorphisms observed in the 35 genes corresponded to previously described ones confirms the quality of the sequences obtained with pyrosequencing technology.
In a specific pyrosequencing experiment combining fragments of the gyrase A gene of the quinolone-resistant strain URFtCIPR and a Rickettsia genome, we were able to detect not only this gene among the Rickettsia contigs, but also its mutations. This result indicated that pyrosequencing can detect introduced (either intentionally or naturally) genes as well as their mutations in microbial genomes, and thus could be useful in the detection of a mutated strain that would be resistant to the reference treatment for pneumonia, following exposure to bioweapon agents (http://www.bt.cdc.gov/agent/tularemia/tularemia-biological-weapon-abstract.asp#4). For the demonstration, it would have been better to determine the complete genome sequence of the quinolone-resistant strain rather than the susceptible one. Unfortunately, we did not have the resistant strain when we started the program. However, as we find it interesting to evaluate the capability of pyrosequening technology to detect the DNA gyrase gene mutations, we added fragments of the amplifiate of this gene to the DNA extract of a Rickettsia. The fragment supporting the mutation was thus treated as part of the rickettsial genome and was retrieved in its assembly.
It is critical to demonstrate the clonal origin of a pathogen during an outbreak. Comparing complete genomes is the ultimate tool to identify strain-specific sequences. F. tularensis strains show high genetic homogeneity, especially in the holarctica subspecies. If several typing methods, such as Insertion Sequence Probed RFLP (Thomas et al. 2003), macrorestriction using pulsed field gel electrophoresis (PFGE) and amplified fragment length polymorphism (Garcia Del Blanco et al. 2002), repetitive element PCR, arbitrarily primed PCR (Johansson et al. 2000), and ribotyping (Grif et al. 2003) were eventually used for typing F. tularensis subspecies, the results would lack high-resolution discrimination between individual strains. Multilocus sequence typing (MLST) failed to separate F. tularensis strains at the strain level (Johansson et al. 2004b). Techniques based on the analysis of variable-number tandem repeats identified F. tularensis at the strain level and also identified epidemic strains (Farlow et al. 2001; Johansson et al. 2004a). This was based on 25 loci and allowed the recognition of 120 genotypes among 192 strains. The F. tularensis Multi Spacer Typing (MST) method used here is one we have previously used to successfully type many human pathogens, including the potential bioterrorism agents Yersinia pestis, Coxiella burnetii, and Rickettsia sp. (Drancourt et al. 2004; Glazunova et al. 2005; Zhu et al. 2005). The advantage of the method we use here is that the typing system is designed in order to choose polymorphic areas in the studied strains, and thus is likely to separate it from all others. In this study, MST exhibited greater discriminatory power and reasonable phylogenetic classification for F. tularensis. We classified the F. tularensis holarctica DNA from strains URFT1, LVS, 74 Swedish clinical isolates, and five clinical samples into 27 genotypes, whereas MLVA represented by six VNTR loci produced only 19 genotypes. Our strain had a unique genotype. Moreover, MST allowed quick direct typing of clinical samples without a culture step and allowed the identification of an additional original genotype.
In conclusion, our work demonstrates that pyrosequencing without finishing is a tool adapted for performing quick bacterial genome sequencing in the context of a sudden outbreak or a bioterrorist attack. If the 2 wk needed to obtain contigs cannot be significantly shortened, the comparison with other genomes, which depends on the number of computer scientists involved in the project, may be reduced to the minimum time necessary. In the present work, as mapping and BLAST comparison was the most time-consuming task (4 wk), the time between DNA extraction and end of analysis could have been a total of 6 wk (Fig. 1).
Methods
F. tularensis strains
The strain of the subspecies holarctica used for genome sequencing was URFT1 (Fournier et al. 1998). Its quinolone-resistant mutant (URFtCIPR) was also studied. The 74 additional isolates studied in the typing procedure were collected from Swedish patients with suspected ulceroglandular tularaemia between 1995 and 2005. All of the isolates except two were from Middle Sweden (defined as the region of Sweden between the city of Jönköping in the south and the village of Sveg in the north); the other two were from the northern part of Sweden. All 74 isolates were obtained from wounds. Samples were collected from the ulcer by use of a cotton swab, which was transported to the laboratory in transport medium. Swabs were cultured on tul agar (Gaspar et al. 1961). Colonies suspected to be F. tularensis were isolated, confirmed by PCR, and stored at −70°C. Template DNA was prepared using a commercial DNA extraction kit (QIAamp DNAmini kit, Qiagen).
