Abstract
Insulin resistance is necessary but not sufficient for the development of type 2 diabetes. Diabetes results when pancreatic beta-cells fail to compensate for insulin resistance by increasing insulin production through an expansion of beta-cell mass or increased insulin secretion. Communication between insulin target tissues and beta-cells may initiate this compensatory response. Correlated changes in gene expression between tissues can provide evidence for such intercellular communication. We profiled gene expression in six tissues of mice from an obesity-induced diabetes-resistant and a diabetes-susceptible strain before and after the onset of diabetes. We studied the correlation structure of mRNA abundance and identified 105 co-expression gene modules. We provide an interactive gene network model showing the correlation structure between the expression modules within and among the six tissues. This resource also provides a searchable database of gene expression profiles for all genes in six tissues in lean and obese diabetes-resistant and diabetes-susceptible mice, at 4 and 10 wk of age. A cell cycle regulatory module in islets predicts diabetes susceptibility. The module predicts islet replication; we found a strong correlation between 2H2O incorporation into islet DNA in vivo and the expression pattern of the cell cycle module. This pattern is highly correlated with that of several individual genes in insulin target tissues, including Igf2, which has been shown to promote beta-cell proliferation, suggesting that these genes may provide a link between insulin resistance and beta-cell proliferation.
Type 2 diabetes is a disorder that involves an increased demand for insulin brought about by insulin resistance, together with a failure to compensate with sufficient insulin production. Although insulin resistance occurs in most obese individuals, diabetes is generally forestalled through compensation with increased insulin. This increase in insulin occurs through an expansion of beta-cell mass and/or increased insulin secretion by individual beta-cells. Failure to compensate for insulin resistance leads to type 2 diabetes.
One way to understand the pathophysiology of diabetes is to examine the coordinate changes in gene expression that occur in insulin-responsive tissues and pancreatic islets in obese animals that either compensate for insulin resistance or progress to type 2 diabetes. In each case, there are groups of genes that undergo changes in expression in a highly correlated fashion. By identifying groups of correlated transcripts (gene expression modules) during the compensation and development of diabetes, we can gain insight into potential pathways and regulatory networks in obesity-induced diabetes.
We study two strains of mice that differ in obesity-induced diabetes susceptibility. In this study, we surveyed gene expression in six tissues of lean and obese C57BL/6 (B6) and BTBR mice aged 4 wk and 10 wk. B6 mice remain essentially non-diabetic at all ages, irrespective of obesity. When obese, BTBR mice become severely diabetic by 10 wk of age.
By analyzing the correlation structure of the genes under three contrast conditions, obesity, strain, and age, we identified gene expression modules associated with the onset of diabetes and provide an interactive co-expression network model of type 2 diabetes. We found a key module that is comprised of cell cycle regulatory genes. In the islet, the expression profile of these transcripts accurately predicts diabetes and is highly correlated with islet cell proliferation.
Results
Diabetes results from the convergence of age, strain, and obesity
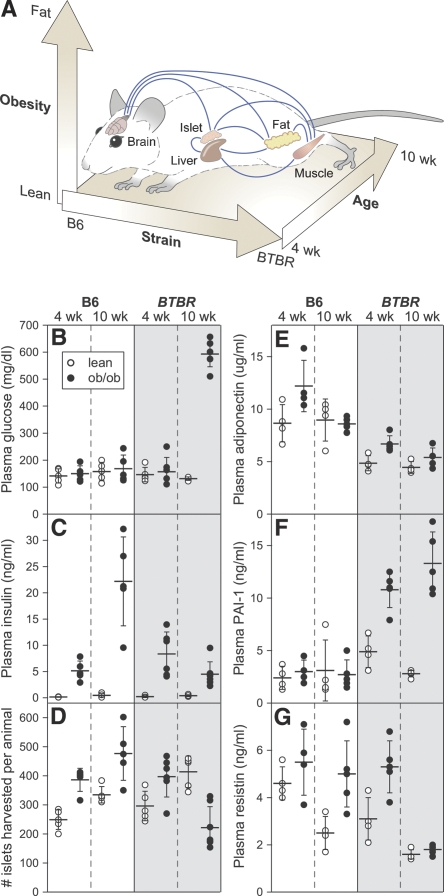
To understand the relative contribution of age, strain, and obesity to the etiology of type 2 diabetes, we compared mice across three primary axes—time: 4 vs. 10 wk of age; body mass: lean vs. obese; and strain: C57BL/6 (B6) vs. BTBR mouse strain (Fig. 1A). B6 and BTBR mice differ in obesity-induced diabetes susceptibility. The B6 strain is essentially nondiabetic when it carries the leptinob/ob (ob) mutation, whereas, by 10 wk of age, the BTBR ob mouse is severely diabetic (Fig. 1B). Since the BTBR ob mouse is not yet diabetic at 4 wk of age, changes in gene expression at this time are potential causes rather than consequences of hyperglycemia. The ability of the B6 mouse to maintain euglycemia when challenged with morbid obesity is due to a >50-fold increase in circulating insulin at 10 wk of age (Fig. 1C). This contrasts with the relative hypoinsulinemia of the 10-wk-old BTBR ob mouse. The difference in insulin at 10 wk of age is correlated with the number of islets harvested from the pancreas (Fig. 1D), suggesting that the ability to continue compensating for insulin resistance is a function of the ability to continue expanding beta-cell mass.
