Abstract
We have developed a method for mapping unmethylated sites in the human genome based on the resistance of TspRI-digested ends to ExoIII nuclease degradation. Digestion with TspRI and methylation-sensitive restriction endonuclease HpaII, followed by ExoIII and single-strand DNA nuclease allowed removal of DNA fragments containing unmethylated HpaII sites. We then used array comparative genomic hybridization (CGH) to map the sequences depleted by these procedures in human genomes derived from five human tissues, a primary breast tumor, and two breast tumor cell lines. Analysis of methylation patterns of the normal tissue genomes indicates that the hypomethylated sites are enriched in the 5′ end of widely expressed genes, including promoter, first exon, and first intron. In contrast, genomes of the MCF-7 and MDA-MB-231 cell lines show extensive hypomethylation in the intragenic and intergenic regions whereas the primary tumor exhibits a pattern between those of the normal tissue and the cell lines. A striking characteristic of tumor cell lines is the presence of megabase-sized hypomethylated zones. These hypomethylated zones are associated with large genes, fragile sites, evolutionary breakpoints, chromosomal rearrangement breakpoints, tumor suppressor genes, and with regions containing tissue-specific gene clusters or with gene-poor regions containing novel tissue-specific genes. Correlation with microarray analysis shows that genes with a hypomethylated sequence 2 kb up- or downstream of the transcription start site are highly expressed, whereas genes with extensive intragenic and 3′ untranslated region (UTR) hypomethylation are silenced. The method described herein can be used for large-scale screening of changes in the methylation pattern in the genome of interest.
DNA methylation is a major epigenetic mechanism of controlling gene expression in mammalian cells (Walsh et al. 1998; Egger et al. 2004; Bernstein et al. 2007). The majority of cytosine methylation occurs in the CpG dinucleotide, and 70%–80% of CpG dinucleotide is methylated in mammalian genome. The remaining unmethylated CpG dinucleotides are mainly associated with the CpG island (Bird 1999; Robertson and Jones 2000; Eckhardt et al. 2006). Genomic DNA methylation undergoes extensive repatterning during embryogenesis and carcinogenesis (Feinberg and Tycko 2004; Esteller 2007; Schaefer et al. 2007), and the genomic DNA methylation pattern may be regulated by environmental perturbation and inherited across generations (Whitelaw and Whitelaw 2006). Although intensive studies have shown that promoter methylation, particularly methylation of the CpG island (CPGI), is involved in regulation of transcription of genes (Herman and Baylin 2003; Stransky et al. 2006), the regulation and meaning of genome-wide epigenetic changes are still unclear at the present time. In cancer cells the genome becomes both globally hypomethylated and locus-specifically hypermethylated relative to the genome derived from normal tissue but with an overall decrease in DNA methylation (Wilson et al. 2007). Investigations of hypomethylation in tumor genome indicate that global hypomethylation occurs in the intragenic and intergenic regions and in repeat sequence elements (Wilson et al. 2007). However, the distribution of the hypomethylated sites and the relationship between global hypomethylation and tumorigenesis have not been investigated.
To investigate this problem, we developed a method to map the hypomethylated sites in breast cancer cell lines and the primary breast tumor genome relative to the normal tissue genome. We took advantage of the unique property of TspRI restriction endonuclease to generate a 3′ 9-base-long single-strand tail, which is resistant to exonuclease III digestion (Henikoff 1984). If a methylation-sensitive restriction endonuclease is used together with TspRI to digest DNA, then the unmethylated sites will become sensitive to ExoIII digestion. This procedure allows biochemical elimination of unmethylated DNA sequences which can then be mapped by using high-resolution comparative genomic hybridization (CGH) between ExoIII-digested and mock-digested DNA.
Using this novel technique, we obtained several novel findings concerning the distribution of hypomethylated sites in MCF-7, MDA-MB-231, and normal tissue genomes. We showed that global hypomethylation in breast cancer cell lines is mainly localized in the megabase-sized hypomethylated zones that coincide with fragile sites, evolutionary breakpoints (defined with respect to mice), and tissue-specific large genes or gene clusters and with tumor suppressor genes. In contrast, the hypomethylated sites in normal human tissue genomes are mainly localized at the 5′ region of widely expressed genes and are mapped near regulatory elements. We analyzed the correlation of genome-wide differential methylation and gene expression in MCF-7 cells and showed that genes with extensive intragenic hypomethylation or hypomethylation at 3′ untranslated region (UTR) showed low expression compared with genes with hypomethylation at 5′ region. Furthermore, we showed that hypomethylation within 2 kb up- or downstream from the transcription start site is correlated with high expression, suggesting that epigenetic regulation of sequences surrounding the initiation site plays a role in gene transcription (Appanah et al. 2007).
Results
Protection of TspRI DNA fragments from exonuclease III digestion
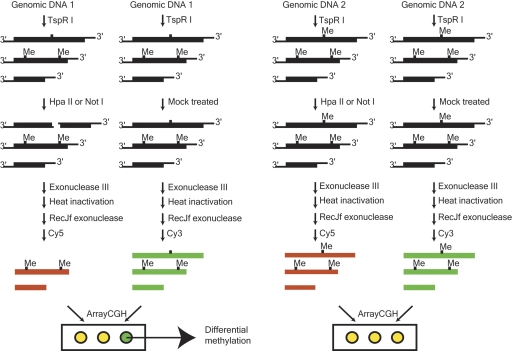
We noticed that TspRI digestion generates an ExoIII-resistant nine-base single-stranded tail at the 3′ ends (Henikoff 1984). If the DNA is further digested with methylation-sensitive restriction endonucleases, the unmethylated DNA fragments will become sensitive to degradation by ExoIII enzyme. This procedure allows biochemical “deletion” of unmethylated DNA and enrichment for methylated DNA sequences. The strategy is illustrated in Figure 1.
Figure 1.
Schematic diagram showing the strategy of mapping hypomethylated sequences in the genome by TspRI–HpaII–ExoIII digestion. Genomic DNA was first digested with TspRI, which created DNA fragments with a nine-base 3′ extension and were resistant for exonuclease III digestion. The DNA fragments were then digested with methylation-sensitive restriction enzyme HpaII. After treatment with exonuclease III and RecJf exonuclease, ExoIII-digested or mock-digested DNA was purified and labeled with Cy3 and Cy5 fluorescence dyes, respectively. Cy3 hybridization intensity was normalized to Cy5 for comparison among samples.
We used plasmid DNA to test this strategy. As shown in Supplemental Figure 1, digestion of plasmid pDsRed1-C1 DNA with TspRI produced 2009-, 1283-, 1110-, 271-, and 13-bp DNA fragments, and the 2009-bp DNA fragment could be further digested with BamHI to produce 1798- and 211-bp fragments with an end that was sensitive to ExoIII degradation. Indeed, the two BamHI fragments were sensitive to ExoIII degradation but not the others. This result demonstrated that TspRI-generated DNA ends were resistant to ExoIII degradation, whereas ends with short single-strand tails generated by other restriction endonucleases were sensitive to ExoIII degradation.
Mapping of genome-wide unmethylated sequences by TspRI–ExoIII–array CGH
The scheme described above could be used to remove DNA fragments containing unmethylated HpaII sites in genomic DNA after HpaII digestion. If the genomic DNA thus depleted of the unmethylated sequences is used to co-hybridize with the ExoIII mock-treated genomic DNA in a CGH scheme, then the unmethylated sequences will show up with reduced signals in the ExoIII-digested DNA. We employed an Agilent 244K array chip for mapping the unmethylated sequences in genomic DNA obtained from two breast cancer cell lines, one primary breast tumor, and five normal human tissues.
