Abstract
Large copy number variants (CNVs) have been recently found as structural polymorphisms of the human genome of still unknown biological significance. CNVs are significantly enriched in regions with segmental duplications or low-copy repeats (LCRs). Williams-Beuren syndrome (WBS) is a neurodevelopmental disorder caused by a heterozygous deletion of contiguous genes at 7q11.23 mediated by nonallelic homologous recombination (NAHR) between large flanking LCRs and facilitated by a structural variant of the region, a ∼2-Mb paracentric inversion present in 20%–25% of WBS-transmitting progenitors. We now report that eight out of 180 (4.44%) WBS-transmitting progenitors are carriers of a CNV, displaying a chromosome with large deletion of LCRs. The prevalence of this CNV among control individuals and non-transmitting progenitors is much lower (1%, n = 600), thus indicating that it is a predisposing factor for the WBS deletion (odds ratio 4.6-fold, P = 0.002). LCR duplications were found in 2.22% of WBS-transmitting progenitors but also in 1.16% of controls, which implies a non–statistically significant increase in WBS-transmitting progenitors. We have characterized the organization and breakpoints of these CNVs, encompassing ∼100–300 kb of genomic DNA and containing several pseudogenes but no functional genes. Additional structural variants of the region have also been defined, all generated by NAHR between different blocks of segmental duplications. Our data further illustrate the highly dynamic structure of regions rich in segmental duplications, such as the WBS locus, and indicate that large CNVs can act as susceptibility alleles for disease-associated genomic rearrangements in the progeny.
Knowledge of the human genome sequence and availability of genome scanning technologies such as microarray-based comparative genomic hybridization, paired-end clone sequencing, computational approaches, and high-throughput single-nucleotide polymorphism genotyping have facilitated detection of previously unrecognized sub-microscopic structural variants of the human genome, including inversions, insertions, and copy number variations (CNVs) which range from 1 kb to >3 Mb. Recent reports have demonstrated the presence of thousands of such sub-microscopic structural variants spanning up to 18% of the human reference DNA sequence (http://projects.tcag.ca/variation/?source=hg18) (Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Tuzun et al. 2005; Conrad et al. 2006; McCarroll et al. 2006; Redon et al. 2006). A high percentage of these variants (25%–50%) are located in close proximity to segmental duplications or low-copy repeat elements (LCRs), large blocks of highly identical sequence (>95%) generated recently during primate evolution (Bailey et al. 2002; Cheung et al. 2003; Zhang et al. 2005). Regions enriched in LCRs are known to show higher genomic instability mediated by nonallelic homologous recombination (NAHR) between the repetitive LCR elements (Inoue and Lupski 2002), which may also explain their association with large structural variation. In fact, locus-specific mutation rates for genomic rearrangements in several regions appear to be up to four orders of magnitude greater than nucleotide-specific rates for base substitutions (Lupski 2007).
Structural variants, jointly with single-nucleotide and other small sequence changes, are thought to be the major contributors to genetic variation among individuals (Feuk et al. 2006). Although structural variants in some genomic regions have no obvious phenotypic consequence, others may have a significant influence either alone or in combination with other genetic or environmental factors. Contribution of CNVs to phenotypic variation and/or disease is thought to occur mainly through altering gene dosage, disrupting genes, perturbing gene expression regulation even at long distances, or unmasking recessive mutations on the other allele (Gonzalez et al. 2005). In addition, normal structural genetic variation could increase the risk of secondary rearrangements associated with disease, either somatic, causing a sporadic disease in the individual, or in the germline, predisposing to disease in the offspring (Eichler et al. 2007; Lupski 2007).
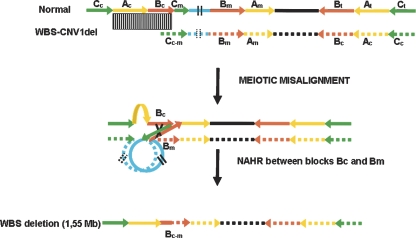
The Williams-Beuren syndrome (WBS) locus is a model region of high genome plasticity. WBS is a rare genetic neurodevelopmental disorder [OMIM 194,050] caused by a heterozygous deletion of multiple contiguous genes at chromosome band 7q11.23 (Pérez Jurado et al. 1996; Morris and Mervis 2000). Although it can be transmitted in an autosomal-dominant fashion, the majority of the cases occur sporadically with an estimated prevalence of the disease as high as 1/7500 newborns (Stromme et al. 2002). Two-thirds of the deletions arise from crossover events between both chromosome 7 homologs during meiosis, while intrachromosomal rearrangements occur in one-third of cases (Urban et al. 1996; Baumer et al. 1998; Bayés et al. 2003). The genomic architecture of the 7q11.23 region is complex with a single-copy interval flanked by three blocks (A, B, and C) of highly homologous (∼99.5%) LCRs with a modular structure shaped during genome evolution (Fig. 1) (Peoples et al. 2000; Valero et al. 2000; Antonell et al. 2005). NAHR between misaligned LCRs is the mechanism that mediates the recurrent WBS deletions (Bayés et al. 2003) which are almost identical size in most cases. Fine mapping of deletion ends has revealed that most WBS individuals (89%) present a common deleted interval of 1.55 Mb containing 26–28 genes between the centromeric and medial LCR blocks B, while ∼8% of cases present a larger recurrent deletion of 1.83 Mb between centromeric and medial blocks A (Del Campo et al. 2006), and only ∼2%–3% carry atypical deletions (Korenberg et al. 2000; Pérez Jurado 2003; Howald et al. 2006).
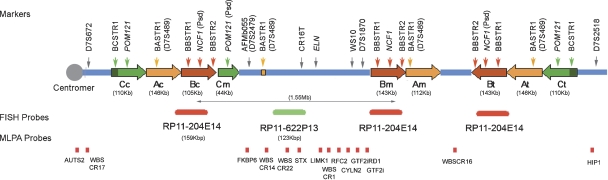
Figure 1.
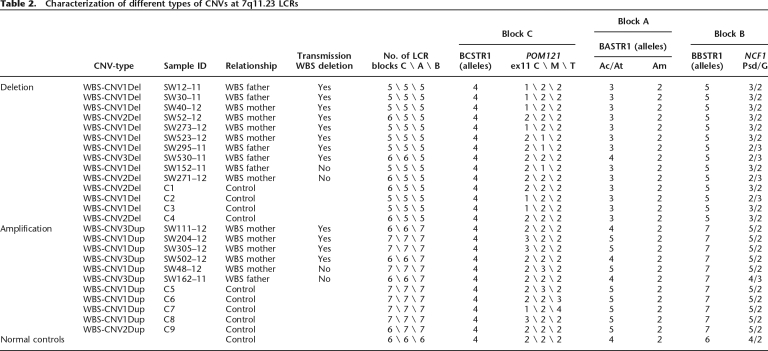
Schematic representation of the genomic structure of the WBS locus at 7q11.23. There are three large segmental duplications or LCRs—centromeric (c), medial (m), and telomeric (t)—each of which is composed of three distinct blocks—A, B and C—represented by colored arrows that indicate their relative orientation with respect to each other. The most common WBS 1.55-Mb deletion arising from NAHR between blocks B is indicated. Note that block Cm is much shorter than blocks Cc and Ct, missing the darker green region that harbors BCSTR1. The locations of the relevant multiple- and single-copy markers used in this study are indicated on top of the scheme, while the MLPA probes and the BACs used as FISH probes are shown at the bottom.
A structural variant consisting on a paracentric ∼2-Mb inversion of the WBS locus has been found with increased frequency in the progenitors transmitting the WBS chromosome (20%–25%) compared to the general population (4%–5%) (Osborne et al. 2001; Bayés et al. 2003). Heterozygosity for this polymorphic chromosomal microinversion in 7q11.23 is thought to lead to abnormal meiotic pairing and therefore an increased susceptibility to unequal recombination causing the WBS deletion. Similar paracentric microinversions have been found as putative susceptibility variants for other recurrent genomic rearrangements, such as the deletions causing Angelman syndrome, Sotos syndrome, and the 17q21 microdeletion syndrome (Gimelli et al. 2003; Koolen et al. 2006; Sharp et al. 2006; Shaw-Smith et al. 2006).
