Abstract
We have developed an optimized array-based approach for customizable allele-specific gene expression (ASE) analysis. The central features of the approach are the ability to select SNPs at will for detection, and the absence of need to PCR amplify the target. A surprisingly long probe length (39–49 nt) was needed for allelic discrimination. Reconstitution experiments demonstrate linearity of ASE over a broad range. Using this approach, we have discovered at least two novel imprinted genes, NLRP2, which encodes a member of the inflammasome, and OSBPL1A, which encodes a presumed oxysterol-binding protein, were both preferentially expressed from the maternal allele. In contrast, ERAP2, which encodes an aminopeptidase, did not show preferential parent-of-origin expression, but rather, cis-acting nonimprinted differential allelic control. The approach is scalable to the whole genome and can be used for discovery of functional epigenetic modifications in patient samples.
The ability to perform genome-wide single nucleotide polymorphism (SNP) analysis has made possible genome-wide association studies for the identification of common disease variants. Whole-genome studies of the epigenome, or nonsequence-based information inherited during cell division, has lagged behind. Part of the reason is the diverse nature of epigenetic control elements, such as DNA methylation and multiple chromatin modifications. Standard array-based allele-indiscriminate gene expression analysis may reveal epigenetic changes at individual genes, or may simply reflect dynamic changes in gene expression mediated by trans-acting regulatory components such as transcription factors. The ability to discriminate allele-specific expression (ASE) of the two alleles of genes can reveal changes in epigenetic control, since the two alleles are affected by the same transcription factors, yet would differ in cis-acting control elements.
Two previous studies examined ASE utilizing a 25-nt microarray (Lo et al. 2003; Pant et al. 2006). Both studies required PCR amplification of the target. The first study analyzed ASE from 602 SNPs utilizing the Affymetrix-defined adaptor-PCR approach to reduce the complexity of the genome, i.e., by digesting with a restriction enzyme, adding adaptors, and PCR-amplifying (Lo et al. 2003). In the other, 1983 heterozygous SNPs were examined using PCR amplification of each target sequence (Pant et al. 2006), using the criteria that SNPs must be at a single location in the human genome and at least 25 bp away from any exon/intron boundary. Both studies showed a surprisingly high frequency of ASE, affecting 54% or 53% of genes, respectively. Another recent report found that 300 out of 4000 genes (∼8%) acquired stochastic monoallelic expression on subcloning lymphoblastoid cell lines (Gimelbrant et al. 2007). This study also specifically excluded genes of interest to us, namely imprinted genes or those showing cis-regulatory allelic imbalance.
Here, we have developed a general array-based strategy for ASE analysis for the purpose of epigenetic target discovery that allows user selection of the SNPs to be examined, and obviates the need for PCR amplification of the target, avoiding potential PCR sequence-specific bias, as well as the cost and restriction on SNPs that PCR imposes. We have validated the approach using reconstitution experiments, allelotyping of cell lines of known genotyping, and pyrosequencing analysis of novel targets, and we have discovered at least two new imprinted genes.
Results
Optimization of feature design for allele-specific discrimination
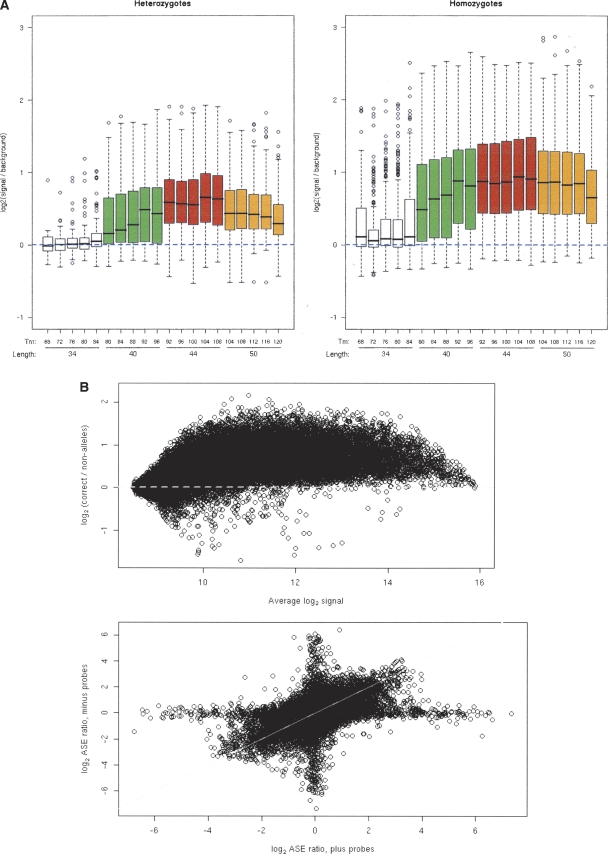
Stringent hybridization of longer oligonucleotides generates specific and sensitive gene-expression analysis, overcoming the cross-hybridization potential of short oligonucleotides. Long oligonucleotides, however, tend to display a muted response to single-base mismatches, which is a requirement for allele discrimination as shown in Figure 1A and Wong et al. (2004). To identify the optimal length and Tm for ASE, we created a single-array design that contained 20 probe sets (five Tm × four lengths) designed to assess ASE of 2194 SNPs in gene coding regions. Each probe set varied the minimum and maximum probe length, as well as the target Tm of each oligonucleotide. Probe lengths ranged from 29 to 55 bases, and relative melting temperatures of 68°C to 120°C (Fig. 1A). This algorithm is not only designed to maintain an approximately equal Tm for all array probes, but also balances the Tm of each probe in either side of the mismatch. This places the mismatch at the thermodynamic center of the oligonucleotide where it is most disruptive, and the approach is preferable to a simple sliding window. To improve hybridization kinetics we hybridized this optimization array design using a small volume hybridization chamber (45 μL) with active mixing. To determine