Clinical samples
Six swab samples collected from patients living in Middle Sweden with suspected ulceroglandular tularaemia were first used to inoculate tul plates and later subjected to DNA extraction using a commercial kit (NucliSens magnetic extraction protocol, Biomerieux).
DNA extraction of URFT1
The strain was grown on chocolate isovitalex agar, harvested, and suspended in TE buffer. DNA was extracted according to the classical lytic treatment using SDS and proteinase K followed by phenol isoamyl alcohol extraction. DNA was resuspended in TE buffer and visualized on an agarose gel stained with ethidium bromide.
Sequencing, assembly, and comparison to other F. tularensis genomes
Genome sequencing of the URFT1 strain was performed using pyrosequencing technology as previously described (Margulies et al. 2005). The contigs were obtained from an assembly using the Newbler software of the 454 Life Science Corp. suite package. The contigs were considered for mapping to the four reference Francisella GenBank genomes: F. tularensis holarctica strains LVS (NC_007880) and OSU18 (NC_008369), as well as F. tularensis tularensis strains SCHU S4 (NC_006570) and FSC 198 (NC_008245). Mapping contigs against the published Francisella genomes was performed using the PROJECTOR 2 software. As F. tularensis genomes are full of repetitive IS elements (Larsson et al. 2005), we performed mapping with and without the RepeatMasker algorithm, which is an option of this software (RepeatMasker can filter automatically repetitive sequences to minimize incorrect mapping events) (van Hijum et al. 2005). For a comparative analysis, the mapped and unmapped contigs were also subjected to alignment using NUCmer of the MUMmer 3.0 program (Kurtz et al. 2004). The contigs that did not map with both tools were then analyzed by the BLASTN algorithm (Altschul et al. 1997) against the four Francisella genomes and/or the nonredundant GenBank database to identify any homologs of other genomes. Sequences of the contigs were deposited in GenBank under Project ID 19645.
Gaps between mapped URFT1 contigs were numbered, and their corresponding genes in the LVS genome were identified by visual inspection of the map and/or confirmed by in silico BLAST analysis. For a gap that was not later identified as a repeated gene, operon, or element, we carried out a PCR reaction to check whether it was a true gap or an artifact due to a mistake in sequencing or assembly in the URFT1 genome. To perform this, a pair of primers was designed upstream and downstream of this gap according to the LVS sequence, corresponding to a 2153-bp predicted segment.
Analysis of a F. tularensis quinolone-resistant strain (URFtCIPR isolate)
In vitro resistance was assessed in a blood agar plate containing a disk of the antibiotic ciprofloxacin (Mast Diagnostics) using the method previously reported for Bartonella henselae (Biswas et al. 2006, 2007). By using PCR amplification and sequencing of the strain URFtCIPR isolate using primers (FtgyrAF, 5′-GTCATCATC TATAAATATCG-3′ and FtgyrAR, 5′-TTAATATATGACCCCGTA GG-3′) targeting the QRDR (Quinolone-Resistant Determining Region) of gyrase A, we have previously observed that a C to T mutation at position 524 of the gene, which leads to a substitution of a threonine residue (T) at position 83 (Escherichia coli numbering) of the protein with isoleucine (I), was associated with the resistance of this strain to fluoroquinolone compounds (J.M. Rolain, unpubl.). Two primers were designed according to the available genome to frame this region and generate a 3-kb PCR product (Ftgyr3000F, 5′-GCCATACATTACACCAGCTA-3′, and Ftgyr3000R, 5′-GACAAGTCTGGAACTACAGC-3′) that was resolved in a different run containing genomic DNA of a Rickettsia, without the URFT1 isolate, by pyrosequencing. After purification, the amplicon was nebulized to a mean fragment size of 400 bp, end repaired, and adapter ligated according to the standard procedures described previously (Margulies et al. 2005). We followed standard post-run bioinformatics processing on the GS20-run assembly to determine contigs that passed various quality filters. These contigs were used in our downstream analysis. In particular, the known mutation was located using a BLAST analysis of a nonmutated short sequence of the QRDR of gyrA (GGTAAATATCACCCTCATGGAGATACAGCTGTTTACGATAC TATTGTCAGA) directly on the contigs obtained during this run.