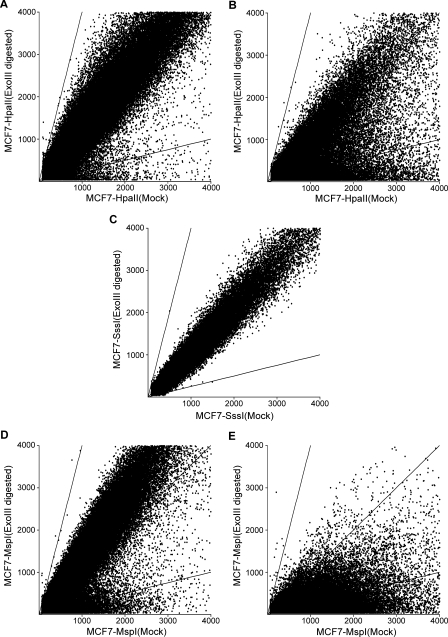
Out of 244K data points in the Agilent chip, 53,728 are flanked by TspRI sites and contain HpaII sites, and the remaining data points are within TspRI fragments that contain no HpaII sites. The DNA hybridization data showed that the hybridization signals of the data points not containing HpaII sites (Fig. 2A) were close to a ratio of one between ExoIII-digested and control mock-digested DNA, as expected. On the other hand, a large fraction of the hybridization signal was lower in the ExoIII-digested DNA that contained HpaII restriction sites (Fig. 2B). Those data points with lower signals were by implication DNA fragments containing unmethylated HpaII sites.
Figure 2.
Scatter plot of TspRI–ExoIII–array CGH data. (A) MCF7 genomic DNA digested with TspRI–HpaII–ExoIII; hybridization signals of the data points not containing HpaII sites flanked by TspRI sites are close to a ratio of one between ExoIII-digested and control mock-digested DNA, as expected since these TspRI fragments should not contain ExoIII-sensitive ends. The two lines correspond to ratios of four and 0.25, respectively, between the Cy3 and Cy5 signals. Some of the HpaII-sensitive background is probably due to sequence polymorphism creating new HpaII sites, as revealed by SssI and MspI experiments. (B) A large fraction of hybridization signal in the 53,728 TspRI fraction containing HpaII sites is lower after digestion with HpaII and ExoIII. (C) After SssI methylation of CpG sites in vitro, the 53,728 TspRI fragments containing HpaII sites became resistant to HpaII–ExoIII digestion with a hybridization ratio of around one in contrast to the result in B. (D,E) MCF7 genomic DNA digested with CpG-methylation–insensitive restriction enzyme MspI results in depletion of signals with a ratio of one in the fraction containing HpaII sites and a lowering of signals, whereas the fragments without HpaII sites are not affected significantly.
To confirm the array CGH analysis, we protected all HpaII sites by methylation in vitro with SssI methylase. Indeed, all the fragments became resistant to ExoIII degradation (Fig. 2C). This result indicated that the sequences with lower hybridization ratios (see Fig. 2B) indeed contain unmethylated HpaII sites. Furthermore, digestion with CpG-methylation–insensitive restriction enzyme MspI resulted in depletion of signals with ratios of one in the fraction containing HpaII sites whereas fragments without HpaII sites were not affected significantly (Fig. 2D,E). To further validate the technique, we analyzed the methylation status of 30 randomly chosen sites by using methylation-sensitive PCR reaction. The methylation status of all 30 sites was confirmed by this technique. We also analyzed the methylation status of 20 sites mapped by array CGH in histone gene clusters with bisulfite sequencing technique, and the methylation status of these sites was confirmed. These analyses validate the TspRI–ExoIII technique as a method for genome-wide screening of unmethylated sites.
Extensive hypomethylation of MCF-7 genomes in intragenic and intergenic regions as compared with genomes of normal human tissues
If a cutoff value of four for the hybridization ratio between mock-digested and ExoIII-digested DNA, then there are 9802 hypomethylated sites in the MCF-7 genome. After SssI treatment, this number is reduced to 48. In contrast, genomes derived from breast, brain, leukocyte, testis, and liver contain only 1376, 2827, 3680, 1197, and 1230 hypomethylated sites, respectively. This result indicates that the MCF-7 genome is highly hypomethylated relative to the normal tissue genomes. There are 4280 and 2565 hypomethylated sites in MDA-MB-231 and a primary breast tumor genome, respectively. These values are between those of normal breast tissue and the MCF-7 genome.
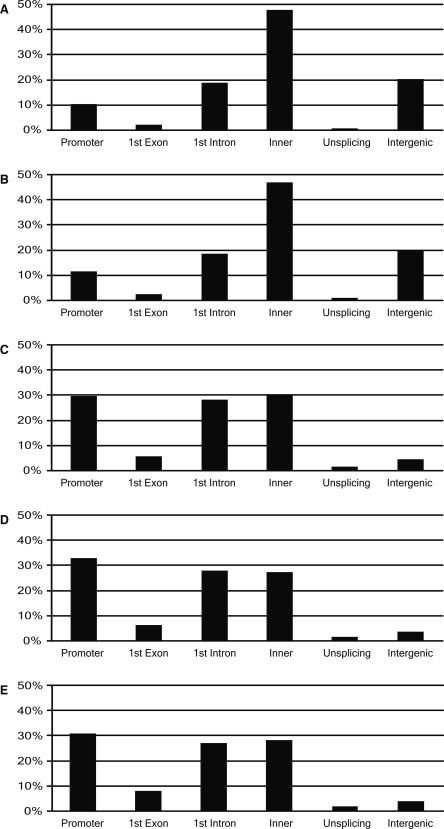
To understand the nature of unmethylated sites in MCF-7 genome relative to those from normal tissues, we analyzed the distribution of positions of oligonucleotide probes associated with hypomethylated sequences in the genome. Figure 3 and Supplemental Figure 2 show the comparative distribution of unmethylated sites with respect to the positions of the genes in MCF-7, MDA-MB-231, and other human tissue genomes. Most strikingly, the extensively hypomethylated sites in the MCF-7 genome are enriched in the intragenic region (excluding exon 1 and intron 1) and in the intergenic regions without genes or EST sequences (P < 0.01). In comparison, normal breast genomes are enriched in the promoter (32.8%), exon 1 (6.3%), and intron 1 (28.1%) close to the 5′ end of the gene (P < 0.01), a region known to contain regulatory sequences. Similar distributions are found in human brain, leukocyte, testis, and liver tissue genomes.
Figure 3.
Distribution of positions of hypomethylated sites in promoter, 1st exon, 1st intron, inner region of gene, and region containing no transcribed sequences: (A) MCF7, (B) MDA-MB-231, (C) tumor breast, (D) normal breast, and (E) normal brain. The extensively hypomethylated sites in MCF7 and MDA-MB-231 genomes are enriched in the intragenic and intergenic regions (P < 0.01, Fisher’s exact test). By contrast, normal tissue genomes are enriched in the promoter, exon 1, and intron 1 close to the 5′ end of the gene (P < 0.01, Fisher’s exact test).
Analysis of the distribution pattern of the hypomethylated sites in MCF-7 genomes shows that there are two major types of hypomethylated sites. The first type is composed of single isolated hypomethylated sites, mainly associated with genes. This type of hypomethylated sites is found in the normal tissue genomes. The second type of distribution of hypomethylated sites is the large megabase-sized hypomethylated zones, ranging in size from one to eight megabases. This type of distribution is unique to the cancer cell lines. These two types of distribution of hypomethylated sites are described below.