the optimum probe length range and Tm, we performed experiments on three cell lines from CEPH individuals with available genotype from the HapMap project using an array that was customized for exactly this family. In Figure 1A, we display box plots of the log2 ratio of a true allele to the average of the two nonalleles. The left panel shows, for heterozygotes, the maximum of the two true alleles versus the average of the two nonalleles. The right panel is for homozygotes, showing the true allele versus the average of the two nonalleles. These data show that we can identify the expressed alleles and so may discriminate heterozygotes from homozygotes. Most importantly, the highest signal-to-noise ratio (i.e., the best data) comes from the longest features (39–49 nt), which are significantly longer than the 25-nt features used in previous studies (Lo et al. 2003; Pant et al. 2006). Long feature lengths are easily achieved by the NimbleGen maskless array synthesis approach, which allows the construction of long-oligo arrays at essentially no added cost. Melting temperature did not appear to make a difference for the majority of different feature lengths. However, the highest melting temperature had detrimental effects on the longest features.
Figure 1.
Optimization and validation of an array-based hybridization approach to allele-specific expression (ASE). (A) Optimization of feature design. (Left) For all cases in which a CEPH sample was heterozygous at a SNP, shown are the log2 ratios of the average of the two alleles at the SNP to the average of the two nonalleles, as a function of the length and Tm of the feature on the array. Probe lengths ranged from 29 bases to 55 bases (white = 29–39 nt, green = 35–45 nt, red = 40–49 nt, yellow = 45–55 nt), and relative melting temperatures of 68°C to 120°C (melting temperature increased from left to right within each color; see equation in design of array features in the Methods). (Right) For all cases in which a CEPH sample was homozygous at a SNP, shown are the log2 ratios of the allele present to the average of the two nonalleles, as a function of the length and Tm of the feature on the array. For both homozygotes and heterozygotes, the highest signal-to-noise was achieved by probe lengths of 40–49 nt. (B) Validation by allelotyping known alleles from cDNA of heterozygotes for a given SNP. (Top) The difference between the average log2 signal for the two alleles at the SNP and the average log2 signal for the two nonalleles, with results for all probes for a SNP combined. (Bottom) The estimated log2 ASE ratio at each SNP, as measured from the probes on the “plus” strand, against that measured from the probes on the “minus” strand.
Proof of principle detection of expressed SNPs in heterozygotes
Our first method of validating ASE was to design a customized array based on known SNPs within a CEPH kindred (family 1341), in which the four grandparents and the two parents of the family have available genotype through HapMap. Figure 1B (top) represents MA plots for all heterozygotes: the Y-axis is the log2 ASE ratio (for an expressed allele vs. the background, control alleles), and the X-axis is the average log2 intensity. The ASE ratio is therefore the log2 ratio of the two bases that we know the individual carries, to the average of the two bases we know the individual does not carry (i.e., nonallele bases). The multiple probes for a particular SNP are averaged. These results demonstrate our ability to measure the expression of the specific alleles at a SNP, as the majority of the measurements are well above 0, particularly for genes of moderate to high expression (points with high intensity, on the right side of each figure), indicating that we are detecting the correct allele in the vast majority of cases, and the ASE estimates are largely concordant, except for cases in which one or the other strand showed no skewing in expression.
Our labeling approach utilizes random priming, which labels both strands. Separate measures were obtained using the probes from the forward (“plus”) DNA strand for a SNP and using the probes from the reverse (“minus”) strand. This gives more precise information for a single SNP. When we compared the measurement of the two strands for each allele present in DNA, by measuring the log2 ASE ratio in heterozygotes (that is, the ratio of the intensities for the two true alleles), the two measures of the log2 ASE for individual SNPs showed great consistency (Fig. 1B, bottom). The cloud of points centered at (0.0) corresponds to SNPs with no difference in the expression of the two alleles. Some SNPs in these CEPH cell lines show dramatic differences in the level of expression of the two alleles, and in these cases, the log2 ASE ratio from the plus probes is generally similar to that from the minus probes, suggesting that we are indeed measuring true allele-specific silencing.