Status of LVS attenuation candidate genes in URFT1
In a recent study, Rohmer et al. (2006) identified a set of LVS genes that may be responsible for the virulence attenuation of this strain. This set was first downloaded from the NCBI database as FASTA files, and then a BLASTN analysis was performed against URFT1 contigs to identify their corresponding homologs. To detect putative nucleic acid differences in sequences of each gene between the avirulent LVS strain and the virulent Francisella genomes (strains OSU18, SCHU S4, U112, and URFT1), multiple sequence alignments were performed using the MUSCLE software (Edgar 2004).
MST typing based on comparative Francisella genomes
To identify homologous multiple spacer sequences between Francisella genomes, we compared URFT1 genomic contigs to LVS (subspecies holartica) and SCHU S4 (subspecies tularensis) genomes that we downloaded from the GenBank database in FASTA format. We constructed two databases containing the intergenic spacers of SCHU S4 and LVS with a size ≥250 bp. This size was arbitrarily chosen in order to provide sufficient information. These spacers were 308/1385 and 245/1666 of the Schu4 and LVS spacers, respectively. The 480 contigs from strain URFT1 were subsequently used as query sequences to search for their corresponding homogenous sequences in the two spacer databases by using BLASTN. Alignment between the three corresponding spacer sequences was performed using the ClustalW program (Thompson et al. 1994). Spacers selected for typing were selected in four successive steps: (1) those flanked with conserved DNA sequences of at least 50 bp at each extremity to allow the design of consensus primers, (2) <600 bp in order to use the same primer pair for amplification and sequencing, (3) at least two polymorphic sites, and (4) the lowest similarity between each other.
Phylogenetic analysis of the studied strains was performed using the Neighbor-Joining and Maximum Parsimony methods within the MEGA 3.1 software (Kumar et al. 2001). For this purpose, sequences of the selected spacers were concatenated and then subjected to multiple sequence alignment using the ClustalW software. To evaluate the discriminative power of the MST method, we also compared this approach to the Multiple-Locus Variable-Number Tandem Repeat Analysis (MLVA) method with the six polymorphic VNTR loci, Ft-M3, Ft-M6, Ft-M20, Ft-M21, Ft-M22, and Ft-M24 (Johansson et al. 2004).
Acknowledgments
We thank Bernadette Giumelli for technical help. These studies were supported by a grant from Sanofi-Aventis France (Sanofi-Aventis Group) and Bayer Pharma as part of a multiorganism call for proposals. K.E. was supported by a grant from the European Commission (NOE EPG LSHB–CT 2005-512061).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.071266.107.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres S.B., Richter E.W., Nagiec M.J., Sumby P., Porcella S.F., Deleo F.R., Musser J.M., Richter E.W., Nagiec M.J., Sumby P., Porcella S.F., Deleo F.R., Musser J.M., Nagiec M.J., Sumby P., Porcella S.F., Deleo F.R., Musser J.M., Sumby P., Porcella S.F., Deleo F.R., Musser J.M., Porcella S.F., Deleo F.R., Musser J.M., Deleo F.R., Musser J.M., Musser J.M. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Raoult D., Rolain J.M., Raoult D., Rolain J.M., Rolain J.M. Molecular characterization of resistance to macrolides in Bartonella henselae. Antimicrob. Agents Chemother. 2006;50:3192–3193. doi: 10.1128/AAC.00263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Raoult D., Rolain J.M., Raoult D., Rolain J.M., Rolain J.M. Molecular mechanisms of resistance to antibiotics in Bartonella bacilliformis. J. Antimicrob. Chemother. 2007;59:1065–1070. doi: 10.1093/jac/dkm105. [DOI] [PubMed] [Google Scholar]