Megabase-sized hypomethylated zones in tumor cells: Association with large genes, fragile sites, evolutionary breakpoints, chromosomal translocation, and newly evolved tissue-specific genes
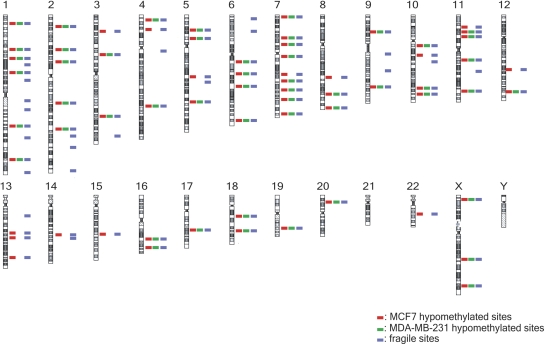
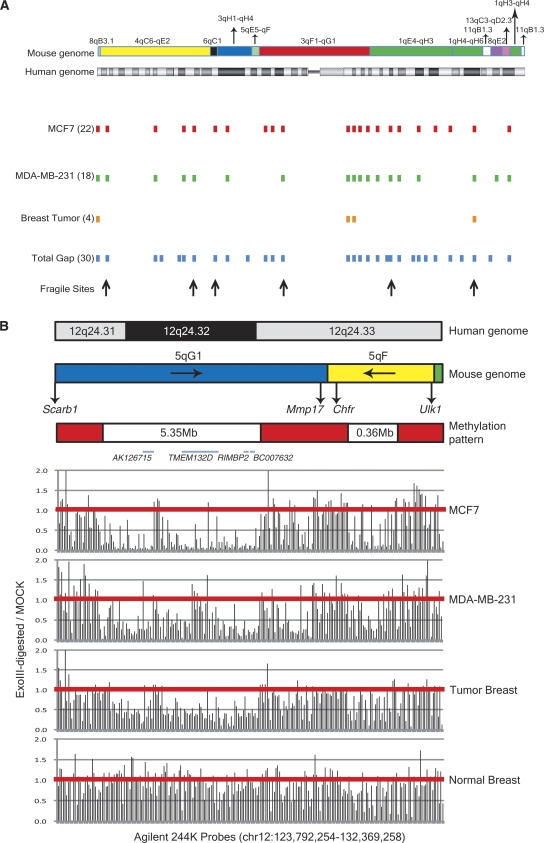
An interesting and intriguing feature of hypomethylation in MCF-7 tumor genome is the presence of megabase-sized, extensively hypomethylated zones. We found that 68 of the 86 known common fragile sites (http://www.genenames.org/) in the human genome were in the hypomethylated zones (P < 0.01) in the MCF-7 genome (Fig. 4). The hypomethylated zones which associate with fragile sites, evolutionary breakpoints, and chromosomal translocation breakpoints are further illustrated for chromosome 7. The gene structural features, such as fragile sites and chromosome rearrangement breakpoints associated with disease, were fully depicted in previous literature (Scherer et al. 2003). There are 18 genes greater than 500 kb in size. Two of the large genes, MAGI2 and EXOC4, are not in the 53,728 TspRI fragments containing HpaII sites in MCF-7. Of the remaining 16 large genes, 15 are extensively hypomethylated in MCF-7 (Supplemental Fig. 3B). We further found that megabase-sized hypomethylated zones in chromosome 7 in MCF-7 correlated with all the fragile sites, FRA7A–FRA7J (P < 0.01), and with 78% of chromosomal rearrangement breakpoints (P < 0.01). These correlations are also significant in MDA-MB-231 genome (P < 0.01).
Figure 4.
Megabase-sized hypomethylated zones in tumor cell genomes associated with fragile sites. Red and green lines represent the hypomethylated zones in MCF7 and MDA-MB-231, respectively. Blue lines represent 86 common fragile sites in human genome. We found that 79% of fragile sites were in the hypomethylated zones in the MCF-7 genome (P < 0.01), and this correlation was also significant in the MDA-MB-231 genome (P < 0.01).
The majority of large hypomethylated zones in MCF-7 genome occur in the genomic region with low gene density, as seen in chromosome 7. This is also shown in other chromosomes, such as chromosome 1 (Fig. 5A). There are 30 megabase-sized low-gene-density regions in chromosome 1 with an average size of 1.8 Mb. The mapped positions of hypomethylated zones in MCF-7 (P < 0.01), in MDA-MB-231 (P < 0.01), and in primary breast tumor genomes are subsets of these low-gene-density regions. Interestingly, these sites are also associated with breakpoints joining different syntenic segments of mouse genome in human genome. Several specific examples are described in more detail below to illustrate these unique features.
Figure 5.
Megabase-sized hypomethylation zones in tumor cells. The majority of large hypomethylated zones in the MCF7 genome occur in the genomic region with low gene density. (A) Hypomethylated zones in breast tumor genomes are located in low-gene-density regions in chromosome 1 (lowest line). In MCF-7 and MDA-MB-231 genomes, the hypomethylated zones are in association with these low-gene-density regions, and the P-values of Fisher’s exact test are <0.01. Arrows represent the hypomethylated fragile sites in MCF-7 cell line genome, FRA1A (1p35–36.1), FRA1B (DAB1), FRA1C (1p31.2), FRA1E (DPYD), FRA1G (1q25.1), and FRA1H (USH2A and ESRRG). (B) The 12q24.31–32 hypomethylated regions (white bars) are found in the genomes of MCF-7 (P < 0.01), MDA-MB-231 (P < 0.01), and primary breast tumor (P < 0.01) as compared with normal breast genome. The red horizontal lines represent ratios of one between signals of ExoIII-digested and mock-digested DNA.
A 5.35-Mb hypomethylated region in the telomeric 12q24.31–32 is found in the genomes of MCF-7 (P < 0.01), MDA-MB-231 (P < 0.01), and primary breast tumor (P < 0.01) (Fig. 5B). This region is near the evolutionary breakpoint joining a segment from mouse 5qG1 to a segment from mouse 5qF and is located in the chromosome fragile site (Ruiz-Herrera et al. 2006), a chromosomal imbalance region in tumors (Rutherford et al. 2005). A large brain-enriched gene, TMEM132D (832 kb), is found in this zone and is flanked by two large, low-gene-density regions. On one side is a large, 2-Mb gene-poor region containing a number of novel tissue-specific cDNA with no mouse homologs (e.g., BC073948, a mammary gland–enriched cDNA).
We found that large genes located at the fragile sites were extensively hypomethylated and silenced in MCF-7, such as CNTNAP2 in the FRA7I region and DMD in the FRAXC region (see Supplemental Fig. 4A,B). In addition, several large tumor suppressor genes are also extensively hypomethylated. For example, the large tumor suppressor genes, OPCML (1.12 Mb), DCC (1.19 Mb), CADM1 (0.33 Mb), CDH4 (0.68 Mb), and NTRK3 (0.38 Mb), are found silenced and extensively hypomethylated intragenically.