Figure 1.
Ten-week-old BTBR ob mice are severely diabetic. (A) Schematic representation of experimental model depicting a gene expression network connecting key tissues in a mouse when examined over three primary axes: obesity, strain, and age. Clinical phenotypes are shown for five to seven animals for each of the eight groups of mice used for study. Plasma glucose (B), insulin (C), total number of islets harvested per pancreas (D), adiponectin (E), PAI-1 (F), and resistin (G) are plotted. Open (lean) and closed (ob/ob) circles represent individual mice. Horizontal bars show mean values for each group (±SD).
We observed significant differences between B6 and BTBR mice in circulating adipose-derived hormones (adipokines). Adiponectin has been shown to regulate peripheral insulin sensitivity. There was a ∼50% decrease in circulating adiponectin in BTBR mice relative to B6 mice, irrespective of age and obesity (Fig. 1E). Plasminogen activator inhibitor-1 (PAI-1), a bio-marker for inflammation, showed an obesity-dependent increase at 4 and 10 wk only in BTBR mice (Fig. 1F). In rodents, resistin is highly expressed in adipose tissue and is thought to negatively regulate hepatic insulin sensitivity (Haluzik and Haluzikova 2006). Resistin showed an obesity-dependent increase in all but the diabetic BTBR mice (Fig. 1G). In summary, three key adipokines showed significant differences between the various groups of mice, consistent with a potential role for adipose tissue in the diabetes susceptibility of BTBR mice.
Differential expression of individual genes among six key tissues
Since age and obesity are necessary to unmask diabetes, we sought to deconstruct their relative contribution in B6 and BTBR mice by gene expression profiling of lean and obese male mice at 4 and 10 wk of age. In each strain, average expression levels from the four groups of mice can be sorted into 15 distinct theoretical patterns (Supplemental Table S1). Using EBarrays, the empirical Bayes method described in Methods, for each transcript in each strain, we calculated the posterior probability for each of the 15 patterns and assigned the transcript to the expression pattern with maximum posterior probability (MPP). Differentially expressed (DE) transcripts are defined as those with MPP > 0.7 (MPP > 0.5 for hypothalamus) for one of the DE patterns (patterns 2–15 in Supplemental Table S1) in at least one mouse strain. Supplemental Figure S1 plots the MPP for DE transcripts in B6 vs. the MPP for DE transcripts in BTBR. We found that >96% of the DE transcripts were confined to seven of the 14 possible DE patterns (shaded in Supplemental Table S1). This approach has enabled us to identify primary and secondary drivers of changes in gene expression in the six tissues profiled. It is important to note that thresholds were chosen to balance false discovery rate (FDR) and number of genes identified (see legend of Supplemental Fig. S1). To ensure that modules identified were robust to more stringent thresholds for DE transcript identification, we varied the MPP cutoff and remapped the modules onto those identified at lower thresholds. For all tissues, except hypothalamus, the modules identified with MPP > 0.9 dendrogram tree were remapped onto the MPP > 0.8 and MPP > 0.7 dendrogram trees, maintaining their color designation. For hypothalamus the modules from the 0.6 tree were remapped onto the 0.5 tree, as the 0.7 tree had only one module (turquoise). Supplemental Figure S7 shows the result from islet (hypothalamus is similar). As shown, most clusters are highly conserved.
Primary vs. secondary drivers of differential gene expression at the individual transcript level
There were three variables in our experiment: obesity, age, and strain. Using a nonsupervised hierarchical algorithm, the 40 mice, five in each of eight groups, were clustered based solely on the DE transcripts for each tissue. Differential gene expression in islet, liver, and adipose tissue was primarily driven by obesity, whereas differential expression in the two muscle tissues and hypothalamus was driven primarily by age (Supplemental Fig. S2). Secondarily, age determined the clustering of the mice in islet, liver, and adipose tissue, whereas obesity drove the secondary changes in muscle and hypothalamus.