In this report we describe novel genomic rearrangements at 7q11.23 that are present as low-frequency polymorphic variants in the general population and behave as susceptibility factors for the WBS deletion. During our investigations, eight out of 180 (4.44%) progenitors transmitting the WBS chromosome were found to carry large deletions of LCRs at 7q11.23, while four (2.22%) carried a duplication of LCRs. The prevalence of these polymorphic CNVs among control individuals was estimated to be 1% and 1.16% (deletion and duplication, respectively). No phenotype other than the increased risk for secondary germline rearrangements was identified in the CNV carriers. We have characterized those alleles and propose specific models for aberrant chromosome pairing and recombination leading to the polymorphic CNVs and high plasticity of this chromosomal region.
Results
Identification of CNVs of the 7q11.23 LCRs in some WBS parents
Genotyping of short tandem repeat markers (STRs) located at the LCRs in WBS families had long ago suggested that some progenitors were carriers of an abnormal number of copies of these LCR blocks (Pérez Jurado et al. 1996). In order to better identify and fully characterize such putative aberrant alleles, we analyzed all WBS available families (n = 180 trios) with multiple assays surrounding the WBS locus (Fig. 1). All 180 WBS patients had fully characterized de novo 7q11.23 deletions 1.55 Mb (160, 89%), 1.83 Mb (14, 8.3%), or atypical (5, 2.7%) in size. The parental origin of the deleted chromosome was determined in all cases by the lack of parental inheritance at polymorphic loci within the critical region (53% maternal, 47% paternal). The parents for which a meiotic NAHR event occurred in their germ cell, resulting in a deleted chromosome present in the WBS offspring, are hereafter defined as transmitting progenitors.
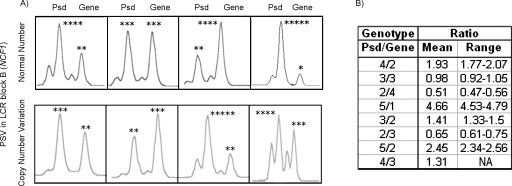
We first analyzed a site-specific paralogous sequence variant (PSV) located in LCR blocks B that detects a GT deletion Δ in the two NCF1 pseudogenes at blocks Bc and Bt with respect to the NCF1 gene at block Bm (Fig. 2). Quantification of this indel PSV had previously been used with high efficiency to map deletion breakpoints in WBS patients, since the ratio of peak heights (pseudogene-to-gene [Psd/Gene] ratio) indicates the relative number of NCF1 pseudogenes and genes within an individual’s genome (Del Campo et al. 2006). Most individuals displayed ratios of 4:2, compatible with four pseudogene and two gene copies (one gene and two pseudogenes per allele). However, some variations not affecting the total number of block copies, generated by gene conversion have been described (Del Campo et al. 2006). We detected a total of 16 parents of WBS patients with abnormal Psd/Gene ratios: 12 WBS transmitters and four non-transmitters (Table 1). Eight transmitting parents (4.44%) and two non-transmitters (1.11%) displayed values in the same range as some WBS patients, either 3:2 or 2:3, indicating a deletion of one NCF1 pseudogene-type copy. Four transmitting parents (2.22%) and two non-transmitters (1.11%) displayed values compatible with the gain of an additional NCF1 copy, either pseudogene-type (5:2) or gene-type (4:3). Representative results of the genotypes at this PSV are shown in Figure 2. No overlapping values were obtained for any of the predicted genotypes.
Figure 2.
Representative genotypes at the indel-type PSV of exon 2 of NCF1 in individuals with different pseudogene/gene copy numbers. (A) Each panel includes a genotype with two peaks corresponding to the PCR amplicons of the NCF1 pseudogene (Psd, ΔGT) and gene (GT) copies from several individuals. The predicted number of NCF1 pseudogene and gene copies is shown with asterisks on top of each peak. Note that some variability of the relative ratios without affecting the total number of copies (upper panel) is observed among individuals in the normal population likely generated by gene conversion between the gene and pseudogenes. The lower panel shows the genotypes of individuals with either deletion-type or duplication-type CNVs affecting block B of the WBS LCRs. (B) The actual Psd/Gene peak ratio values (mean and range) obtained for each predicted genotype.
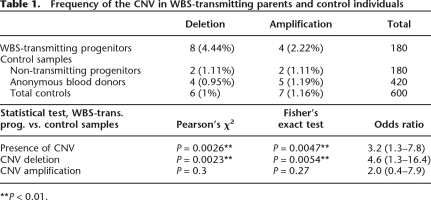
Table 1.
Frequency of the CNV in WBS-transmitting parents and control individuals
**P < 0.01.
In order to determine the population frequencies of these CNVs affecting block B, we analyzed the NCF1 PSV in additional 420 population individuals. We found four individuals with dosage values indicative of deletion (five loci instead of six) and five with dosage values indicative of amplification (seven loci). Including non-transmitting WBS parents in the normal control group, the population frequencies for the deletion and duplication CNVs were 1% (6/600) and 1.16% (7/600), respectively (Table 1). Statistical analysis using χ2 and Fisher’s exact tests showed that the prevalence of the deletion-type CNV was significantly increased among WBS-transmitting parents (P = 0.0023/P = 0.0054), while a slight increase in the prevalence of the duplication-type CNV was not significant (P = 0.3/P = 0.27) (Table 1).
We analyzed two additional block B loci to verify the previous findings and define whether the entire block B was included in the CNV in those samples. Genotyping of the 180 WBS trios and additional 420 controls at the multiple-copy microsatellites BBSTR1 (Fig. 3A) and BBSTR2 ratified the presence of either five or seven alleles in the same samples defined by the NCF1-PSV (Table 2; Supplemental Table 1). The remaining parental and control samples displayed the expected normal number of six block B alleles, while WBS patients, as expected, displayed either five (1.55-Mb deletion) or four alleles (1.83-Mb deletion). Therefore, at least the fragment between BBSTR1 and BBSTR2 (∼70 kb), if not the entire block B (100–140 kb), is included in all CNVs found.
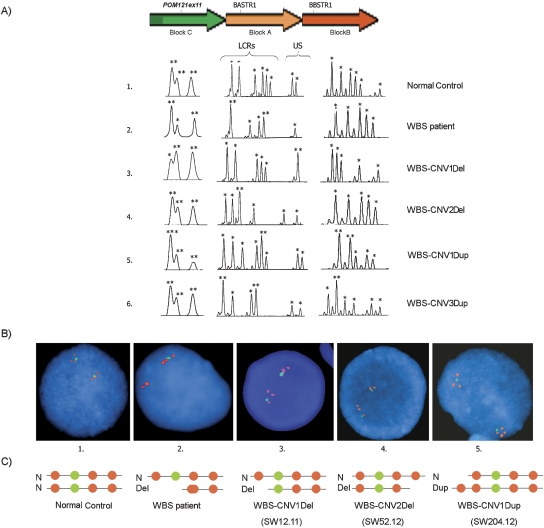
Figure 3.