- Broekhuijsen M., Larsson P., Johansson A., Bystrom M., Eriksson U., Larsson E., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Larsson P., Johansson A., Bystrom M., Eriksson U., Larsson E., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Johansson A., Bystrom M., Eriksson U., Larsson E., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Bystrom M., Eriksson U., Larsson E., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Eriksson U., Larsson E., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Larsson E., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Prior R.G., Sjostedt A., Titball R.W., Forsman M., Sjostedt A., Titball R.W., Forsman M., Titball R.W., Forsman M., Forsman M. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 2003;41:2924–2931. doi: 10.1128/JCM.41.7.2924-2931.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Johnson J.A., Pusch G.D., Morris J.G., Stine O.C., Johnson J.A., Pusch G.D., Morris J.G., Stine O.C., Pusch G.D., Morris J.G., Stine O.C., Morris J.G., Stine O.C., Stine O.C. The genome of non-O1 Vibrio cholerae NRT36S demonstrates the presence of pathogenic mechanisms that are distinct from those of O1 Vibrio cholerae. Infect. Immun. 2007;75:2645–2647. doi: 10.1128/IAI.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis D.T., Inglesby T.V., Henderson D.A., Bartlett J.G., Ascher M.S., Eitzen E., Fine A.D., Friedlander A.M., Hauer J., Layton M., Inglesby T.V., Henderson D.A., Bartlett J.G., Ascher M.S., Eitzen E., Fine A.D., Friedlander A.M., Hauer J., Layton M., Henderson D.A., Bartlett J.G., Ascher M.S., Eitzen E., Fine A.D., Friedlander A.M., Hauer J., Layton M., Bartlett J.G., Ascher M.S., Eitzen E., Fine A.D., Friedlander A.M., Hauer J., Layton M., Ascher M.S., Eitzen E., Fine A.D., Friedlander A.M., Hauer J., Layton M., Eitzen E., Fine A.D., Friedlander A.M., Hauer J., Layton M., Fine A.D., Friedlander A.M., Hauer J., Layton M., Friedlander A.M., Hauer J., Layton M., Hauer J., Layton M., Layton M., et al. Tularemia as a biological weapon: Medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Diep B.A., Gill S.R., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Gill S.R., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Lin F., Lin J., Carleton H.A., Mongodin E.F., Lin J., Carleton H.A., Mongodin E.F., Carleton H.A., Mongodin E.F., Mongodin E.F., et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Drancourt M., Roux V., Dang L.V., Tran-Hung L., Castex D., Chenal-Francisque V., Ogata H., Fournier P.E., Crubezy E., Raoult D., Roux V., Dang L.V., Tran-Hung L., Castex D., Chenal-Francisque V., Ogata H., Fournier P.E., Crubezy E., Raoult D., Dang L.V., Tran-Hung L., Castex D., Chenal-Francisque V., Ogata H., Fournier P.E., Crubezy E., Raoult D., Tran-Hung L., Castex D., Chenal-Francisque V., Ogata H., Fournier P.E., Crubezy E., Raoult D., Castex D., Chenal-Francisque V., Ogata H., Fournier P.E., Crubezy E., Raoult D., Chenal-Francisque V., Ogata H., Fournier P.E., Crubezy E., Raoult D., Ogata H., Fournier P.E., Crubezy E., Raoult D., Fournier P.E., Crubezy E., Raoult D., Crubezy E., Raoult D., Raoult D. Genotyping, Orientalis-like Yersinia pestis, and plague pandemics. Emerg. Infect. Dis. 2004;10:1585–1592. doi: 10.3201/eid1009.030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow J., Smith K.L., Wong J., Abrams M., Lytle M., Keim P., Smith K.L., Wong J., Abrams M., Lytle M., Keim P., Wong J., Abrams M., Lytle M., Keim P., Abrams M., Lytle M., Keim P., Lytle M., Keim P., Keim P. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 2001;39:3186–3192. doi: 10.1128/JCM.39.9.3186-3192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund A.L., Kuoppa K., Svensson K., Salomonsson E., Johansson A., Bystrom M., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Kuoppa K., Svensson K., Salomonsson E., Johansson A., Bystrom M., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Svensson K., Salomonsson E., Johansson A., Bystrom M., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Salomonsson E., Johansson A., Bystrom M., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Johansson A., Bystrom M., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Bystrom M., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Oyston P.C., Michell S.L., Titball R.W., Noppa L., Michell S.L., Titball R.W., Noppa L., Titball R.W., Noppa L., Noppa L., et al. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 2006;59:1818–1830. doi: 10.1111/j.1365-2958.2006.05061.x. [DOI] [PubMed] [Google Scholar]
- Fournier P.E., Bernabeu L., Schubert B., Mutillod M., Roux V., Raoult D., Bernabeu L., Schubert B., Mutillod M., Roux V., Raoult D., Schubert B., Mutillod M., Roux V., Raoult D., Mutillod M., Roux V., Raoult D., Roux V., Raoult D., Raoult D. Isolation of Francisella tularensis by centrifugation of shell vial cell culture from an inoculation eschar. J. Clin. Microbiol. 1998;36:2782–2783. doi: 10.1128/jcm.36.9.2782-2783.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier P.E., Drancourt M., Raoult D., Drancourt M., Raoult D., Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infect. Dis. 2007;7:711–723. doi: 10.1016/S1473-3099(07)70260-8. [DOI] [PubMed] [Google Scholar]