Each tissue contains modules of highly correlated differentially expressed transcripts enriched for specific biological functions
To create a framework to explore strain-, obesity-, and age-dependent determinants of gene expression, we restricted ourselves to the DE transcripts in each tissue and calculated the correlation coefficient among all transcripts, and partitioned them into color-coded modules by the method of Zhang and Horvath (2005) (Supplemental Table S2). Such modules often contain transcripts of related biological function (Carlson et al. 2006; Gargalovic et al. 2006; Ghazalpour et al. 2006; Horvath et al. 2006). Each tissue yielded 19 distinct co-expression network modules, except hypothalamus, which had 10, for a total of 105 modules (Supplemental Fig. S3). The modules were largely cohesive, as quantified by measuring the pairwise correlations of transcripts within each module. Average absolute pairwise correlations exceeded 0.7 for 84%, 100%, 79%, 95%, 90%, and 30% of the modules in islet, adipose, liver, solues, gastrocnemius, and hypothalamus, respectively (data not shown).
To assess the biological relevance of the co-expression network modules we asked if modules contained genes enriched for specific biological processes. A substantial number of modules in each tissue were enriched for genes with specific gene ontology (GO) classifications. For example, in each tissue, except hypothalamus, we identified a single module significantly enriched with genes involved in cell cycle regulation (P < 10−14 for each tissue) (Supplemental Table S4). Thus, unsupervised clustering of genes with highly correlated expression profiles yields modules enriched for biologically relevant processes (Horvath et al. 2006).
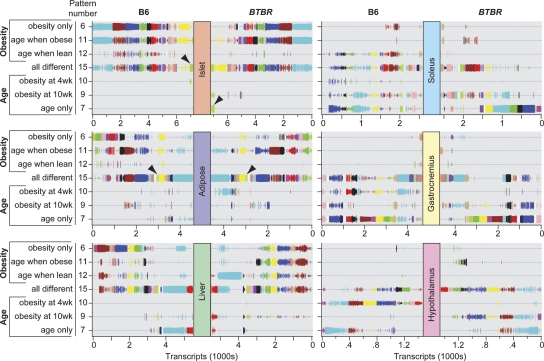
We examined the contrast condition(s) responsible for differential expression for all DE transcripts in each of the six tissues: obesity, age, and/or strain (Fig. 2; see also http://diabetes.wisc.edu/kelleretal2008/fig2.php for hyperlinked data page). Strain-dependent differences are evident when the pattern distribution for a particular color-coded module is shifted in the two strains. For example, the cell cycle module in islets (Fig. 2, green-yellow, arrowhead) is predominantly “all different” in B6, reflecting a combination of obesity and age as primary drivers of DE for this module. However, a large fraction of these genes change to an “age only” pattern in BTBR, resulting from the loss in an obesity-dependent signal present in B6. In contrast to the regulation of cell cycle genes observed in the islets, a similar set of genes in adipose tissue (Fig. 2, light green, arrowhead), also enriched in cell cycle regulatory transcripts, showed an “all different” expression pattern in both mouse strains. Supplemental Table S3 lists the number of transcripts contained within each co-expression module as a function of expression pattern, strain, and tissue (see also http://diabetes.wisc.edu/kelleretal2008/tabs3.php). In short, an unsupervised analysis of expression modules identified a key change in cell cycle gene expression in islets that distinguishes a diabetes-susceptible from a non-diabetes-susceptible mouse strain.
Figure 2.
Co-expression modules can be deconstructed to show strain-dependent changes in transcript expression patterns. The strain-specific expression pattern for each co-expression module is illustrated for all six tissues profiled. The color of a particular module within one tissue is not related to that same color for a module of another tissue but is preserved across strains. The vertical size of the lines used to illustrate the module transcripts is proportional to the strain-specific posterior probability determination illustrated in Supplemental Figure S1. A decrease in the size of the symbols is evident in the hypothalamus compared to the other tissues, reflecting the decreased posterior probability cutoff (0.5) that was used for DE transcript identification in hypothalamus. For each strain and all tissues, every transcript has a unique expression pattern. Filled arrowheads highlight the cell cycle regulatory modules in islet and adipose tissue. Strain-dependent differences in expression pattern are evident when the pattern distribution for a particular color-coded module is shifted in the two strains. For example, the cell cycle regulatory gene set in islets largely shifts from pattern 15 in B6 to pattern 7 in BTBR (see arrowheads). This figure is hyperlinked to our microarray gene expression database at http://diabetes.wisc.edu/kelleretal2008/fig2.php.