Characterization of the 7q11.23 CNVs by quantitative PCR-based methods and FISH. (A) Representative genotypes at the multiple-copy PSV (indel at exon 11 of POM121, block C) and microsatellites BASTR1 (block A) and BBSTR1 (block B) in individuals with different CNVs. Individual 1 is a normal control, carrying six alleles per LCR block and two alleles at the WBS region. BASTR1 shows eight alleles because it recognizes four loci, one deleted in WBS patients (D7S489B), and three located in blocks A. BBSTR1 recognizes three loci, one from each block B. Individual 2 is a WBS patient with the most common 1.55-Mb deletion that leads to the loss of one block B and the entire single-copy region in between. Individuals 3 and 4 carry deletion-type CNVs resulting in deletion of the centromeric-type blocks C, A, and B (3, WBS-CNV1Del) or of only the telomeric-type blocks A and B (4, WBS-CNV2Del). Individuals 5 and 6 carry duplication-type CNVs including all three blocks sequences (5, WBS-CNV1Dup) or just block B (6, WBS-CNV3Dup). Asterisks over each peak indicate the number of alleles, as predicted by dosage analysis (US, unique sequence). The predicted number of alleles is indicated by asterisks on top of each peak. (B) Dual-color interphase FISH in nuclei from the same individuals 1–5 with BACs RP11–204E14 (BZ724405) (block B), and RP11–622P13 (AC073846) (WBS region), giving red and green signals, respectively. Individual 1 shows the normal pattern in both chromosomes. Individual 2 shows a deletion of the green probe and a red signal in one chromosome due to a deletion mediated by inter-block B recombination. In individual 3, a red spot in one chromosome is missing due to deletion of block Bc, while in individual 4 the missing signal corresponds to a block B located at telomeric side. Individual 5 shows an extra signal in one chromosome corresponding to an additional block B present at the centromeric side. (C) Schematic representation of the hypothetical structure of the region as shown by FISH signals.
Table 2.
Characterization of different types of CNVs at 7q11.23 LCRs
Characterization of different types of CNVs
To further define the extension of the CNVs, we analyzed additional loci located within the LCRs (blocks A and C) as well as in the surrounding single copy regions by using microsatellites, PSVs and multiple ligation probe amplification (MLPA) (Fig. 1).
The multiple-copy microsatellite BASTR1 (D7S489) recognizes four loci: one within the WBS commonly deleted region (allele range, 166–180 bp), one in block Am (allele range, 136–144 bp), and two in blocks Ac and At (allele range, 144–160 bp) (Pérez Jurado et al. 1996; Robinson et al. 1996). In 13/14 individuals with the block B deletion CNV (Table 2), only three alleles in the 144- to 160-bp range were detected, indicating that they also missed one copy of this part of either block Ac or block At. In 8/11 individuals with gain of one block B, a gain of one block A allele in the 144- to 160-bp range was also observed. All individuals independently of the type of CNV presented the normal two alleles in the size range of WBS commonly deleted region (Fig. 3A). In addition, all single-copy loci analyzed (STR and MLPA probes) surrounding the LCRs, either proximal, within, or distal to the WBS-critical region, also showed normal biallelic or diploid dosage in all cases (Supplemental Table 1).
We then performed a MegaBLAST alignment of the entire length of all three block C paralog sequences (∼110 kb for Cc and Ct, 44 kb for Cm) and defined microsatellites and site-specific PSVs that could be genotyped either directly or following restriction enzyme digestion. The microsatellite BCSTR1 recognizes two loci located at the beginning of blocks Cc and Ct but absent in block Cm (Fig. 1). All individuals analyzed revealed a normal pattern with four alleles at BCSTR1 (Table 2), implicating that this portion of the blocks C was not included in the CNVs found. An indel-type PSV located on exon 11 of the POM121 genes distinguishes a 3-bp deletion (block Cc) and a 6-bp insertion (block Ct) with respect to block Cm, then allowing the relative quantification of copy number at each of the Cc, Cm, and Ct loci in a single assay (Fig. 3A). The Cc/Cm/Ct ratios obtained in most control individuals corresponded to the estimated 2:2:2 copies; however, some variations not affecting the total number of block copies have been detected. All tested WBS patients with 1.55 or 1.83 deletions displayed a relative reduction of the middle peak compatible with the expected 2:1:2 ratio due to deletion of Cm block sequences. Nine of the 14 individuals with deletion-type CNV showed ratios compatible with the loss of a C block: either Cc, with a 1:2:2 ratio, or Cm, with a 2:1:2 ratio. However, five individuals with a deletion of block B sequences showed a normal 2:2:2 ratio, indicating that this locus of block C was not affected by their CNV. In a similar manner, 7/11 individuals with duplication-type CNV at block B displayed ratios compatible with a gain of block C sequences, either Cc or Cm type, while four out of the 11 individuals showed normal 2:2:2 ratios, indicating a normal number of blocks C. Figure 3A shows representative genotypes at three loci, one per LCR block, of individuals with the different variants found, while Table 2 contains the relevant genotypic data set in all cases (full data set can be seen in the Supplementary Table 1).
To confirm the number of LCRs and define the chromosomal organization of these CNVs, we performed dual-color interphase FISH analysis in the available samples. We combined a single-copy probe from the WBS region (RP11–622E21, in green) with a probe for the block B LCR copies (RP11–204E11, in red) (Fig. 3B,C). Control samples displayed the expected disposition of signals per chromosome, with one LCR block on the centromeric side (Bc), then the single-copy probe, and two additional LCR blocks (Bm and Bt). WBS individuals with a 1.55-Mb deletion showed one normal chromosome and the other with only two red signals corresponding to blocks Bt and the recombinant Bc–Bm. In the WBS-transmitting father SW12.11, we found the normal disposition of signals in one chromosome while the signal corresponding to block Bc was missing on the other chromosome, confirming the LCR deletion at the centromeric side (Fig. 3B,C, sample 3). In sample SW52.12, the signal corresponding to block Bt was missing on one chromosome, indicating a deletion on the telomeric side. Out of the duplication-type CNVs, only sample SW204.12 was available for molecular cytogenetic analysis. We observed four block B signals on the variant chromosome, two at either side of the green signal, confirming a duplication of LCR blocks on the centromeric side.
Chromosomal organization and predicted mutational mechanism of the variant regions
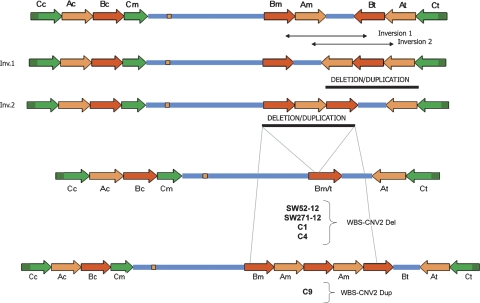
Therefore, at least three types of rearrangements were defined by quantitative analysis of the different LCR block loci and FISH (Table 2): (1) a deletion of ∼300 kb including all three blocks (WBS-CNV1Del, nine cases) and the reciprocal duplication (WBS-CNV1Dup, seven cases) affecting centromeric-type sequences; (2) a deletion of ∼250 kb (WBS-CNV2Del, four cases) and the reciprocal duplication (WBS-CNV2Dup, one case) including just blocks A and B, most likely at the telomeric side; and (3) a deletion (WBS-CNV3Del, one case) or duplication (WBS-CNV3Dup, three cases) of only block B sequences, at least from BBSTR1 to BBSTR2 (∼70 kb).
Based on the known genomic structure of the region, we predicted that the deletion and duplication CNV affecting the three LCR blocks (WBS-CNV1Del and WBS-CNV1Dup) should have arisen as reciprocal products of NAHR between misaligned blocks Cc and Cm (Fig. 4). For the other variants identified, predictions were less obvious, although PSV data suggested that the rearrangements most likely included telomeric LCRs. Given the opposite orientation of the medial and telomeric blocks of LCRs, small paracentric inversions in between those LCRs are likely to occur. This architecture might facilitate the generation of deletions and duplications of only two blocks (A and B) by NAHR between the blocks now situated in tandem (Fig. 5), consistent with the WBS-CNV2Del and WBS-CNV2Dup. WBS-CNV3 could have been generated by rearrangements between smaller LCR modules flanking the telomeric block B.
Figure 4.