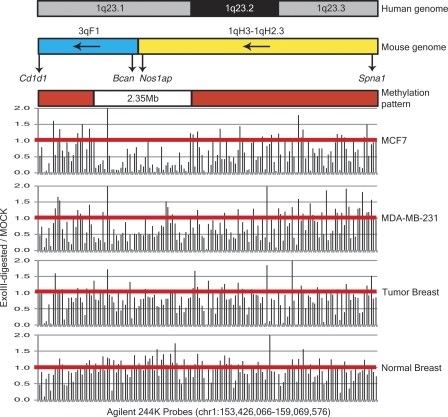
The second type of hypomethylated zone contains clusters of tissue-specific genes. As shown in Figure 6, a 2.3-Mb hypomethylated zone containing immune- and brain-specific gene cluster (OR, FCRL, and CD genes) is found at 1q23.1 in MCF-7 genome (P < 0.01) and primary tumor genome (P < 0.01) and less prominently in MDA-MB-231 genome (P = 0.60). An evolutionary breakpoint is located in the middle of this hypomethylated zone. There are many novel genes created in this region when compared with mouse genome, including FCRL genes and some OR genes in a 97.3-kb region from 155.75 to 156.79 Mb of the human genome. BLAT analysis shows that human FCER1A, DARC, AIM2, PYHIN1, MNDA, IFI16, and APCS genes have undergone extensive sequence evolution, in that little homology is found between the mRNA sequences of human and mouse genomes. Interestingly, AIM2, PYHIN1, IFI16, and DARC all have been shown to have antitumor or tumor-suppressor activity (Ding et al. 2004; Chen et al. 2006; Zijlstra and Quigley 2006; Zhang et al. 2007). New mRNA species not found in mouse genome are also present in this region (e.g., thymus-specific AK057554). The chromosomal translocation breakpoint fusing MEF2D and DAZAP1 in chromosome 19 (Prima et al. 2005) is located at the border of this hypomethylated zone.
Figure 6.
Megabase-sized hypomethylation zones which contain clusters of tissue-specific genes in tumor cells. As compared with normal breast genome, the 1q23 hypomethylated region is found in the genomes of MCF-7 and primary breast tumor (P < 0.01) and less prominently in MDA-MB-231 genome (P = 0.60). The red horizontal lines represent ratios of one between signals of ExoIII-digested and mock-digested DNA.
Another example of a hypomethylated zone is the 8-Mb centromeric zone covering 11p11–11q11 (Supplemental Fig. 5A) containing olfactory receptor gene clusters with recent sequence evolution. This region has been suggested as a tumor suppressor locus for liver cancer (Ricketts et al. 2002) and is involved in translocation in lymphoma (Cuneo et al. 2001). One end of this region coincides with an insertion of prostate surface antigen and prostate cancer prognostic marker gene FOLH1 (Maraj and Markham 1999) found in mouse chromosome 7qD3. This gene together with its flanking newly evolved human-specific genes, FGCP, is located at the breakpoint joining mouse 2qE1 and 2qD. The FOLH1 sequence is also partially duplicated as a PSMAL gene at another hypomethylated zone located at an evolutionary breakpoint at a 3-Mb hypomethylated zone in 11q14.3 (Supplemental Fig. 5B). The novel centromeric gene, TRIM48, which has no mouse homolog, has two homologous copies as TRIM49 in the 11q14.3 breakpoint next to PSMAL. Centromeric copies of FOLH1 and TRIM48 are also found in chimpanzee but not in Rhesus monkey, suggesting that translocation to the centromeric region is a relatively recent event. A novel mammary gland enriched gene, SPRYD5, was found buried within the OR gene cluster in the centromeric region of chromosome 11. This gene has at least eight copies in the p- and q-sides of the centromere and is duplicated at the 11q14.3 hypomethylated zone. It is also present at the centromere of human chromosome 2 at the evolutionary breakpoint joining DNA sequences from mouse chromosome 1 and chromosome 2. BLAT analysis also shows that this is a newly evolved gene with a few partially duplicated sequences in Rhesus monkey genome corresponding to the human 11q14.3 breakpoint but not at the centromere of chromosome 11.
Correlation between DNA methylation and gene expression: High expression of genes with hypomethylation 2 kb up- or downstream from transcription start site and low expression of genes with extensive intragenic unmethylation and hypomethylation at the 3′ UTR
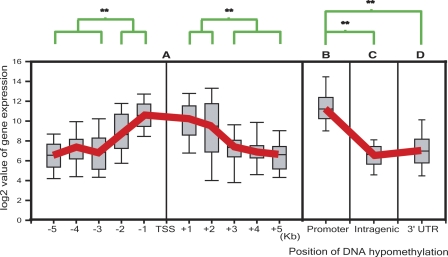
Since promoter demethylation has been shown to be associated with active genes, we tested whether genes with the unmethylated promoter region exhibited higher levels of expression. There are a total of 1423 genes in MCF-7 with unmethylated sites located within 5 kb up- or downstream from the transcription starting site. We divided the unmethylated sites every kb up or downstream from the transcription start site and plotted the gene expression level obtained from Affymetrix expression microarray analysis for each group of genes. The expression level for each group is rather heterogeneous. However, the average value for each group showed a gradient from the transcription start site. Genes with unmethylated sites within 2 kb upstream or downstream from a transcription start site in general show high levels of expression (Fig. 7). This result suggests that sequences involved in epigenetic regulation reside within 2 kb up- and downstream from a transcription initiation site (P < 0.01, t-test).
Figure 7.
Box plot of correlation between position of DNA hypomethylation and gene expression. Gene expression level obtained from Affymetrix analysis is correlated with hypomethylated sites at every kb up- or downstream from transcription initiation site. (A) Hypomethylation 2 kb up- or downstream from transcription start site is correlated with higher gene expression (**P < 0.01, t-test). The red line represents the median expression level for each group of genes with hypomethylated sites located 1–5 kb up- or downstream from the transcription start site. (B) Stand-alone promoter has a high level of expression. (C,D) Genes with extensive intragenic hypomethylation and hypomethylation at 3′ UTR show low expression (**P < 0.01, t-test).
Sequences upstream from a transcription start site often overlap with adjacent genes. To avoid the possibility of transcription interference from adjacent genes, we analyzed the relationship between promoter hypomethylation and gene expression in genes with stand-alone upstream sequences. As shown in Figure 7, genes with a hypomethylated promoter region indeed show a very high level of expression (average relative expression value 5421). In contrast, genes with extensive intragenic hypomethylated sites (more than three sites) are in general expressed at a low level (average expression value 156). In particular, all the 273 large genes (size from 0.1 to 2.3 Mb) with extensive intragenic hypomethylation are expressed at background level (P < 0.01, t-test). We also noticed that genes with hypomethylation at 3′ UTR (excluding those with extensive intragenic methylation) were also expressed at a low level (average expression level 642) with a few exceptions.
Strong association of unmethylated sites in normal human tissue genomes with CpG island, nuclease-hypersensitive sites, and transcription factor binding sites
The hypomethylated sequences near the oligonucleotide probes in the tissue genomes are found frequently associated with CpG island (CPGI), nuclease hypersensitive sites, or transcription factor binding sites as indicated in the UCSC genome browser. About 70%–86% of the probes associated with hypomethylated sequences in normal tissue genomes are within 1.5 kb from CPGI (P < 0.01), and about 20% of the sites are within 100 bp of CPGI (P < 0.01). In contrast, only 19% and 4% of hypomethylated sites in MCF-7 genome are located within 1.5 kb and 100 base pairs of CPGI, respectively. The results showed that promoter region, exon 1, and intron 1 are highly enriched in hypomethylated sites near CPGI in normal human tissue genomes (P < 0.01).