- Garcia Del Blanco N., Dobson M.E., Vela A.I., De L.P.V., Gutierrez C.B., Hadfield T.L., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., Dobson M.E., Vela A.I., De L.P.V., Gutierrez C.B., Hadfield T.L., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., Vela A.I., De L.P.V., Gutierrez C.B., Hadfield T.L., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., De L.P.V., Gutierrez C.B., Hadfield T.L., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., Gutierrez C.B., Hadfield T.L., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., Hadfield T.L., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., Kuhnert P., Frey J., Dominguez L., Rodriguez Ferri E.F., Frey J., Dominguez L., Rodriguez Ferri E.F., Dominguez L., Rodriguez Ferri E.F., Rodriguez Ferri E.F. Genotyping of Francisella tularensis strains by pulsed-field gel electrophoresis, amplified fragment length polymorphism fingerprinting, and 16S rRNA gene sequencing. J. Clin. Microbiol. 2002;40:2964–2972. doi: 10.1128/JCM.40.8.2964-2972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar A.J., Tresselt H.B., Ward M.K., Tresselt H.B., Ward M.K., Ward M.K. New solid medium for enhanced growth of Pasteurella tularensis. J. Bacteriol. 1961;82:564–569. doi: 10.1128/jb.82.4.564-569.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazunova O., Roux V., Freylikman O., Sekeyova S., Fournous G., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Roux V., Freylikman O., Sekeyova S., Fournous G., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Freylikman O., Sekeyova S., Fournous G., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Sekeyova S., Fournous G., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Fournous G., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Tyczka J., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Tokarevich N., Kovacava E., Marrie T.J., Raoult D., Kovacava E., Marrie T.J., Raoult D., Marrie T.J., Raoult D., Raoult D. Coxiella burnetii genotyping. Emerg. Infect. Dis. 2005;11:1211–1217. doi: 10.3201/eid1108.041354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grif K., Dierich M.P., Much P., Hofer E., Allerberger F., Dierich M.P., Much P., Hofer E., Allerberger F., Much P., Hofer E., Allerberger F., Hofer E., Allerberger F., Allerberger F. Identifying and subtyping species of dangerous pathogens by automated ribotyping. Diagn. Microbiol. Infect. Dis. 2003;47:313–320. doi: 10.1016/s0732-8893(03)00095-6. [DOI] [PubMed] [Google Scholar]
- Hodges L., Penn R.L., Penn R.L. Tularemia and bioterrorism. In: Fong I.W., Alibek K., Alibek K., editors. Bioterrorism and infectious agents: A new dilemma for the 21st century. Springer; New York: 2005. pp. 71–98. [Google Scholar]
- Johansson A., Ibrahim A., Goransson I., Eriksson U., Gurycova D., Clarridge J.E., Sjostedt A., Ibrahim A., Goransson I., Eriksson U., Gurycova D., Clarridge J.E., Sjostedt A., Goransson I., Eriksson U., Gurycova D., Clarridge J.E., Sjostedt A., Eriksson U., Gurycova D., Clarridge J.E., Sjostedt A., Gurycova D., Clarridge J.E., Sjostedt A., Clarridge J.E., Sjostedt A., Sjostedt A. Evaluation of PCR-based methods for discrimination of Francisella species and subspecies and development of a specific PCR that distinguishes the two major subspecies of Francisella tularensis. J. Clin. Microbiol. 2000;38:4180–4185. doi: 10.1128/jcm.38.11.4180-4185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A., Farlow J., Larsson P., Dukerich M., Chambers E., Bystrom M., Fox J., Chu M., Forsman M., Sjostedt A., Farlow J., Larsson P., Dukerich M., Chambers E., Bystrom M., Fox J., Chu M., Forsman M., Sjostedt A., Larsson P., Dukerich M., Chambers E., Bystrom M., Fox J., Chu M., Forsman M., Sjostedt A., Dukerich M., Chambers E., Bystrom M., Fox J., Chu M., Forsman M., Sjostedt A., Chambers E., Bystrom M., Fox J., Chu M., Forsman M., Sjostedt A., Bystrom M., Fox J., Chu M., Forsman M., Sjostedt A., Fox J., Chu M., Forsman M., Sjostedt A., Chu M., Forsman M., Sjostedt A., Forsman M., Sjostedt A., Sjostedt A., et al. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 2004a;186:5808–5818. doi: 10.1128/JB.186.17.5808-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A., Forsman M., Sjostedt A., Forsman M., Sjostedt A., Sjostedt A. The development of tools for diagnosis of tularemia and typing of Francisella tularensis. APMIS. 