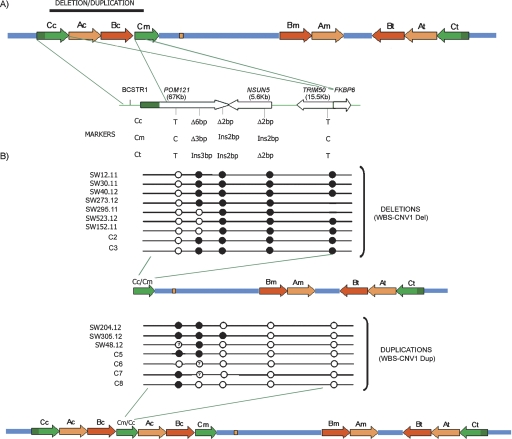
Schematic representation of the WBS-CNV1 deletion and duplication variants and characterization of breakpoints. (A) Scheme of the 7q11.23 genomic region with a more-detailed structure of the entire block C and its gene content represented by arrows that define the transcriptional direction (POM121, NSUN5, TRIM50, and FKBP6). The location and sequence differences of the specific PSVs analyzed in this study are shown below. A complete alignment of blocks Cc and Cm is shown in Supplemental Figure 1. (B) Mapping the region of exchange in block C in the individuals with the ∼300-kb deletion or duplication WBS-CNV1. Each row represents the inferred recombinant block Cc–Cm in the indicated individual, with predicted genotypes at each locus depicted by circles: (open circles) Cc-type; (solid circles) Cm-type. A question mark in the circle indicates that the genotype at that specific locus could not be predicted due to polymorphism/gene conversion. The resulting chromosomes with deletion or duplication CNVs are shown.
Figure 5.
Predicted schematic representation of the WBS-CNV2 deletion and duplication variants and mechanisms of generation. Based on the known architecture of the WBS locus, inversions between the medial and telomeric blocks of LCRs are likely to occur. In fact, such inversions are already present in the regional maps obtained by different assemblies of the human genome (Hillier et al. 2003; Scherer et al. 2003). Chromosomes with such inversion types would present two blocks of the medial and telomeric LCRs, A or B, depending on the inversion breakpoint, arranged in tandem with the other block between. This structure might facilitate the generation of deletions and duplication of only two blocks (excluding block C) by NAHR between the blocks in tandem, consistent with the WBS-CNV2 variants found.
Junction fragment mapping of WBS-CNV1
We decided to further characterize the most common and apparently reciprocal WBS-CNV1Del/Dup, by mapping the Cc–Cm junction fragments in the CNV-bearing recombinant chromosome (Fig. 4). We genotyped five of the identified site-specific PSVs distributed along the block C sequence, quantified the relative number of Cc- and Cm-type sequences, and then inferred the copy type at the recombinant chromosome.
In addition to the indel-type PSV located at exon 11 of the POM121 genes, we analyzed PSVs at exons 6 and 16 of POM121, exon 4 of NSUN5/NSUN5C/NSUN5B, and exon 2 of TRIM50/TRIM74/TRIM73, along with a site-specific nucleotide at the border between blocks C and A. The location and the copy-specific nucleotides at the five PSVs are shown in Figure 4, and the detection method and numerical results are summarized in Supplemental Tables 1 and 2. All recombination breakpoints leading to the deletion and duplication CNVs were predicted to occur within the first half of the misaligned Cc–Cm blocks with no unique hotspot, although most deletion-type CNVs had occurred in the ∼5-kb interval between POMex11 and POMex16 (Fig. 4). In terms of gene copy number, individuals with the WBS-CNV1 displayed a deletion or duplication of the centromeric NSUN5C and TRIM74 genes, the medial copy related with POM121 that is not expressed due to the lack of the first five exons, along with the pseudogenes located in the centromeric blocks B (GTF2IP1, NCF1P1, and GTF2IRD2P1) and A (STAG3L1 and PMS2L). Therefore, none of the ancestral functional copies of the genes present in the WBS LCRs was found to be affected by the CNVs in these individuals.
Haplotype analysis
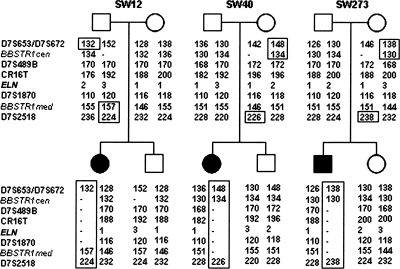
In order to investigate whether the deletions mediated by parental CNVs occurred through either intra- or interchromosomal events and whether the polymorphic CNVs had originated in a common ancestor or through recurrent genomic mutations, we determined the CNV-associated haplotypes by genotyping eight microsatellites distributed along the 7q11.23 region located either centromeric (D7S672/D7S653), within (BBSTR1c/m, D7S489B, CR16T, ELN, D7S1870), or telomeric (D7S2518/D7S2490) to the common WBS deletion (Fig. 6). Genotype analysis in all the studied samples showed that there was not a single haplotype shared by the chromosomes identified with each of the structural variants, consistent with the finding of different breakpoints and suggesting that most regional CNVs occurred through independent mutational events in multiple founders. Haplotype analysis of the unaffected siblings in three families also enabled us to establish the phase of the alleles on the parental chromosomes, showing that all three WBS deletions were the result of interchromosomal NAHR events.
Figure 6.
Haplotype analysis in three WBS families with parental CNVs We have performed the haplotype using polymorphic markers located centromeric (D7S672/653), within (BBSTR1c, D7S489B, CR16T, ELN, D7S1870, BBSTR1m), and telomeric (D7S2158) to the WBS locus. Parental haplotypes were inferred from the siblings’ genotypes, assuming no recombination. The presence of recombination between centromeric and telomeric in all three WBS cases indicates that the deletions arose as a result of interchromosomal meiotic NAHR.
Lack of phenotypic consequences of the CNVs
Family history and medical records were available from all the WBS parents with the CNVs, and five of them underwent a formal medical evaluation including a more detailed medical history and physical exam. No significant medical disorder or family history of disease susceptibility was prevalent among the CNV carriers. Medical records were not available from the other population individuals carrying CNVs.
Deletion-type CNVs at 7q11.23 predispose to unequal crossing-over, leading to WBS deletion
The significantly higher frequency of deletion-type CNVs at 7q11.23 among WBS-transmitter parents supports the hypothesis that heterozygosity for such alleles is a susceptibility factor for the WBS deletion (4.6-fold increased risk; range, 1.3–16.4) by facilitating the misalignment and unequal recombination in meiotic prophase. All but one of the WBS children of CNV carriers had deletions of 1.55 Mb in size, mediated by NAHR between blocks Bc and Bm. The only exception was patient SW530, whose deletion was 1.83 Mb in size mediated by NAHR between blocks A.
We also tested the possibility that the regional CNVs could be related with the previously described paracentric inversion. In order to determine whether an inversion had mediated the WBS deletions in these cases, we obtained data from three site-specific PSVs within block B in the WBS patients born to CNV carriers (SSNs 8, 10, and 11 reported in Bayés et al. 2003). At these positions there should always be a gain of a Bt-type sequence and loss of a Bc-type sequence if the rearranged WBS chromosome was originated by interchromosomal unequal exchange in an inversion carrier, as we previously predicted (Bayés et al. 2003). We identified a single case, SW52.01, with a deletion that had occurred through this mechanism. Heterozygosity for the inversion polymorphism was documented by three-color FISH in interphase nuclei in the transmitting parent, also carrying the CNV (SW52.12), and was discarded in the remaining 5/5 WBS-transmitting parents available for molecular cytogenetic testing (data not shown). Therefore, the single association found is likely due to chance, and the large CNVs described here appear not to be related with the paracentric inversions.
Based on the observed breakpoints and documented interchromosomal exchange, we propose a model for the chromosomal pairing between normal and polymorphic chromosomes in carriers of deletion-type CNVs at 7q11.23 that leads to a WBS deletion in the offspring (Fig. 7). Interchromosomal misalignment and unequal crossing-over between blocks Bc and Bm due to the absence of Bc in the WBS-CNV1Del chromosome would lead to an allele missing the WBS-critical region (1.55 Mb) or the reciprocal duplication.