Analysis of the nature of hypomethylated sites outside transcribed sequences in normal tissue genomes indicated that they are enriched in promoter sequences. For example, in breast genome, there are 321 such sites, and 222 sites (69.2%) are mapped within 5 kb from transcription sites near CPGI (83%) or nuclease-hypersensitive sites (28.8%). Of the remaining hypomethylated intergenic sites with no genes nearby or expressed sequence tags (ESTs), 30% are associated with CPGI and 42% are associated with transcription factor binding sites. These results indicate that the majority of hypomethylated sites in normal tissue genomes are associated with transcription-regulatory elements.
Histogram analysis reveals similar hypomethylated patterns in human tissue genomes
Analysis of a number of unmethylated sites in different chromosomes of normal human tissue genomes shows a differential hypomethylation density (number of unmethylated sites per Mb) among the chromosomes. Chromosomes 17 and 19 are consistently the most hypomethylated in all the five tissue genomes and in MCF-7 genome. The X chromosome is consistently the most hypermethylated. Chromosomes 4, 13, and 18 are also relatively hypermethylated in the genome of brain, breast, testis, and liver but not in leukocytes (Supplemental Fig. 6A).
Because hypomethylated sites are widely and sparsely distributed in the tissue genomes, it is difficult to examine the overall distribution pattern of these sites in each chromosome. To visualize the distribution pattern of unmethylated sites in the human chromosomes of different tissues, we plotted the number of hypomethylated sites per 100 kb (ratio < 0.25) in each chromosome. This analysis reveals some interesting hypomethylated domains in human tissue genomes. For example, in chromosome 6 there is a hypomethylated region mapped in the histone gene clusters and the HLA gene region (Supplemental Fig. 6B). This region is found in the genomes of leukocytes, brain, liver, and testis but less prominently in normal breast. A survey of the hypomethylated sites indicates that all of the HOX gene clusters are hypomethylated in all five of the tissue genomes analyzed.
The overall distribution of hypomethylated sites in human tissue genomes is remarkably similar. The similar distribution pattern of hypomethylated sites in different tissue genomes is mainly the result of extensive sharing of hypomethylated sites among the tissue genomes. For example, 80% and 84% of hypomethylated sites in brain and breast genome, respectively, are also found in the leukocyte genome. These extensively shared hypomethylated sites are associated with genes that are widely expressed, as seen in the GEPIS and Stanford SOURCE tissue expression databases. On the other hand, tissue-specific genes could be found with hypomethylated 5′ region sequences (promoter/exon1/intron1) after performing subtraction between the unmethylated sites of two different tissues. For example, brain-enriched or brain-specific genes, such as ELAVL2, POU3F4, GRIA2, GRIA3, OLFM3, NRG3, etc., are found hypomethylated in the 5′ region of the genes in the brain genome.
Discussion
In this study we presented a new method for scanning hypomethylated sites in the human genome based on the resistance of TspRI ends to ExoIII degradation. This method is not dependent on PCR amplification and therefore will not be subjected to differential amplification by PCR reaction. The method was validated by SssI methylation protection, by sensitivity to MspI digestion, and by confirmation using methylation-sensitive PCR reaction and bisulfite sequencing. The fact that more than 96% of hypomethylated sites are not altered after trichostatin A treatment further supports the robustness of this method.
Although the mapped hypomethylated sites are only a subset of the hypomethylated sites in the genome under study due to the limited number of oligonucleotide probes on the chip and to the limited number of sites probed by HpaII digestion, this method allows a rapid whole-genome scan to reveal the novel features of distribution of hypomethylated sites. Indeed, this method allows us to uncover several interesting features of hypomethylation sequences in both tumor cell lines and normal genomes.
The most intriguing finding of this study is the discovery of megabase-sized hypomethylated zones that are co-localized with breakpoints (Murphy et al. 2005; Shimada et al. 2005; Schibler et al. 2006), chromosomal rearrangement breakpoints, fragile sites, tumor suppressors, large genes, and novel tissue-specific genes without mouse homologs in gene-poor regions. These large hypomethylated regions are also AT-rich, low in CPGI, and show low sequence conservation with other genomes. Several extensively hypomethylated large genes have also been shown to reside at the chromosomal breakpoints of chromosomal rearrangement/deletion, such as PRDM16 (Stevens-Kroef et al. 2006), AKT3 (Boland et al. 2007), MDS1 (Cherry et al. 2001), PTPRZ1 (Mulholland et al. 2006), CPNE8 (Ramsey et al. 2003), NKAIN2 (Bocciardi et al. 2005), NTRK3 (Jin et al. 2007), and NRG1 (Adelaide et al. 2000). These results support the hypothesis that hypomethylation is involved in genome instability (Dodge et al. 2005) through recombination of unmethylated sequences. Hypomethylation has been shown to be a necessary, but not sufficient, condition for V(D)J recombination (Engler and Storb 1999). The increased recombination rate of hypomethylated DNA has been suggested as the cause of genomic instability in tumor genomes (Eden et al. 2003). It is possible that hypomethylation zones may serve to regulate higher-order chromatin organization to allow differential expression of genes involved in tumorigenesis.
The existence of large hypomethylated zones suggests aberrations in the DNA methylation process in tumor cells. However, although we have found such zones in a primary breast tumor genome, we cannot exclude the possibility that the extensive hypomethylation observed in tumor cell lines may be the result of in vitro culture. Further analysis of primary tumors is needed to assess the biological role of these extensively demethylated zones. Our gene expression analysis shows that all the DNMT genes except TRDMT1 are expressed in MCF-7 cells. It remains to be shown whether TRDMT1 gene product is involved in the maintenance of methylation of regions containing large genes, evolutionary breakpoints, and fragile sites.
The second major finding in this study is the correlation between intragenic hypomethylation and gene silencing. Current research efforts focus on the relationship between methylation status in the promoter region with gene expression. Few studies address the role of intragenic hypomethylation in gene expression. It is still unclear whether intragenic hypomethylation is a cause of gene inactivation and, if so, how it could lead to silencing of a gene. It is interesting also to note that the majority of genes with hypomethylation at 3′ UTR are also expressed at very low level. The role of intragenic and 3′ UTR hypomethylation in gene expression deserves further investigation.
Our study shows that the hypomethylation in MCF-7 tumor cell genome is mainly due to extensive intragenic hypomethylation of large genes, in tissue gene cluster and in regions with low gene density or devoid of genes but with novel tissue-specific mRNA and EST. Hypomethylation seems to occur in regions containing tissue-specific genes, especially genes highly expressed in brain and lymphoid tissues. The extensively hypomethylated genes are all silenced, as shown by microarray analysis.
In contrast to the MCF-7 and MDA-MB-231, the genomes derived from normal tissues contain dispersed hypomethylated sites mainly in the regulatory region of the gene. The hypomethylated sites are extensively shared among the five tissues studied. The similarity of the overall distribution pattern of hypomethylated sites in different human tissue genomes is mainly the result of hypomethylation of the 5′ regulatory region of genes that are widely expressed among tissues. Subtraction analysis of the unmethylated sites between two genomes can allow one to examine tissue-specific or tumor-specific hypomethylated sites. We illustrate this principle with detailed gene-by-gene analysis of expression patterns of subtracted data from the tissue expression databases. However, we found that hypomethylation of the regulatory region of tissue-specific genes may also be found in other tissues. The significance of this observation is not known at the present time.
In conclusion, we have made the novel observation that there are megabase-sized hypomethylation zones in tumor cell-line genomes, and these structures are not found in the five normal tissue genomes analyzed. The novel findings should provide new paradigms for understanding long-range organization of DNA methylation in normal, MCF-7, and MDA-MB-231 genomes.