Validation by quantitative allele-specific expression reconstitution
In order to determine whether ASE detection by hybridization to oligonucleotide arrays is quantitative, we created a set of oligonucleotide mixtures. For each of 65 SNPs (covering 10 genes), we obtained oligonucleotides corresponding to the two alleles and mixed them together in different proportions. For this experiment, the 45-nt oligonucleotides were synthetically biotinylated to allow their detection. To simulate the real situation in the complexity of the target in these experiments, we mixed the oligonucleotide mixture with cDNA obtained from total RNA from a single placental sample. We varied both the ratios of alleles of the synthetic targets, as well as the total concentration, testing 21 conditions (see Supplemental Tables 1, 2). We chose 1 pm to 200 pM concentration, because this range falls within the linear detection range for microarrays (Albert et al. 2003) and encompasses the range used in previous resequencing experiments (Wong et al. 2004).
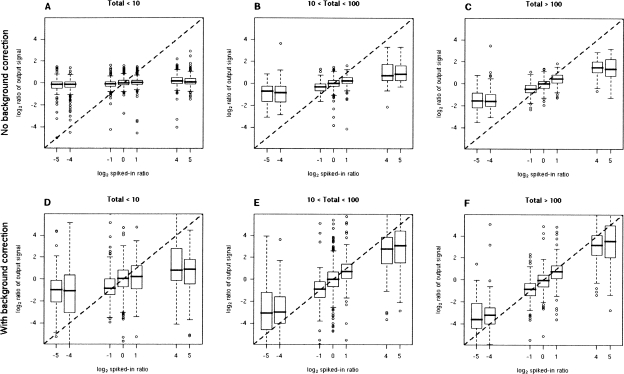
Figure 2 shows box plots of the log2 ratio of the output signals against the log2 spiked-in ratio, as a function of the total amount of spiked-in DNA. A nearly linear relationship is seen, at least in the case that the total amount of DNA > 10 μM, indicating that we are able to recover the ASE ratios from the microarray intensity data. In the upper panels of Figure 2, no background correction was applied, which results in an attenuation of the signal: instead of a ratio like a/b, we display (a + x)/(b + x), where x is background. For a/b > 1, (a + x)/(b + x) < a/b. In the lower panels of Figure 2, we correct for background by subtracting the average intensity of the probes for two nonalleles at each SNP; the background correction results in reduced attenuation of the signal and so more accurate measures of the ASE ratios, though it also results in somewhat greater variability in the estimates.
Figure 2.
Reconstitution experiment measuring output ASE as a function of varying input amounts and allelic ratios. Log2 ratio of observed signal against the log2 ratios spiked-in, as a function of the total amount of DNA (in micromolars) spiked-in. (A–C) Without background for three input concentrations; (D–F) with background correction, with background estimated by the average signal from the probes of the two nonalleles.
Distribution of ASE in the sample set
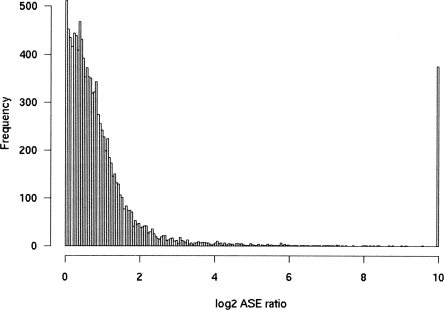
We redesigned the array based on the results from our optimization of probe lengths and temperatures (39–49 nt, average length 40 nt, calculated Tm of 100°C). We then analyzed six CEPH cell lines on an array composed of 12,000 SNPs in 5770 genes. Approximately 50% of genes are expressed in lymphocytes (Cheung et al. 2003), and we had found that the ability to discriminate alleles is improved with higher concentrations of input signal (Fig. 2). Because of this, we chose to focus on probes with log2 signal in the top quartile to obtain an estimate of the level of allele-specific expression in our sample set. Among these SNPs, ∼10% showed an absolute log2 ratio > 2 (i.e., ASE ratio >4 or <1/4, Fig. 3). Inclusion of the top half of signals rather than top quartile increased the fraction of SNPs with ASE ratio above 4%–18%.
Figure 3.
Observed frequency of ASE skewing in six CEPH cell lines. Histogram of the absolute of log2 ASE ratio for SNPs in the top quartile of gene-expression level. Among these, 10% showed an ASE ratio >4.