2004b;112:898–907. doi: 10.1111/j.1600-0463.2004.apm11211-1212.x. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M., Tamura K., Jakobsen I.B., Nei M., Jakobsen I.B., Nei M., Nei M. MEGA2: Molecular Evolutionary Genetics Analysis software. Arizona State University edition; Tempe, AZ: 2001. [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L., Shumway M., Antonescu C., Salzberg S.L., Antonescu C., Salzberg S.L., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P., Oyston P.C., Chain P., Chu M.C., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Oyston P.C., Chain P., Chu M.C., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Chain P., Chu M.C., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Chu M.C., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Duffield M., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Fuxelius H.H., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Garcia E., Halltorp G., Johansson D., Isherwood K.E., Halltorp G., Johansson D., Isherwood K.E., Johansson D., Isherwood K.E., Isherwood K.E., et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.D., Kling-Backhed H., Giannakis M., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Kling-Backhed H., Giannakis M., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Giannakis M., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Cordum H.S., Wang C., Elliott G., Edwards J., Wang C., Elliott G., Edwards J., Elliott G., Edwards J., Edwards J., et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: Evolution during disease progression. Proc. Natl. Acad. Sci. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Brittnacher M., Svensson K., Buckley D., Haugen E., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Brittnacher M., Svensson K., Buckley D., Haugen E., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Svensson K., Buckley D., Haugen E., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Buckley D., Haugen E., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Haugen E., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Zhou Y., Chang J., Levy R., Hayden H., Forsman M., Chang J., Levy R., Hayden H., Forsman M., Levy R., Hayden H., Forsman M., Hayden H., Forsman M., Forsman M., et al. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect. Immun. 2006;74:6895–6906. doi: 10.1128/IAI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M., Karamohamed S., Pettersson B., Uhlen M., Nyren P., Karamohamed S., Pettersson B., Uhlen M., Nyren P., Pettersson B., Uhlen M., Nyren P., Uhlen M., Nyren P., Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Ronaghi M., Uhlén M., Nyrén P., Uhlén M., Nyrén P., Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Svensson K., Larsson P., Johansson D., Bystrom M., Forsman M., Johansson A., Larsson P., Johansson D., Bystrom M., Forsman M., Johansson A., Johansson D., Bystrom M., Forsman M., Johansson A., Bystrom M., Forsman M., Johansson A., Forsman M., Johansson A., Johansson A. Evolution of subspecies of Francisella tularensis. J. Bacteriol. 2005;187:3903–3908. doi: 10.1128/JB.187.11.3903-3908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R., Johansson A., Neeson B., Isherwood K., Sjostedt A., Ellis J., Titball R.W., Johansson A., Neeson B., Isherwood K., Sjostedt A., Ellis J., Titball R.W., Neeson B., Isherwood K., Sjostedt A., Ellis J., Titball R.W., Isherwood K., Sjostedt A., Ellis J., Titball R.W., Sjostedt A., Ellis J., Titball R.W., Ellis J., Titball R.W., Titball R.W. Discrimination of human pathogenic subspecies of Francisella tularensis by using restriction fragment length polymorphism. J. Clin. Microbiol. 2003;41:50–57. doi: 10.1128/JCM.41.1.50-57.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hijum S.A., Zomer A.L., Kuipers O.P., Kok J., Zomer A.L., Kuipers O.P., Kok J., Kuipers O.P., Kok J., Kok J. Projector 2: Contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res. 2005;33:W560–W566. doi: 10.1093/nar/gki356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Fournier P.E., Ogata H., Raoult D., Fournier P.E., Ogata H., Raoult D., Ogata H., Raoult D., Raoult D. Multispacer typing of Rickettsia prowazekii enabling epidemiological studies of epidemic typhus. J. Clin. Microbiol. 2005;43:4708–4712. doi: 10.1128/JCM.43.9.4708-4712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]