Methods
Cell culture
MCF-7 cells were cultured in RPMI1640 medium (GIBCO/BRL) supplemented with 10% (v/v) fetal bovine serum (GIBCO/BRL), 2.0 g/L sodium bicarbonate. MDA-MB-231 cells were cultured in L15 medium (GIBCO/BRL) supplemented with 10% (v/v) fetal bovine serum (GIBCO/BRL). All cell lines were incubated in a humidified 37°C incubator with 5% CO2.
Genomic DNA extraction
Cells were washed with 1× PBS and resuspended with cell lysis buffer. Cells were treated with 0.1 mg/mL of RNase A for an hour at 37°C and with 0.3 mg/mL proteinase K for 12–16 h at 55°C. Samples were extracted with equal volumes of phenol/chloroform/isoamyl alcohol mixture (24:25:1); the extraction procedure was repeated until the interface was clean. An equal volume of chloroform was then added, and the solution was centrifuged for 10 min at 13,000g. The aqueous phase was ethanol-precipitated, and the DNA pellet was washed with 70% ethanol, air-dried, and dissolved in d3H2O. In addition, samples of genomic DNA of human brain, breast, liver, testis, leukocytes, and breast tumor tissues, were purchased from BioChain.
Preparation of RNA
Cells were rinsed twice with 1× PBS. Total RNA was extracted according to the RNeasy Mini Kit Spin Protocol (QIAGEN). The integrity of the RNA extract was checked by agarose gel electrophoresis, and the concentration of RNA was estimated by ultraviolet spectrophotometry.
Protected from exonuclease III digestion by TspRI ends
DNA (10–15 μg) was digested with a methylation-sensitive restriction enzyme, such as HpaII, in a 100-μL total volume solution for 3 h, 5 μL (50 units) of TspRI was then added, and the resulting mixture was reacted at 65°C for 3 h using a Hot Top PCR machine. Exonuclease III (100 units) was then added. Reaction was carried out at 30°C for 1 h. Exonuclease III was heat-inactivated by heating at 70°C for 20 min. RecJf (50 units) was added to remove single-strand DNA, and the enzyme was inactivated by heating at 65°C for 20 min. DNA was then phenol/chloroform-extracted and ethanol-precipitated.
Array-CGH protocols
Array-CGH was performed using the Agilent Human Genome CGH microarrays 185K and 244K (Agilent Technologies), high-resolution 60-mer oligonucleotide-based microarray sets containing 184,672 and 243,431 probes, respectively. Labeling and hybridization were performed according to the protocol provided by Agilent. MCF-7, MDA-MB-231, and the tissue genomes were processed as shown in Figure 1, and HpaII was selected as the methylation-sensitive restriction enzyme. Briefly, 2–3 μg of the undigested and digested DNA were double-digested with AluI and RsaI for 2 h at 37°C. The digested DNA was labeled by random priming using the Agilent Genome DNA Labeling Kit PLUS. ExoIII-digested and control mock-digested DNA were pooled and hybridized with human Cot I DNA at 65°C. Washing was performed according to the Agilent protocol. Arrays were analyzed using the Agilent AA DNA Microarray Scanner and Agilent Feature Extraction software (v.9.1). Results were displayed using Agilent CGH Analytics software (v.3.4). The Cy3 hybridization intensity was normalized to Cy5 for comparison among samples. The log2 ratios (log2 Cy5/Cy3) were calculated and compared. The array CGH data including raw data and normalized data were deposited at the NCBI GEO website (GEO accession no. GSE9015).
Affymetrix analysis
Affymetrix microarray was performed using Human U133 2.0 (Affymetrix). Details of the methods for RNA quality, sample labeling, hybridization, and expression analysis were according to the manual of Affymetrix Microarray Kit. The microarray data were deposited at the NCBI GEO website (GEO accession number GSE9015).
Sodium bisulfite reaction
The sodium bisulfite reaction was performed according to the EZ DNA Methylation-Gold Kit instruction manual (Zymo Research).
Statistical evaluation
Statistical analysis was carried out using SPSS15.0. The hypomethylated zones in breast tumor cell line genomes correlated with fragile sites; evolutionary and rearrangement breakpoints were mainly evaluated by using the Fisher’s exact test. The comparative distribution of unmethylated sites with respect to the positions of the genes among MCF-7, MDA-MB-231, and human tissue genomes was also evaluated by Fisher’s exact test; Student’s t-test was employed to evaluate the correlation between the position of DNA hypomethylation and gene expression. All reported P values were considered significant for *P < 0.05 and **P < 0.01.
Acknowledgments
We thank Wan-Chun Chang and Chia-Ling Hua from the National Research Program for Genomic Medicine, who provided excellent help with the experiments. This work is supported by the Program for Promoting Academic Excellence of Universities (Phase II, NSC 94-2752-B-010-001-PAE), by a grant from the Ministry of Education, Aim for the Top University Plan, and by the National Program and a grant from the National Research Program of Genomic Medicine (NSC94-3112-B-010-003 and NSC95-3112-B-010-012) from the National Science Council of Taiwan.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.070961.107.
References
- Adelaide J., Chaffanet M., Mozziconacci M.J., Popovici C., Conte N., Fernandez F., Sobol H., Jacquemier J., Pebusque M., Ron D., Chaffanet M., Mozziconacci M.J., Popovici C., Conte N., Fernandez F., Sobol H., Jacquemier J., Pebusque M., Ron D., Mozziconacci M.J., Popovici C., Conte N., Fernandez F., Sobol H., Jacquemier J., Pebusque M., Ron D., Popovici C., Conte N., Fernandez F., Sobol H., Jacquemier J., Pebusque M., Ron D., Conte N., Fernandez F., Sobol H., Jacquemier J., Pebusque M., Ron D., Fernandez F., Sobol H., Jacquemier J., Pebusque M., Ron D., Sobol H., Jacquemier J., Pebusque M., Ron D., Jacquemier J., Pebusque M., Ron D., Pebusque M., Ron D., Ron D., et al. Translocation and coamplification of loci from chromosome arms 8p and 11q in the MDA-MB-175 mammary carcinoma cell line. Int. J. Oncol. 2000;16:683–688. doi: 10.3892/ijo.16.4.683. [DOI] [PubMed] [Google Scholar]
- Appanah R., Dickerson D.R., Goyal P., Groudine M., Lorincz M.C., Dickerson D.R., Goyal P., Groudine M., Lorincz M.C., Goyal P., Groudine M., Lorincz M.C., Groudine M., Lorincz M.C., Lorincz M.C. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3:e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Meissner A., Lander E.S., Meissner A., Lander E.S., Lander E.S. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation de novo. Science. 1999;286:2287–2288. doi: 10.1126/science.286.5448.2287. [DOI] [PubMed] [Google Scholar]