Parent of origin analysis in a replication set distinguishes two classes of genes with ASE imbalance: Imprinted genes and genes under sequence-dependent cis-regulation
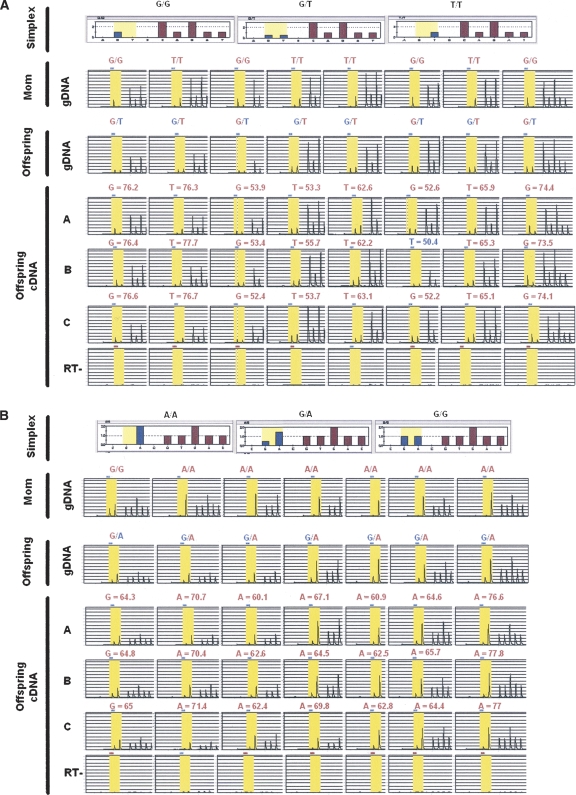
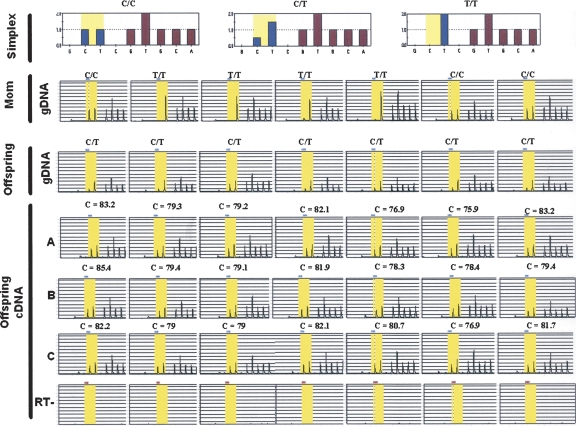
Our goal was to show that an array-based PCR-independent approach could identify known and novel imprinted genes. One of the genes showing allelic skewing with a log2 ratio ≥ 1.5 was PEG10, a known imprinted gene (Supplemental Figs. 1, 2). KCNQ1, another known imprinted gene, also demonstrated a high level of allelic skewing. Two known imprinted genes, GRB10 and HTR2A, were on the array, but not expressed in lymphoblasts, and thus we could not determine the ASE ratio. In addition, several genes on the array not expected to be imprinted did not demonstrate allelic skewing, e.g., TUBGCP6 (Supplemental Fig. 3). In the case of two genes, we were able to demonstrate that they are novel imprinted genes by analyzing allele-specific expression in fetal tissues and maternal samples, which we confirmed by pyrosequencing analysis (Alderborn et al. 2000). The first of these was NLRP2, which encodes a member of the inflammasome (Petrilli et al. 2005). For eight matched maternal (decidua) and offspring pairs in which the maternal sample was homozygous and the offspring sample was heterozygous at the assayed SNP, pyrosequencing analysis showed that ASE of cDNA of all eight cases was preferentially skewed toward the maternal allele (Fig. 4A). Furthermore, NLRP2 also showed allelic skewing in primary tissues, fetal heart, and kidney (Supplemental Fig. 4).
Figure 4.
Parent-of-origin analysis of genes showing ASE in a replicate set. (A) NLRP2 demonstrates preferential expression of the maternal allele in placentas of eight different individuals. (B) OSBPL1A demonstrates preferential expression of the maternal allele in placentas of seven (shown) out of 10 different individuals. In both A and B, the first row demonstrates the simplex files for the three possible genotypes of the pyrosequencing assay. The next two rows demonstrate the pyrosequencing raw data files for the gDNA of the mother (homozygous) and the offspring (heterozygous), respectively. The final rows demonstrate the ASE (in triplicate) from cDNA of the cases and the corresponding RT− control for each assay. In all cases, the maternal allele (red) is preferentially expressed compared with the paternal allele (blue). The columns represent individual sample sets.