Figure 7.
Predicted mechanism for unequal crossing-over leading to the WBS deletion. Heterozygosity for a large CNV in the region, such as the represented WBS-CNV1del, can facilitate the misalignment of chromosome 7 homologs in meiosis I and the unequal crossing-over (black X) between blocks Bc and Bm, leading to the most common 1.55-Mb WBS deletion. The normal allele is represented with continuous lines and arrows, while the chromosome carrying the CNV is represented by dotted lines and arrows.
Discussion
Several submicroscopic paracentric inversions have been associated with increased risk of disease-associated rearrangements in the progeny. A common paracentric inversion in 8p23 present in 26% of individuals of European descent increases susceptibility to the formation of unbalanced chromosome rearrangements such as invdup(8p) or +der(8p) (Giglio et al. 2001). Other relatively common small paracentric inversions mediated by segmental duplications have been found with higher prevalence among the parents of recurrent microdeletion syndromes, including WBS at 7q11.23, Angelman/Prader-Willi syndromes at the 15q11-q13 region, Sotos syndrome at 5q35.1, and the recently described 17q21.31 microdeletion syndrome (Osborne et al. 2001; Bayés et al. 2003; Gimelli et al. 2003; Sharp et al. 2006; Shaw-Smith et al. 2006). Heterozygosity for paracentric inversions is known to lead to the formation of a loop for the correct sequence alignment during pairing of homologous chromosomes, resulting in either acentric or dicentric, usually nonviable, chromosomes when a recombination event takes place in the loop. However, if the region contains segmental duplications, misalignment and NAHR between those segmental duplications within the loop or in the flanking regions may cause either deletion or duplication of the intervening genomic interval, causing a genomic disorder (Bayés et al. 2003).
We have identified and characterized a variety of structural copy number polymorphisms in the 7q11.23 region that may also confer increased susceptibility to the formation of germline deletions causing WBS. We found that 4.44% WBS-transmitting progenitors that are carriers of an allele with only two LCRs at 7q11.23. Similar genomic variants were found in six out of 600 (1%) control individuals including non-transmitter parents. The significantly increased prevalence of this CNV among WBS-transmitting parents (P = 0.002) suggests that heterozygosity for this allele is a susceptibility factor for the WBS deletion (4.6-fold increased risk) by facilitating the misalignment and unequal recombination in meiotic prophase and thus the generation of the WBS deletion. On the other hand, we have found 2.22% WBS-transmitter parents as carriers of duplication-type CNVs in the region have a slight but not significant (P = 0.3) increased prevalence with respect to that of the general population (1.16%). Most likely, heterozygosity for the duplication-type CNV could also facilitate misalignment in the region, but NAHR between misaligned LCRs would not involve the single-copy WBS locus in most cases.
We have also shown that the most common CNV alleles, WBS-CNV1Del and WBS-CNV1Dup, originated as predicted by unequal crossing-over within the ∼39-kb region of homology between blocks Cc and Cm, which are in the same orientation at the centromeric side of the WBS locus. Interphase FISH documented the deletion or duplication of LCRs at the centromeric side, and we were able to infer the haplotype of the recombinant Cc–Cm block from the genotype at several site-specific PSVs. Interestingly, all WBS-CNV1Del junction fragments were located centromeric to the POMex16 PSV (16 kb of Cc–Cm alignment), while WBS-CNV1Dup junction fragments were more variable but all had occurred centromeric to the NSUN5ex4 PSV. This clustering of WBS-CNV1 junction fragments in the first part of the block Cc–Cm alignment could be due to the higher sequence identity of the first 17 kb compared with the last 22 kb (98.8% vs. 95.2%, Supplemental Figure 1), since NAHR preferentially occurs in regions of very high sequence identity (Stankiewicz and Lupski 2002). Alternatively, the observed clustering could be an ascertainment bias caused by the fact that we have screened phenotypically normal individuals. Rearrangements with more distal breakpoints would interrupt the ancestral copies of functional genes in the Cm block (such as NSUN5 and TRIM50) and could then be associated with phenotypic consequences. The blocks C involved in the rearrangement contain two partial, POM121 and FKBP6, and two complete transcriptional units, NSUN5 and TRIM50. NSUN5, TRIM50, and FKBP6 appear to encode the ancestral and functional full-size protein only in the Cm locus, while the Cc and Ct blocks encode truncated, shorter, or chimeric predicted proteins (NSUN5C/5B, TRIM74/73, and FKBP6T). However, POM121 is expressed from two copies both coding for potentially functional proteins (Cc and Ct) while the Cm copy is not expressed due to the lack of the promoter region and first five exons. The other two LCR blocks completely deleted or duplicated in the WBS-CNV1 polymorphisms, Ac and Bc, contain only pseudogenes (either not expressed or not translated) and multiple-copy transcriptional units of unknown functional relevance. Therefore, deletion or duplication WBS-CNV1 in all cases does not appear to affect functional genes, in agreement with the observed lack of phenotype in CNV carriers.
Given the opposite orientation of the medial and telomeric blocks A and B, small paracentric inversions mediated by those blocks are likely to occur. In fact, such inversion was already reported in one of the reference human genome assemblies of human chromosome 7 as well as chimpanzee (Hillier et al. 2003; Scherer et al. 2003; Antonell et al. 2005). The chromosomes with that type of inversions would present two blocks of LCRs, A or B, depending on the inversion breakpoint, arranged in tandem with the other block in between. This structure might facilitate the generation of deletions and duplication of only two blocks (A and B) mediated by NAHR between the blocks in tandem, consistent with the WBS-CNV2 polymorphisms (Fig. 5). Since blocks Bm and Bt contain apparently functional copies of the GTF2IRD2 gene, individuals with WBS-CNV2 or WBS-CNV3 present with either a deletion or duplication of one of those copies along with other pseudogenes and transcriptional units, but no significant phenotypic consequences were evident in the CNV carriers.
Analysis of the deposited sequences in genome databases from several individuals along with recent experimental data indicates that the degree of structural polymorphism of this region is likely more frequent than reported here. The modular structure of some of the segmental duplications with internal repeats predisposes the region to further variability, especially at blocks A and the distal portions of blocks Cc and Ct. In fact, comparative analysis of BACs from different genomic libraries unambiguously assigned to specific loci revealed a 28-kb CNV (1–3 repeats) containing PMS2L sequences within block A. Blocks A and C are composed of smaller modules of segmental duplications that are also located in other genomic regions mainly of chromosome 7, with up to 20 copies for some portions of block A in the reference genome. Copy number variation at some of those repetitive loci has been obtained by comparative genomic hybridization experiments with BAC microarrays, affecting multiple HapMap individuals (Redon et al. 2006; Wong et al. 2007). However, given the multiple copy nature of those LCRs, the specific location, size, and copy number of these CNVs cannot be easily determined by high-throughput experimental procedures. On the contrary, our data indicates that, although variability mediated by smaller modules of the LCRs can be frequent, the large CNVs in 7q11.23 are not common (∼1%) and the CNV boundaries defined in the human genome variation database are not accurate.
WBS is mostly a sporadic condition with a few cases of documented vertical transmission and only two families reported with affected siblings from normal parents (Kara-Mostefa et al. 1999; Scherer et al. 2005). Out of those families, a set of WBS siblings was likely due to maternal gonadal mosaicism for a premeiotic deletion event, while the other set was indeed caused by recurrent meiotic deletion events in an inversion-carrier father. Given the rare finding of WBS recurrence in families, the recurrence risk in the sibship of a proband with WBS appears to be very low. Assuming the reported 1:7500 prevalence of WBS in newborns, the 4.6-fold increased risk for progenitors with “predisposing” alleles would imply an estimated global risk of 1:1500, similar to that of paracentric inversion carriers. In theory, a similar risk for the 7q11.23 duplication syndrome associated with language impairment should also be considered, given the reciprocal deletion/duplication nature (Somerville et al. 2005). Despite the numbers, the implications for genetic counseling in population carriers in the absence of family history are uncertain.