- Bocciardi R., Giorda R., Marigo V., Zordan P., Montanaro D., Gimelli S., Seri M., Lerone M., Ravazzolo R., Gimelli G., Giorda R., Marigo V., Zordan P., Montanaro D., Gimelli S., Seri M., Lerone M., Ravazzolo R., Gimelli G., Marigo V., Zordan P., Montanaro D., Gimelli S., Seri M., Lerone M., Ravazzolo R., Gimelli G., Zordan P., Montanaro D., Gimelli S., Seri M., Lerone M., Ravazzolo R., Gimelli G., Montanaro D., Gimelli S., Seri M., Lerone M., Ravazzolo R., Gimelli G., Gimelli S., Seri M., Lerone M., Ravazzolo R., Gimelli G., Seri M., Lerone M., Ravazzolo R., Gimelli G., Lerone M., Ravazzolo R., Gimelli G., Ravazzolo R., Gimelli G., Gimelli G. Molecular characterization of a t(2;6) balanced translocation that is associated with a complex phenotype and leads to truncation of the TCBA1 gene. Hum. Mutat. 2005;26:426–436. doi: 10.1002/humu.20235. [DOI] [PubMed] [Google Scholar]
- Boland E., Clayton-Smith J., Woo V.G., McKee S., Manson F.D., Medne L., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., Clayton-Smith J., Woo V.G., McKee S., Manson F.D., Medne L., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., Woo V.G., McKee S., Manson F.D., Medne L., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., McKee S., Manson F.D., Medne L., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., Manson F.D., Medne L., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., Medne L., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., Zackai E., Swanson E.A., Fitzpatrick D., Millen K.J., Swanson E.A., Fitzpatrick D., Millen K.J., Fitzpatrick D., Millen K.J., Millen K.J., et al. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am. J. Hum. Genet. 2007;81:292–303. doi: 10.1086/519999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.F., Ou-Yang F., Hung J.Y., Liu J.C., Wang H., Wang S.C., Hou M.F., Hortobagyi G.N., Hung M.C., Ou-Yang F., Hung J.Y., Liu J.C., Wang H., Wang S.C., Hou M.F., Hortobagyi G.N., Hung M.C., Hung J.Y., Liu J.C., Wang H., Wang S.C., Hou M.F., Hortobagyi G.N., Hung M.C., Liu J.C., Wang H., Wang S.C., Hou M.F., Hortobagyi G.N., Hung M.C., Wang H., Wang S.C., Hou M.F., Hortobagyi G.N., Hung M.C., Wang S.C., Hou M.F., Hortobagyi G.N., Hung M.C., Hou M.F., Hortobagyi G.N., Hung M.C., Hortobagyi G.N., Hung M.C., Hung M.C. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol. Cancer Ther. 2006;5:1–7. doi: 10.1158/1535-7163.MCT-05-0310. [DOI] [PubMed] [Google Scholar]
- Cherry A.M., Bangs C.D., Jones P., Hall S., Natkunam Y., Bangs C.D., Jones P., Hall S., Natkunam Y., Jones P., Hall S., Natkunam Y., Hall S., Natkunam Y., Natkunam Y. A unique AML1 (CBF2A) rearrangement, t(1;21)(p32;q22), observed in a patient with acute myelomonocytic leukemia. Cancer Genet. Cytogenet. 2001;129:155–160. doi: 10.1016/s0165-4608(01)00439-3. [DOI] [PubMed] [Google Scholar]
- Cuneo A., Bardi A., Wlodarska I., Selleslag D., Roberti M.G., Bigoni R., Cavazzini F., De Angeli C., Tammiso E., del Senno L., Bardi A., Wlodarska I., Selleslag D., Roberti M.G., Bigoni R., Cavazzini F., De Angeli C., Tammiso E., del Senno L., Wlodarska I., Selleslag D., Roberti M.G., Bigoni R., Cavazzini F., De Angeli C., Tammiso E., del Senno L., Selleslag D., Roberti M.G., Bigoni R., Cavazzini F., De Angeli C., Tammiso E., del Senno L., Roberti M.G., Bigoni R., Cavazzini F., De Angeli C., Tammiso E., del Senno L., Bigoni R., Cavazzini F., De Angeli C., Tammiso E., del Senno L., Cavazzini F., De Angeli C., Tammiso E., del Senno L., De Angeli C., Tammiso E., del Senno L., Tammiso E., del Senno L., del Senno L., et al. A novel recurrent translocation t(11;14)(p11;q32) in splenic marginal zone B cell lymphoma. Leukemia. 2001;15:1262–1267. doi: 10.1038/sj.leu.2402191. [DOI] [PubMed] [Google Scholar]
- Ding Y., Wang L., Su L.K., Frey J.A., Shao R., Hunt K.K., Yan D.H., Wang L., Su L.K., Frey J.A., Shao R., Hunt K.K., Yan D.H., Su L.K., Frey J.A., Shao R., Hunt K.K., Yan D.H., Frey J.A., Shao R., Hunt K.K., Yan D.H., Shao R., Hunt K.K., Yan D.H., Hunt K.K., Yan D.H., Yan D.H. Antitumor activity of IFIX, a novel interferon-inducible HIN-200 gene, in breast cancer. Oncogene. 2004;23:4556–4566. doi: 10.1038/sj.onc.1207592. [DOI] [PubMed] [Google Scholar]
- Dodge J.E., Okano M., Dick F., Tsujimoto N., Chen T., Wang S., Ueda Y., Dyson N., Li E., Okano M., Dick F., Tsujimoto N., Chen T., Wang S., Ueda Y., Dyson N., Li E., Dick F., Tsujimoto N., Chen T., Wang S., Ueda Y., Dyson N., Li E., Tsujimoto N., Chen T., Wang S., Ueda Y., Dyson N., Li E., Chen T., Wang S., Ueda Y., Dyson N., Li E., Wang S., Ueda Y., Dyson N., Li E., Ueda Y., Dyson N., Li E., Dyson N., Li E., Li E. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Burger M., Burton J., Cox T.V., Davies R., Down T.A., Burton J., Cox T.V., Davies R., Down T.A., Cox T.V., Davies R., Down T.A., Davies R., Down T.A., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden A., Gaudet F., Waghmare A., Jaenisch R., Gaudet F., Waghmare A., Jaenisch R., Waghmare A., Jaenisch R., Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P.A., Liang G., Aparicio A., Jones P.A., Aparicio A., Jones P.A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Engler P., Storb U., Storb U. Hypomethylation is necessary but not sufficient for V(D)J recombination within a transgenic substrate. Mol. Immunol. 1999;36:1169–1173. doi: 10.1016/s0161-5890(99)00124-8. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Tycko B., Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Herman J.G., Baylin S.B., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Jin W., Yun C., Hobbie A., Martin M.J., Sorensen P.H., Kim S.J., Yun C., Hobbie A., Martin M.J., Sorensen P.H., Kim S.J., Hobbie A., Martin M.J., Sorensen P.H., Kim S.J., Martin M.J., Sorensen P.H., Kim S.J., Sorensen P.H., Kim S.J., Kim S.J. Cellular transformation and activation of the phosphoinositide-3-kinase-Akt cascade by the ETV6-NTRK3 chimeric tyrosine kinase requires c-Src. Cancer Res. 2007;67:3192–3200. doi: 10.1158/0008-5472.CAN-06-3526. [DOI] [PubMed] [Google Scholar]
- Maraj B.H., Markham A.F., Markham A.F. Prostate-specific membrane antigen (FOLH1): Recent advances in characterising this putative prostate cancer gene. Prostate Cancer Prostatic Dis. 1999;2:180–185. doi: 10.1038/sj.pcan.4500325. [DOI] [PubMed] [Google Scholar]