The second novel imprinted gene identified in our screen was OSBPL1A, or oxysterol-binding protein-like 1A, which encodes an oxysterol-binding protein, a family involved in a wide range of metabolic processes (Jaworski et al. 2001). In a replicate set of samples, seven out of 10 placental samples demonstrated ASE skewing, in a range from 61% to 77% (Fig. 4B). In all cases with allelic skewing, the more abundantly expressed allele was always of maternal origin (Fig. 4B), indicating that OSBPL1A is imprinted, with variation in stringency in the population.
A third gene showing allelic skewing was ERAP2, encoding a leukocyte-derived aminopeptidase that is a member of the oxytocinase subfamily (Tsujimoto and Hattori 2005). In a replicate set of seven placental samples, pyrosequencing demonstrated strong allelic skewing, but in this case, the skewing was not due to the parental origin but to the C allele itself (Fig. 5). Interestingly, we found that ERAP2 lies within the same linkage disequilibrium group as rs2287988 (Supplemental Fig. 5), a SNP previously demonstrated to show association with expression of ERAP2 (Cheung et al. 2005). Likewise, IL16 showed allelic skewing on the array, and IL16 expression has also been shown to be regulated in cis (Cheung et al. 2005).
Figure 5.
ERAP2 demonstrates preferential expression of the C allele in seven cases, but is not affected by parent of origin. Images are assembled as in Figure 4, with cDNA shown in the bottom four rows. Note that the sequence has TT following the (C/T) SNP. Thus, the C/C allele is 2 × C, followed by 2 × T in the simplex, C/T is 1 × C followed by 3 × T, and T/T is 4 × T.
A fourth gene, SCRN1, showed monoallelic expression (Supplemental Fig. 6). We were not able to determine parent of origin, however, because the gene did not demonstrate allelic skewing in placenta. In addition to these confirmed examples of ASE imbalance, a roughly equal number of sequences on the array did not show ASE imbalance by pyrosequencing analysis. At least five of these were due to HapMap genotyping errors rather than our method. Thus, the SNPs showed monoallelic expression on the array, but they were genomic homozygotes rather than the reported heterozygotes on the HapMap database: COLEC11, LOC144983, HK2, ADRA2B, and ZRANB1. In addition, 10 SNPs showed allelic skewing on the array, but not by pyrosequencing analysis. However, in all of these cases, the mismatched allele was not only present within the genotype of the target gene, but was also present in nonunique sequences throughout the genome. Thus, the false positivity was likely due to cross-hybridization from other targets, rather than problems with allele discrimination at a given target, a problem that can easily be overcome in the next iteration of probe design. An exception to this rule was ERBB3, which showed false positivity on the array, but did not cross-hybridize to other sequences.
In summary, array-based PCR-independent ASE analysis can discover allelic skewing in gene expression on patient samples in a customizable, cost-effective manner, and thus identify novel epigenetic targets for normal development and disease. The sensitivity and specificity of the assay can be improved with larger numbers of samples and probes per gene. However, even in this proof of principle stage, it was able to identify two new imprinted genes, two genes showing cis-regulating genetic variation, and a fifth gene that falls into one of those classes, which could not be determined with the tissue samples currently at hand, as well as five genotyping errors in HapMap.
Discussion
In summary, we have developed an array hybridization-based approach to high-throughput allele-specific gene expression analysis that is customizable, in that one can design any set of SNPs of human or model organism for analysis. While we used the NimbleGen platform for these experiments, there is no reason in principle why other array platforms would not be equally useful if the average feature length of 40 nt can be maintained. In addition, we do not need to reduce the complexity of the target by PCR, thus avoiding amplification bias. While here we performed a proof of principle experiment on 12,000 SNPs on a 285-K feature array, a 2.2-M array will soon be available, and we can reduce the number of probes per SNP by at least half (two of the Tm windows were experimentally found to be unnecessary), allowing an average of eight SNPs per gene scalable to the entire genome.
Using this approach, we found two novel imprinted genes on further studies of ∼20 SNPs, showing ASE imbalance with a log2 ratio ≥ 1.5 in at least one sample, as well as two nonimprinted genes under sequence-dependent cis regulation. The threshold for further screening is arbitrary and will likely change as the technology improves. A simple criterion would be to require an ASE imbalance in a larger number of samples or a larger number of SNPs per gene. In addition, currently, all probes are treated equally, but with a larger data set, we can exclude poorly performing probes, just as is done in conventional gene-expression arrays. Furthermore, in the next generation of arrays, we will exclude probes that cross-hybridize to other targets, e.g., using a fuzzy match algorithm (http://www.nimblegen.com/products/chip/index.html).
A current drawback of our approach is that it does not offer genotyping on the same platform. However, genotyping is not needed for experiments on isogenic tissues, for example, comparing multiple tissues in search of tissue-specific imprinted genes or comparing tumor-normal tissue pairs. Indeed, in principle, this approach could be used to distinguish tumor loci with epigenetic inactivation of single genes, loss of heterozygosity affecting large numbers of contiguous genes, or loss of imprinting with monoallelic expression in normal tissue and biallelic expression in the tumor from which it was derived. It should be noted that methods for genome-wide genotyping of targeted sequences (including the expressed SNPs on which we rely) will be available soon, using array-based targeted region genomic purification platforms under development.