In summary, genomic structural features of the 7q11.23 region render this genomic region unstable and prone to structural variation mediated by misalignment and NAHR. In addition to the WBS deletions, we observed a variety of genomic rearrangements mediated by NAHR between different LCR blocks leading to deletions and duplications of LCRs. Our data further illustrate the highly dynamic structure of regions rich in segmental duplications such as the WBS locus and demonstrate that large CNVs can act as susceptibility alleles for disease-associated genomic rearrangements in the progeny. Since thousands of submicroscopic CNVs spanning a large proportion of the human genome have been found, heterozygosity for large CNV could be a rather frequent finding in several regions of any given diploid genome. Although with a lower mutation rate, rearrangements mediated by similar mechanisms may also be responsible for human somatic disorders.
Methods
Subjects
We studied parental samples of a total 180 WBS patients with defined 7q11.23 deletions under institutional review board-approved informed consent. WBS deletions had been previously characterized, and the parental origin of the rearranged chromosome had been established as reported elsewhere (Bayés et al. 2003; Del Campo et al. 2006). Non-transmitting WBS progenitors (n = 180) and Spanish anonymous blood donors (n = 420) were used as controls. Genomic DNA was extracted from peripheral blood by the salting-out method using a Puragene DNA purification Kit (Gentra Systems).
Copy number quantification in LCR blocks
Sequence analysis
We assembled and aligned all sequence data available from genomic BAC and PAC clones spanning blocks Cc (RP11–313p13, RP11–780k20, RP11–450o3), Cm (RP11–483g21, CTB-23i15, RP11–225k6), and Ct (RP5–953a4, RP5–1188a4, RP11–776g16, CTD-2562f8). Site-specificity of the clones was defined by differential anchoring points at least at one of their ends. Consensus sequences for LCRs were established by using the SeqMan program (DNASTAR). Identity and homology distribution between the sequences were determined by PipMaker software (http://bio.cse.psu.edu/pipmaker). A MegaBLAST alignment of the entire length of all three block C paralog sequences (∼110 kb for Cc and Ct, 44 kb for Cm) was done to define microsatellites and site-specific PSVs that could be genotyped.
Quantification of multicopy microsatellite markers
Short tandem repeats (STRs) located on block A (D7S489 or BASTR1), block B (BBSTR1 and BBSTR2), and block C (BCSTR1) of the LCRs flanking the WBS locus were amplified in order to identify the number of copies of each block in WBS progenitors and control individuals. Primers and PCR conditions have been previously reported for the block A and B amplicons and were the same for the novel block C STR (Supplementary Table 2; Bayés et al. 2003). Estimation of the number of alleles at multiloci microsatellites was performed by comparing the relative ratios of the area under the peaks from alleles of the same size in different samples, using the GeneScan 3.1 software (PE Applied Biosystems) as previously described (Bayés et al. 2003).
Quantification of paralogous sequence variants
Site-specific nucleotides
To detect the junction between the blocks C and A we analyzed a site-specific PSV that corresponds to a single nucleotide change located at close to C-A junction, detected using the restriction enzyme HinfI (New England Biolabs), followed by size fractionation on a 3% MetaPhor agarose gel (Bayés et al. 2003). To identify the specific block C (Cc/Cm/Ct), we performed two additional SSN studies by PCR amplification of (1) POM121ex6: the 262-bp PCR product contained a single-nucleotide change, T/C, in the centromeric block C and it was studied using restriction enzyme RsaI (New England Biolabs) followed by size fractionation on 2% MetaPhor agarose gel, and (2) the TRIM50 gene followed by restriction analysis of NgoMIV. The TRIM50 copies located in Ct and Cc correspond to pseudogene copies containing a non-digested product, while a Cm TRIM50 copy contains the functional gene that was digested by the NgoMIV enzyme. Primers and estimation of the number of TRIM50 pseudogene/gene ratio have been described previously (Antonell et al. 2005). In brief, for each assay, a digital image of the gel was captured at varying exposure times to ensure that the bands were not saturated. Then, intensities of bands corresponding to presumed blocks Cc, Cm, and/or Ct were quantified using the Volume Tool from the Quantity One software package (Bio-Rad). Relative intensities were calculated by means of a dosage quotient for the block that can be distinguished relative to the others, using several controls as reference values in the same experiment. Reproducibility of results was evaluated by repeating each experiment at least twice with a different number of PCR cycles.
Indel-type paralogous sequence variants
A PSV at the NCF1 locus that consists of a 2-bp deletion at the Bc and Bt blocks (99 bp) (corresponding to NCF1 pseudogenes) with respect to the Bm block (101 bp) (containing the functional copy) was analyzed for study of block B by previously described protocols (Del Campo et al. 2006).
Additional assays to quantify PSVs that distinguish among all three blocks C were also designed: (1) POM121ex11: a 3-bp deletion is present in the Cm copy (256-bp PCR product) and a 6-bp deletion in the Cc copy (253-bp PCR product) with respect to the Ct copy that harbors the ancestral gene copy (262-bp PCR product); (2) POM121ex16: a 2-bp deletion is present in Cc copy (150-bp amplicon) with respect to the Cm and Ct copies (152-bp amplicon); and (3) NSUN5: a 4-bp deletion exists in the Ct and Cc blocks with respect to Cm block (242-pb) containing the ancestral functional gene.
Genotyping of indel-type PSVs was performed on an ABI 3100 sequencer, and results were analyzed using the GeneScan 3.1 software (Applied Biosystems). Pseudogene/gene ratio quantification was performed by comparing the peak heights corresponding to each variant. Reference values for the most common genotypes (mean and range) were obtained from at least 10 individuals. Deletion or duplication of a specific copy type was predicted when the ratio values fell outside the normal range (Supplemental Table 2). Reproducibility of the results was evaluated by repeating each experiment at least twice.
Fluorescence in situ hybridization (FISH)
Selected BAC clones for FISH analyses were purchased from the HGMP Resource Center (Cambridge, UK). Chromosome preparations were generated from cultures of peripheral blood lymphocytes. Cultures were exposed to Colcemid (GIBCO) in order to obtain metaphase chromosomes in addition to interphase nuclei. Slides were prepared by hypotonic treatment and then fixed and air-dried. Hybridization signals were analyzed and counted on interphase nuclei, while metaphase chromosomes were also analyzed to test hybridization efficiency and to confirm probe specificity.
Dual-color FISH was performed with DNA from BACs RP11–204E14 (GenBank accession no. BZ724405), containing mostly block B sequences, and RP11–622P13 (GenBank accession no. AC073846), located in the WBS single-copy region (Fig. 3). BAC DNA was fluorescently labeled using Spectrum-Red-dUTP (RP11-204E14) and Spectrum-Green-dUTPs (RP11-622P13) by nick translation (Vysis/Abbott). Nuclei were counterstained with DAPI/Antifade (Q-biogene), and slides were visualized under a fluorescent microscope (AH3, Olympus). The images were analyzed with Cytovision 3.1 software (Applied Imaging Ltd). At least 20 interphase nuclei wherein all probe signals could be identified in close alignment were scored in each case.
Microsatellite and MLPA analyses
Six microsatellite markers distributed along the 7q11.23 WBS region (D7S672, BBSTR1, CR16T, D7S2472, D7S1870, and D7S2518) were genotyped in all cases. Two additional microsatellites, D7S653 and D7S2490, were analyzed only if D7S672 and D7S2518, respectively, were not informative. Unaffected siblings of WBS patients were used to establish the phase. Primers, PCR conditions, and detection methods were as previously described (Bayés et al. 2003). A total of 100 ng of genomic DNA from each sample was subjected to MLPA using specific synthetic probes of several single-copy genes in the WBS region (Supplemental Table 2). The MLPA reactions were analyzed on an ABI PRISM 3100 genetic analyzer according to the manufacturer’s instructions. Each MLPA signal was normalized and compared to the corresponding peak height obtained from a control DNA sample.