- Mulholland P.J., Fiegler H., Mazzanti C., Gorman P., Sasieni P., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Fiegler H., Mazzanti C., Gorman P., Sasieni P., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Mazzanti C., Gorman P., Sasieni P., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Gorman P., Sasieni P., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Sasieni P., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Adams J., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Jones T.A., Babbage J.W., Vatcheva R., Ichimura K., Babbage J.W., Vatcheva R., Ichimura K., Vatcheva R., Ichimura K., Ichimura K., et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–791. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Chowdhary B.P., Galibert F., Gatzke L., Galibert F., Gatzke L., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Prima V., Gore L., Caires A., Boomer T., Yoshinari M., Imaizumi M., Varella-Garcia M., Hunger S.P., Gore L., Caires A., Boomer T., Yoshinari M., Imaizumi M., Varella-Garcia M., Hunger S.P., Caires A., Boomer T., Yoshinari M., Imaizumi M., Varella-Garcia M., Hunger S.P., Boomer T., Yoshinari M., Imaizumi M., Varella-Garcia M., Hunger S.P., Yoshinari M., Imaizumi M., Varella-Garcia M., Hunger S.P., Imaizumi M., Varella-Garcia M., Hunger S.P., Varella-Garcia M., Hunger S.P., Hunger S.P. Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19)(q23;p13.3) in acute lymphoblastic leukemia. Leukemia. 2005;19:806–813. doi: 10.1038/sj.leu.2403684. [DOI] [PubMed] [Google Scholar]
- Ramsey H., Zhang D.E., Richkind K., Burcoglu-O’Ral A., Hromas R., Zhang D.E., Richkind K., Burcoglu-O’Ral A., Hromas R., Richkind K., Burcoglu-O’Ral A., Hromas R., Burcoglu-O’Ral A., Hromas R., Hromas R. Fusion of AML1/Runx1 to copine VIII, a novel member of the copine family, in an aggressive acute myelogenous leukemia with t(12;21) translocation. Leukemia. 2003;17:1665–1666. doi: 10.1038/sj.leu.2403048. [DOI] [PubMed] [Google Scholar]
- Ricketts S.L., Garcia N.F., Betz B.L., Coleman W.B., Garcia N.F., Betz B.L., Coleman W.B., Betz B.L., Coleman W.B., Coleman W.B. Identification of candidate liver tumor suppressor genes from human 11p11.2-p12. Genes Chromosomes Cancer. 2002;33:47–59. doi: 10.1002/gcc.1210. [DOI] [PubMed] [Google Scholar]
- Robertson K.D., Jones P.A., Jones P.A. DNA methylation: Past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A., Castresana J., Robinson T.J., Castresana J., Robinson T.J., Robinson T.J. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006;7:R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Hampton G.M., Frierson H.F., Moskaluk C.A., Hampton G.M., Frierson H.F., Moskaluk C.A., Frierson H.F., Moskaluk C.A., Moskaluk C.A. Mapping of candidate tumor suppressor genes on chromosome 12 in adenoid cystic carcinoma. Lab. Invest. 2005;85:1076–1085. doi: 10.1038/labinvest.3700314. [DOI] [PubMed] [Google Scholar]
- Schaefer C.B., Ooi S.K., Bestor T.H., Bourc'his D., Ooi S.K., Bestor T.H., Bourc'his D., Bestor T.H., Bourc'his D., Bourc'his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- Scherer S.W., Cheung J., MacDonald J.R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Cheung J., MacDonald J.R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., MacDonald J.R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Parker-Katiraee L., Skaug J., Khaja R., Skaug J., Khaja R., Khaja R., et al. Human chromosome 7: DNA sequence and biology. Science. 2003;300:767–772. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler L., Roig A., Mahe M.F., Laurent P., Hayes H., Rodolphe F., Cribiu E.P., Roig A., Mahe M.F., Laurent P., Hayes H., Rodolphe F., Cribiu E.P., Mahe M.F., Laurent P., Hayes H., Rodolphe F., Cribiu E.P., Laurent P., Hayes H., Rodolphe F., Cribiu E.P., Hayes H., Rodolphe F., Cribiu E.P., Rodolphe F., Cribiu E.P., Cribiu E.P. High-resolution comparative mapping among man, cattle and mouse suggests a role for repeat sequences in mammalian genome evolution. BMC Genomics. 2006;7:194. doi: 10.1186/1471-2164-7-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M.K., Kim C.G., Kitano T., Ferrell R.E., Kohara Y., Saitou N., Kim C.G., Kitano T., Ferrell R.E., Kohara Y., Saitou N., Kitano T., Ferrell R.E., Kohara Y., Saitou N., Ferrell R.E., Kohara Y., Saitou N., Kohara Y., Saitou N., Saitou N. Nucleotide sequence comparison of a chromosome rearrangement on human chromosome 12 and the corresponding ape chromosomes. Cytogenet. Genome Res. 2005;108:83–90. doi: 10.1159/000080805. [DOI] [PubMed] [Google Scholar]
- Stevens-Kroef M.J., Schoenmakers E.F., van Kraaij M., Huys E., Vermeulen S., van der Reijden B., van Kessel A.G., Schoenmakers E.F., van Kraaij M., Huys E., Vermeulen S., van der Reijden B., van Kessel A.G., van Kraaij M., Huys E., Vermeulen S., van der Reijden B., van Kessel A.G., Huys E., Vermeulen S., van der Reijden B., van Kessel A.G., Vermeulen S., van der Reijden B., van Kessel A.G., van der Reijden B., van Kessel A.G., van Kessel A.G. Identification of truncated RUNX1 and RUNX1-PRDM16 fusion transcripts in a case of t(1;21)(p36;q22)-positive therapy-related AML. Leukemia. 2006;20:1187–1189. doi: 10.1038/sj.leu.2404210. [DOI] [PubMed] [Google Scholar]
- Stransky N., Vallot C., Reyal F., Bernard-Pierrot I., de Medina S.G., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., Vallot C., Reyal F., Bernard-Pierrot I., de Medina S.G., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., Reyal F., Bernard-Pierrot I., de Medina S.G., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., Bernard-Pierrot I., de Medina S.G., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., de Medina S.G., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., de Rycke Y., Elvin P., Cassidy A., Spraggon C., Elvin P., Cassidy A., Spraggon C., Cassidy A., Spraggon C., Spraggon C., et al. Regional copy number-independent deregulation of transcription in cancer. Nat. Genet. 2006;38:1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., Bestor T.H., Chaillet J.R., Bestor T.H., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Whitelaw N.C., Whitelaw E., Whitelaw E. How lifetimes shape epigenotype within and across generations. Hum. Mol. Genet. 2006;15:R131–R137. doi: 10.1093/hmg/ddl200. [DOI] [PubMed] [Google Scholar]
- Wilson A.S., Power B.E., Molloy P.L., Power B.E., Molloy P.L., Molloy P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Howell R.D., Alfonso D.T., Yu J., Kong L., Wittig J.C., Liu C.J., Howell R.D., Alfonso D.T., Yu J., Kong L., Wittig J.C., Liu C.J., Alfonso D.T., Yu J., Kong L., Wittig J.C., Liu C.J., Yu J., Kong L., Wittig J.C., Liu C.J., Kong L., Wittig J.C., Liu C.J., Wittig J.C., Liu C.J., Liu C.J. IFI16 inhibits tumorigenicity and cell proliferation of bone and cartilage tumor cells. Front. Biosci. 2007;12:4855–4863. doi: 10.2741/2433. [DOI] [PubMed] [Google Scholar]
- Zijlstra A., Quigley J.P., Quigley J.P. The DARC side of metastasis: Shining a light on KAI1-mediated metastasis suppression in the vascular tunnel. Cancer Cell. 2006;10:177–178. doi: 10.1016/j.ccr.2006.08.012. [DOI] [PubMed] [Google Scholar]