What are the biological implications of the genes identified in this proof of principle experiment? NLRP2 is regulated by interferon and lipopolysaccharides and suppresses activation of I-κB kinases (Bruey et al. 2004). To date, no gene involved primarily in immune response has been proven to be imprinted, despite the fact that one of the principal evolutionary drives to epigenetic modification is thought to be host defense (Bestor et al. 1994). The second gene, OSBPL1A, is a member of a family of sterol sensors (Olkkonen et al. 2006), and thus, could play a role in lipid metabolism. Another key role of epigenetic modification is thought to involve nutrient sensing (Jirtle and Skinner 2007). OSBPL1A is located 20 kb from the imprinted gene IMPACT, but it has been reported not to be imprinted in mouse brain (Okamura et al. 2004). Our data would indicate that the gene is imprinted in humans in at least some tissues. The fact that imprinting was variable is consistent with the idea that genomic regions can be susceptible, but not exclusively imprinted across all tissues and mammalian species.
Finally, the identification of SNP-dependent parent of origin-independent ASE imbalance is a simple way to identify cis-acting regulatory sequences for gene expression without having to perform genome-wide association studies linking SNPs to total levels of individual gene expression. Thus, using genome-wide ASE analysis, one can compare the two SNPs and the two transcripts of the same gene within the same cell.
Methods
Array design
In order to take advantage of known genotypic information, we designed a custom array design based on available genotype from HapMap (International HapMap Consortium 2005). We chose expressed SNPs from 5′ UTR, 3′ UTR, and exons, based on the web-based search function of the HapMap web-browser (Thorisson et al. 2005) (http://hapmart.hapmap.org). We then sorted the available expressed SNPs based on the heterozygosity of family members and selected for this array (385,000 features) the 12,000 SNPs with greatest heterozygosity, and to test Tm of the platform (32 probes/SNP).
Design of array features
Previous resequencing strategies using NimbleGen arrays interrogated SNP positions with intermediate G/C content; GC content <20% or >65%, which typically produced low-quality data (Wong et al. 2004). To overcome this limitation, we have developed an algorithm for resequencing and genotyping array probe selection that optimizes the oligonucleotide length, mismatch position, and melting temperature. The algorithm accepts input from the user to specify the minimum and maximum probe lengths and target Tm for all probes on the array. The target Tm of the probe is then divided in half, and the portion on each side of the mismatch is varied (within specified length parameters) to reach half of the specified target Tm, calculated as follows: Probe Tm = 5 × (Gn + Cn) + 1 × (An + Tn), where Gn is the number of Gs, Cn is the number of Cs, and so on for the other bases in the probe. This is an empirical modification of the Wallace rule (Wallace et al. 1979), made to better reflect surface probe behavior. This results in array probes with varying length, similar melting temperatures, and with mismatches near the thermodynamic center of each oligonucleotide, where they are most destabilizing, rather than the physical center. Initially we chose 1097 SNPs, which were used for an array designed to optimize the probe length and melting temperature. This array tested probe lengths varying from 29 to 39 nt up to 45–55 nt, with melting temperatures calculated using the formula above between 68°C and 120°C. This array was used to determine that the probe set that gave the best balance between sensitivity and allele discrimination contained probes that varied in length from 39 to 49 nt, with a calculated target Tm of 100°C and an average length of 40 nt. We then designed a 380,000 feature array from NimbleGen with features representing 12,000 SNPs in ∼5800 genes for all possible nucleotides (i.e., A, T, C, G) in both orientations with four sliding windows, using the experimentally determined optimal probe length and Tm. Four sliding windows were achieved by varying the location of the SNP within the feature four individual times.
Cell culture
Six lymphoblast cell lines comprising two trios from CEPH family 1341 were ordered from Coriell. Trio 1 is composed of GM06991, GM06993, GM06985; Trio 2 is composed of GM07034, GM07055, and GM07048. The cell lines were cultured at 37°C in RPMI 1640, 2 mM L-glutamine, 15% heat-inactivated fetal bovine serum.