Statistical analysis
Allele frequencies in WBS-transmitting parents and control individuals were compared using Pearson's χ2 and Fisher’s exact tests. Statistical analysis was performed using R-platform software. A P-value < 0.05 denoted the presence of statistically significant differences.
Acknowledgments
We thank the WBS families for their continuous support and David Comas for providing control samples. I.C. is supported by a Juan de la Cierva postdoctoral fellowship, and V.C. is a FIS investigator. This work was supported by grants from the Spanish Ministries of Science and Education (SAF2004-6382) and Health (FIS PI042016), Genome Spain, and the VI Framework Programme of the European Commission (LSHG-CT-2006-037627).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.073197.107.
References
- Antonell A., de Luis O., Domingo-Roura X., Pérez-Jurado L.A., de Luis O., Domingo-Roura X., Pérez-Jurado L.A., Domingo-Roura X., Pérez-Jurado L.A., Pérez-Jurado L.A. Evolutionary mechanisms shaping the genomic structure of the Williams-Beuren syndrome chromosomal region at human 7q11.23. Genome Res. 2005;15:1179–1188. doi: 10.1101/gr.3944605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Baumer A., Dutly F., Balmer D., Riegel M., Tukel T., Krajewska-Walasek M., Schinzel A.A., Dutly F., Balmer D., Riegel M., Tukel T., Krajewska-Walasek M., Schinzel A.A., Balmer D., Riegel M., Tukel T., Krajewska-Walasek M., Schinzel A.A., Riegel M., Tukel T., Krajewska-Walasek M., Schinzel A.A., Tukel T., Krajewska-Walasek M., Schinzel A.A., Krajewska-Walasek M., Schinzel A.A., Schinzel A.A. High level of unequal meiotic crossovers at the origin of the 22q11. 2 and 7q11.23 deletions. Hum. Mol. Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- Bayés M., Magano L.F., Rivera N., Flores R., Pérez Jurado L.A., Magano L.F., Rivera N., Flores R., Pérez Jurado L.A., Rivera N., Flores R., Pérez Jurado L.A., Flores R., Pérez Jurado L.A., Pérez Jurado L.A. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum. Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J., Wilson M., Zhang J., Khaja R., MacDonald J., Heng H., Koop B., Scherer S., Wilson M., Zhang J., Khaja R., MacDonald J., Heng H., Koop B., Scherer S., Zhang J., Khaja R., MacDonald J., Heng H., Koop B., Scherer S., Khaja R., MacDonald J., Heng H., Koop B., Scherer S., MacDonald J., Heng H., Koop B., Scherer S., Heng H., Koop B., Scherer S., Koop B., Scherer S., Scherer S. Recent segmental and gene duplications in the mouse genome. Genome Biol. 2003;4:R47. doi: 10.1186/gb-2003-4-8-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Carter N.P., Hurles M.E., Pritchard J.K., Hurles M.E., Pritchard J.K., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Del Campo M., Antonell A., Magano L.F., Muñoz F.J., Flores R., Bayes M., Pérez Jurado L.A., Antonell A., Magano L.F., Muñoz F.J., Flores R., Bayes M., Pérez Jurado L.A., Magano L.F., Muñoz F.J., Flores R., Bayes M., Pérez Jurado L.A., Muñoz F.J., Flores R., Bayes M., Pérez Jurado L.A., Flores R., Bayes M., Pérez Jurado L.A., Bayes M., Pérez Jurado L.A., Pérez Jurado L.A. Hemizygosity at the NCF1 gene in patients with Williams-Beuren syndrome decreases their risk of hypertension. Am. J. Hum. Genet. 2006;78:533–542. doi: 10.1086/501073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E.E., Nickerson D.A., Altshuler D., Bowcock A.M., Brooks L.D., Carter N.P., Church D.M., Felsenfeld A., Guyer M., Lee C., Nickerson D.A., Altshuler D., Bowcock A.M., Brooks L.D., Carter N.P., Church D.M., Felsenfeld A., Guyer M., Lee C., Altshuler D., Bowcock A.M., Brooks L.D., Carter N.P., Church D.M., Felsenfeld A., Guyer M., Lee C., Bowcock A.M., Brooks L.D., Carter N.P., Church D.M., Felsenfeld A., Guyer M., Lee C., Brooks L.D., Carter N.P., Church D.M., Felsenfeld A., Guyer M., Lee C., Carter N.P., Church D.M., Felsenfeld A., Guyer M., Lee C., Church D.M., Felsenfeld A., Guyer M., Lee C., Felsenfeld A., Guyer M., Lee C., Guyer M., Lee C., Lee C., et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., Carson A.R., Scherer S.W., Carson A.R., Scherer S.W., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Giglio S., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Ohashi H., Voullaire L., Larizza D., Giorda R., Voullaire L., Larizza D., Giorda R., Larizza D., Giorda R., Giorda R., et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelli G., Pujana M.A., Patricelli M.G., Russo S., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., Pujana M.A., Patricelli M.G., Russo S., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., Patricelli M.G., Russo S., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., Russo S., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., Cheung J., Armengol L., Schinzel A., Estivill X., Armengol L., Schinzel A., Estivill X., Schinzel A., Estivill X., Estivill X., et al. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum. Mol. Genet. 2003;12:849–858. doi: 10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., Freedman B.I., Quinones M.P., Bamshad M.J., Quinones M.P., Bamshad M.J., Bamshad M.J., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Fulton R.S., Fulton L.A., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Fulton R.S., Fulton L.A., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Fulton L.A., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Graves T.A., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Pepin K.H., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Wagner-McPherson C., Layman D., Maas J., Jaeger S., Walker R., Layman D., Maas J., Jaeger S., Walker R., Maas J., Jaeger S., Walker R., Jaeger S., Walker R., Walker R., et al. The DNA sequence of human chromosome 7. Nature. 2003;424:157–164. doi: 10.1038/nature01782. [DOI] [PubMed] [Google Scholar]
- Howald C., Merla G., Digilio M.C., Amenta S., Lyle R., Deutsch S., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Merla G., Digilio M.C., Amenta S., Lyle R., Deutsch S., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Digilio M.C., Amenta S., Lyle R., Deutsch S., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Amenta S., Lyle R., Deutsch S., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Lyle R., Deutsch S., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Deutsch S., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Choudhury U., Bottani A., Antonarakis S.E., Fryssira H., Bottani A., Antonarakis S.E., Fryssira H., Antonarakis S.E., Fryssira H., Fryssira H., et al. Two high-throughput technologies to detect segmental aneuploidies identify new Williams-Beuren syndrome patients with atypical deletions. J. Med. Genet. 2006;43:266–273. doi: 10.1136/jmg.2005.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Qi Y., Scherer S.W., Lee C., Scherer S.W., Lee C., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Inoue K., Lupski J.R., Lupski J.R. Molecular mechanisms for genomic disorders. Annu. Rev. Genomics Hum. Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- Kara-Mostefa A., Raoul O., Lyonnet S., Amiel J., Munnich A., Vekemans M., Magnier S., Ossareh B., Bonnefont J.P., Raoul O., Lyonnet S., Amiel J., Munnich A., Vekemans M., Magnier S., Ossareh B., Bonnefont J.P., Lyonnet S., Amiel J., Munnich A., Vekemans M., Magnier S., Ossareh B., Bonnefont J.P., Amiel J., Munnich A., Vekemans M., Magnier S., Ossareh B., Bonnefont J.P., Munnich A., Vekemans M., Magnier S., Ossareh B., Bonnefont J.P., Vekemans M., Magnier S., Ossareh B., Bonnefont J.P., Magnier S., Ossareh B., Bonnefont J.P., Ossareh B., Bonnefont J.P., Bonnefont J.P. Recurrent Williams-Beuren syndrome in a sibship suggestive of maternal germ-line mosaicism. Am. J. Hum. Genet. 1999;64:1475–1478. doi: 10.1086/302362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Kooy R.F., Reyniers E., Romano C., Fichera M., Reyniers E., Romano C., Fichera M., Romano C., Fichera M., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Korenberg J.R., Chen X.N., Hirota H., Lai Z., Bellugi U., Burian D., Roe B., Matsuoka R., Chen X.N., Hirota H., Lai Z., Bellugi U., Burian D., Roe B., Matsuoka R., Hirota H., Lai Z., Bellugi U., Burian D., Roe B., Matsuoka R., Lai Z., Bellugi U., Burian D., Roe B., Matsuoka R., Bellugi U., Burian D., Roe B., Matsuoka R., Burian D., Roe B., Matsuoka R., Roe B., Matsuoka R., Matsuoka R. VI. Genome structure and cognitive map of Williams syndrome. J. Cogn. Neurosci. 2000;12:89–107. doi: 10.1162/089892900562002. (Suppl. 1) [DOI] [PubMed] [Google Scholar]
- Lupski J.R. Structural variation in the human genome. N. Engl. J. Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Gabriel S.B., Lee C., Daly M.J., Lee C., Daly M.J., Daly M.J., et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Morris C.A., Mervis C.B., Mervis C.B. Williams syndrome and related disorders. Annu. Rev. Genomics Hum. Genet. 2000;1:461–484. doi: 10.1146/annurev.genom.1.1.461. [DOI] [PubMed] [Google Scholar]
- Osborne L.R., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Costa T., Grebe T., Cox S., Tsui L.C., Grebe T., Cox S., Tsui L.C., Cox S., Tsui L.C., Tsui L.C., et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples R., Franke Y., Wang Y.K., Pérez-Jurado L., Paperna T., Cisco M., Francke U., Franke Y., Wang Y.K., Pérez-Jurado L., Paperna T., Cisco M., Francke U., Wang Y.K., Pérez-Jurado L., Paperna T., Cisco M., Francke U., Pérez-Jurado L., Paperna T., Cisco M., Francke U., Paperna T., Cisco M., Francke U., Cisco M., Francke U., Francke U. A physical map, including a BAC/PAC clone contig, of the Williams-Beuren syndrome–deletion region at 7q11.23. Am. J. Hum. Genet. 2000;66:47–68. doi: 10.1086/302722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Jurado L.A. Williams-Beuren syndrome: A model of recurrent genomic mutation. Horm. Res. 2003;59:106–113. doi: 10.1159/000067836. [DOI] [PubMed] [Google Scholar]
- Pérez Jurado L.A., Peoples R., Kaplan P., Hamel B.C., Francke U., Peoples R., Kaplan P., Hamel B.C., Francke U., Kaplan P., Hamel B.C., Francke U., Hamel B.C., Francke U., Francke U. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am. J. Hum. Genet. 1996;59:781–792. [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fiegler H., Shapero M.H., Carson A.R., Chen W., Shapero M.H., Carson A.R., Chen W., Carson A.R., Chen W., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson W.P., Waslynka J., Bernasconi F., Wang M., Clark S., Kotzot D., Schinzel A., Waslynka J., Bernasconi F., Wang M., Clark S., Kotzot D., Schinzel A., Bernasconi F., Wang M., Clark S., Kotzot D., Schinzel A., Wang M., Clark S., Kotzot D., Schinzel A., Clark S., Kotzot D., Schinzel A., Kotzot D., Schinzel A., Schinzel A. Delineation of 7q11.2 deletions associated with Williams-Beuren syndrome and mapping of a repetitive sequence to within and to either side of the common deletion. Genomics. 1996;34:17–23. doi: 10.1006/geno.1996.0237. [DOI] [PubMed] [Google Scholar]
- Scherer S.W., Cheung J., MacDonald J.R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Cheung J., MacDonald J.R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., MacDonald J.R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Osborne L.R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Nakabayashi K., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Herbrick J.A., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Carson A.R., Parker-Katiraee L., Skaug J., Khaja R., Parker-Katiraee L., Skaug J., Khaja R., Skaug J., Khaja R., Khaja R., et al. Human chromosome 7: DNA sequence and biology. Science. 2003;300:767–772. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S.W., Gripp K.W., Lucena J., Nicholson L., Bonnefont J.P., Perez-Jurado L.A., Osborne L.R., Gripp K.W., Lucena J., Nicholson L., Bonnefont J.P., Perez-Jurado L.A., Osborne L.R., Lucena J., Nicholson L., Bonnefont J.P., Perez-Jurado L.A., Osborne L.R., Nicholson L., Bonnefont J.P., Perez-Jurado L.A., Osborne L.R., Bonnefont J.P., Perez-Jurado L.A., Osborne L.R., Perez-Jurado L.A., Osborne L.R., Osborne L.R. Observation of a parental inversion variant in a rare Williams-Beuren syndrome family with two affected children. Hum. Genet. 2005;117:383–388. doi: 10.1007/s00439-005-1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Clark R.A., Schwartz S., Segraves R., Schwartz S., Segraves R., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Stewart H., Price S.M., Blair E., Hennekam R.C., Price S.M., Blair E., Hennekam R.C., Blair E., Hennekam R.C., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Cumming S., Dunn C., Kalaitzopoulos D., Dunn C., Kalaitzopoulos D., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Somerville M.J., Mervis C.B., Young E.J., Seo E.-J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Mervis C.B., Young E.J., Seo E.-J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Young E.J., Seo E.-J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Seo E.-J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Bamforth S., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Peregrine E., Loo W., Lilley M., Perez-Jurado L.A., Loo W., Lilley M., Perez-Jurado L.A., Lilley M., Perez-Jurado L.A., Perez-Jurado L.A., et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stromme P., Bjornstad P.G., Ramstad K., Bjornstad P.G., Ramstad K., Ramstad K. Prevalence estimation of Williams syndrome. J. Child Neurol. 2002;17:269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., Haugen E., Hayden H., Albertson D., Pinkel D., Hayden H., Albertson D., Pinkel D., Albertson D., Pinkel D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Urban Z., Helms C., Fekete G., Csiszar K., Bonnet D., Munnich A., Donis-Keller H., Boyd C.D., Helms C., Fekete G., Csiszar K., Bonnet D., Munnich A., Donis-Keller H., Boyd C.D., Fekete G., Csiszar K., Bonnet D., Munnich A., Donis-Keller H., Boyd C.D., Csiszar K., Bonnet D., Munnich A., Donis-Keller H., Boyd C.D., Bonnet D., Munnich A., Donis-Keller H., Boyd C.D., Munnich A., Donis-Keller H., Boyd C.D., Donis-Keller H., Boyd C.D., Boyd C.D. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am. J. Hum. Genet. 1996;59:958–962. [PMC free article] [PubMed] [Google Scholar]
- Valero M.C., de Luis O., Cruces J., Perez Jurado L.A., de Luis O., Cruces J., Perez Jurado L.A., Cruces J., Perez Jurado L.A., Perez Jurado L.A. Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: The low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s) Genomics. 2000;69:1–13. doi: 10.1006/geno.2000.6312. [DOI] [PubMed] [Google Scholar]
- Wong K.K., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Ng R.T., Brown C.J., Eichler E.E., Brown C.J., Eichler E.E., Eichler E.E., et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lu H.H.S., Chung W.-y., Yang J., Li W.-H., Lu H.H.S., Chung W.-y., Yang J., Li W.-H., Chung W.-y., Yang J., Li W.-H., Yang J., Li W.-H., Li W.-H. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 2005;22:135–141. doi: 10.1093/molbev/msh262. [DOI] [PubMed] [Google Scholar]