RNA isolation and mRNA enrichment
For array experiments, total cellular RNA was isolated using RNA STAT-60 (TEL-TEST). Genomic DNA (gDNA) was isolated from the interphase of the RNA STAT-60 preparation using the protocol from TRIzol Reagent (Invitrogen). RNA quality was monitored using an Agilent Bioanalyzer (Agilent Technologies, Inc.). We enriched for mRNA using the Straight A's mRNA Isolation System from Novagen (EMD Biosciences). For pyrosequencing validation experiments, matched decidual and fetal sample sets were acquired from the University of Washington Fetal Tissue Bank. For these samples, total cellular RNA was isolated using the RNeasy kit (Qiagen). Total DNA from corresponding samples was isolated using the DNeasy kit (Qiagen).
cDNA synthesis and array hybridization
cDNA was synthesized using the Superscript II double-strand synthesis kit (Invitrogen), according to the manufacturer’s protocol using random hexamers for first-strand cDNA synthesis. cDNA (1 μg) was mixed with 1 O.D. of 5′-Cy3 labeled random nonamer (TriLink Biotechnologies) in 62.5 mM Tris-HCl, 6.25 mM MgCl2, and 0.0875% β-mercaptoethanol, denatured at 98°C for 5 min, chilled on ice, and incubated with 100 U Klenow fragment (NEB) and dNTP mix (6 mM each in TE) for 2 h at 37°C. Reactions were terminated with 0.5 M EDTA (pH 8.0), precipitated with isopropanol, and resuspended in water. A total of 13 μg of labeled cDNA was hybridized to each microarray in 1× NimbleGen hybridization buffer (NimbleGen) in a MAUI hybridization apparatus (Biomicro) in a final volume of 45 μL. Arrays were hybridized overnight at 42°C. The next morning, arrays were washed with nonstringent wash buffer (6× SSPE, 0.01% [v/v] Tween-20) for 2 min, and then twice in stringent wash buffer (100 mM MES, 0.1 M NaCl, 0.01% [v/v] Tween-20) for 5 min, all at 47.5°C. Finally, arrays were washed again in nonstringent wash buffer (min) and rinsed twice for 30 sec in 0.05× SSC. Arrays were spun dry in a custom centrifuge.
Pyrosequencing assays
Quantitative ASE analysis was performed on a PSQ HS96 Pyrosequencer (Biotage) according to the manufacturer’s protocol. All allele-specific expression analysis was performed in triplicate (Supplemental Table 3).
Oligonucleotide reconstitution experiment
We synthesized 130 biotinylated 45-nt oligonucleotides (Integrated DNA technology), which were composed of the two alleles the 65 SNP locations tested. We used a TECAN EVO 75 robot (Tecan Group Ltd.) to make ratios of 1:32, 1:16, 1:2, 1:1, 2:1, 16:1, 32:1 in either low (1–8 μM), medium (10–80 μM), or high (100–300 μM) total mass (Supplemental Tables 1, 2). We ran the oligo reconstitution experiment on an array that was designed to be opposite in orientation of mRNA. The synthetic target, which is at low total mass given the limited number of genes, was then mixed with a cellular cDNA sample to approximate a normal complex target, but by design, the cellular cDNA cannot hybridize to the array.
Statistical analysis
All statistical analyses were performed with R version 2.4.1(Ihaka and Gentleman 1996). Multiple arrays in an experiment were quantile normalized (Bolstad et al. 2003) to reduce the between-array variation. Signals were then transformed to the log2 scale. The results for multiple probes for a particular SNP were combined using median polish (Tukey 1977): this gives a single measure of log2 expression for each of the four possible bases at a SNP. Background correction was performed on the original scale by subtracting the average signal for the two bases not listed in dbSNP from the signal for each of the bases listed in dbSNP, and then transforming back to the log2 scale.
Acknowledgments
This work was supported by NIH grant P50HG003233 (A.P.F.). H.T.B. was supported by grants from the Fulbright and the American-Scandinavian Foundation.
Footnotes
[Supplemental material is available online at www.genome.org. Full allele-specific expression array data is available at www.biostat.wisc.edu/~kbroman/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073254.107.
References
- Albert T.J., Norton J., Ott M., Richmond T., Nuwaysir K., Nuwaysir E.F., Stengele K.P., Green R.D., Norton J., Ott M., Richmond T., Nuwaysir K., Nuwaysir E.F., Stengele K.P., Green R.D., Ott M., Richmond T., Nuwaysir K., Nuwaysir E.F., Stengele K.P., Green R.D., Richmond T., Nuwaysir K., Nuwaysir E.F., Stengele K.P., Green R.D., Nuwaysir K., Nuwaysir E.F., Stengele K.P., Green R.D., Nuwaysir E.F., Stengele K.P., Green R.D., Stengele K.P., Green R.D., Green R.D. Light-directed 5′→3′ synthesis of complex oligonucleotide microarrays. Nucleic Acids Res. 2003;31:e35. doi: 10.1093/nar/gng035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderborn A., Kristofferson A., Hammerling U., Kristofferson A., Hammerling U., Hammerling U. Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res. 2000;10:1249–1258. doi: 10.1101/gr.10.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T.H., Chandler V.L., Feinberg A.P., Chandler V.L., Feinberg A.P., Feinberg A.P. Epigenetic effects in eukaryotic gene expression. Dev. Genet. 1994;15:458–462. doi: 10.1002/dvg.1020150603. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P., Irizarry R.A., Astrand M., Speed T.P., Astrand M., Speed T.P., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bruey J.M., Bruey-Sedano N., Newman R., Chandler S., Stehlik C., Reed J.C., Bruey-Sedano N., Newman R., Chandler S., Stehlik C., Reed J.C., Newman R., Chandler S., Stehlik C., Reed J.C., Chandler S., Stehlik C., Reed J.C., Stehlik C., Reed J.C., Reed J.C. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-κB and caspase-1 activation in macrophages. J. Biol. Chem. 2004;279:51897–51907. doi: 10.1074/jbc.M406741200. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Arcaro M., Jen K.Y., Morley M., Spielman R.S., Jen K.Y., Morley M., Spielman R.S., Morley M., Spielman R.S., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T., Ewens K.G., Weber T.M., Morley M., Burdick J.T., Weber T.M., Morley M., Burdick J.T., Morley M., Burdick J.T., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A., Hutchinson J.N., Thompson B.R., Chess A., Thompson B.R., Chess A., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R., Gentleman R. R: A language for data analysis and graphics. J. Comput. Graphical Statist. 1996;5:299–314. [Google Scholar]
- International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski C.J., Moreira E., Li A., Lee R., Rodriguez I.R., Moreira E., Li A., Lee R., Rodriguez I.R., Li A., Lee R., Rodriguez I.R., Lee R., Rodriguez I.R., Rodriguez I.R. A family of 12 human genes containing oxysterol-binding domains. Genomics. 2001;78:185–196. doi: 10.1006/geno.2001.6663. [DOI] [PubMed] [Google Scholar]
- Jirtle R.L., Skinner M.K., Skinner M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P., Yang H.H., Gere S., Buetow K.H., Lee M.P., Gere S., Buetow K.H., Lee M.P., Buetow K.H., Lee M.P., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Yamada Y., Sakaki Y., Ito T., Yamada Y., Sakaki Y., Ito T., Sakaki Y., Ito T., Ito T. An evolutionary scenario for genomic imprinting of Impact lying between nonimprinted neighbors. DNA Res. 2004;11:381–390. doi: 10.1093/dnares/11.6.381. [DOI] [PubMed] [Google Scholar]
- Olkkonen V.M., Johansson M., Suchanek M., Yan D., Hynynen R., Ehnholm C., Jauhiainen M., Thiele C., Lehto M., Johansson M., Suchanek M., Yan D., Hynynen R., Ehnholm C., Jauhiainen M., Thiele C., Lehto M., Suchanek M., Yan D., Hynynen R., Ehnholm C., Jauhiainen M., Thiele C., Lehto M., Yan D., Hynynen R., Ehnholm C., Jauhiainen M., Thiele C., Lehto M., Hynynen R., Ehnholm C., Jauhiainen M., Thiele C., Lehto M., Ehnholm C., Jauhiainen M., Thiele C., Lehto M., Jauhiainen M., Thiele C., Lehto M., Thiele C., Lehto M., Lehto M. The OSBP-related proteins (ORPs): Global sterol sensors for co-ordination of cellular lipid metabolism, membrane trafficking and signalling processes? Biochem. Soc. Trans. 2006;34:389–391. doi: 10.1042/BST0340389. [DOI] [PubMed] [Google Scholar]
- Pant P.V., Tao H., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A., Tao H., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A., Ballinger D.G., Cox D.R., Frazer K.A., Cox D.R., Frazer K.A., Frazer K.A. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli V., Papin S., Tschopp J., Papin S., Tschopp J., Tschopp J. The inflammasome. Curr. Biol. 2005;15:R581. doi: 10.1016/j.cub.2005.07.049. [DOI] [PubMed] [Google Scholar]
- Thorisson G.A., Smith A.V., Krishnan L., Stein L.D., Smith A.V., Krishnan L., Stein L.D., Krishnan L., Stein L.D., Stein L.D. The International HapMap Project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto M., Hattori A., Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim. Biophys. Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Tukey J. Exploratory data analysis. Addison-Wesley; Reading, MA: 1977. [Google Scholar]
- Wallace R.B., Shaffer J., Murphy R.F., Bonner J., Hirose T., Itakura K., Shaffer J., Murphy R.F., Bonner J., Hirose T., Itakura K., Murphy R.F., Bonner J., Hirose T., Itakura K., Bonner J., Hirose T., Itakura K., Hirose T., Itakura K., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: The effect of single base pair mismatch. Nucleic Acids Res. 1979;6:3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.W., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D., Stanton L.W., Liu E.T., Miller L.D., Liu E.T., Miller L.D., Miller L.D. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]