We identified a module of highly correlated genes in liver (turquoise module) that had a strain-specific expression pattern. Within this module is the glucagon receptor (Gcgr), which shows an age-dependent increase in lean and obese B6 and BTBR mice and an obesity-dependent decrease in all mice except 10-wk B6. Hepatic glucagon receptor mRNA expression has been shown to increase in diabetic animals and in fasting conditions, and its expression may be regulated by serum glucose levels via glucokinase (Burcelin et al. 1998). Inhibition of glucagon receptor mRNA expression in liver in db/db animals improves glucose tolerance and normalizes serum glucose, so it is clear that hepatic glucagon signaling plays a crucial role in the pathophysiology of diabetes (Liang et al. 2004). As glucagon signaling in the liver leads to increased nutrient mobilization, we looked in the same module for genes involved in these processes. We identified genes involved in carbohydrate metabolism (isocitrate dehydrogenase 2, aldolase 1A, phosphoglycerate mutase-2, and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3), ketone body production (aldehyde dehyrogenase 1b1), and oxidative phosphorylation (succinate dehydrogenase A, mitochondrial F0 complex, subunit c). This suggests that, by examining correlated genes within a module, we can identify strain-specific differences in key metabolic signals and their downstream effects on cellular metabolism.
The cell cycle modules predict adipose and beta-cell replication and obesity and diabetes
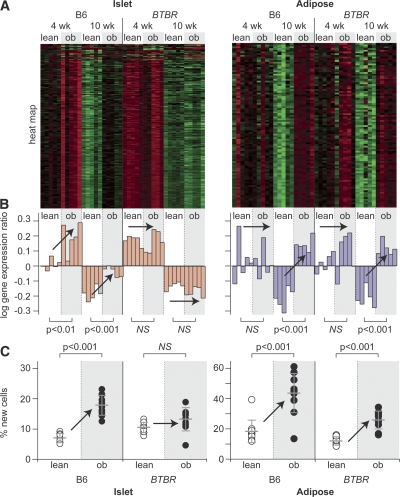
To maintain euglycemia, insulin-resistant animals must increase insulin production, which can occur through an expansion of pancreatic beta-cell mass. We focus here on two of the five tissue-specific modules that were highly enriched in genes controlling the cell cycle. The islet cell cycle module had 217 transcripts, which showed an age-dependent decrease in expression in both mouse strains and an obesity-dependent increase only in B6 mice (Fig. 3A). We calculated the first principle component (PC1) as a single descriptor of the expression pattern for the entire module (Fig. 3B). The PC1 shows an obesity-dependent increase in cell cycle gene expression in the islets of B6 mice at 4 and 10 wk of age. This induction fails to occur in BTBR mice. The same strain-specific differences in obesity-dependent induction of the islet cell cycle module were seen at 4 wk, when the animals were still euglycemic. Thus, the islet cell cycle module changes precede the onset of diabetes.
Figure 3.
Co-expression modules enriched with cell cycle regulation accurately predict diabetes and obesity. Expression heat maps (A) and the PC1 on log10 scale (B) of the cell cycle regulatory modules in islets (217 transcripts) and adipose (96 transcripts) are shown. For the heat maps, red shows increased expression, green shows decreased expression, and black is neutral. Bar plots in B show the PC1 for individual mice and correspond to an expressed decrease for negative values and increased expression for positive values. The percentage of new cells, derived from an in vivo measure of 2H incorporation into newly synthesized DNA, is shown for islets and adipose tissue (C). Where significant obesity-dependent differences were observed, P-values are shown. Arrows are used to show influence of obesity. NS, not significant.
The cell cycle module profile predicts that obesity induces islet cellular proliferation in B6 but not BTBR mice. To measure islet cellular proliferation, we exposed the mice to 8% 2H2O in the drinking water for two weeks and measured the enrichment in 2H in DNA extracted from their islets. By normalizing the islet enrichment values to 2H enrichment in bone marrow DNA, which undergoes complete turnover during this period, we estimated the percent new cells in the islets (Neese et al. 2002). B6 ob mice showed a 2.6-fold increase in the percent new islet cells relative to B6 lean mice (Fig. 3C). In contrast, BTBR islets showed no significant increase in the proportion of new islet cells in response to obesity.
In contrast to islets, the cell cycle module in adipose did not show a strain difference in expression pattern: both B6 and BTBR show an obesity-dependent up-regulation of expression for these genes at 10 wk of age (Fig. 3A,B). Consistent with the gene expression data, cellular proliferation measured in adipose tissue showed no strain difference in the fraction of newly replicated cells, with mice in both strains showing an obesity-dependent increase in cellular growth (Fig. 3C). Thus, the adipose cell cycle module PC1 correctly predicted adipose tissue proliferation and obesity, just as the islet cell cycle module predicted islet proliferation and diabetes.
Gene expression modules are highly correlated with glucose and insulin levels
Modules in islet, adipose, and soleus most strongly correlated with plasma glucose (Supplemental Fig. S4) contained genes previously shown to have a role in glucose homeostasis. For example, the cyan module in islet contains the branched chain aminotransferase 1 enzyme (Bcat1), which on its own had a 0.85 correlation with plasma glucose. Bcat1 catalyzes the transamination of alpha-ketoisocaproate (KIC) and glutamate to yield leucine and alpha-ketoglutarate (alpha-KG). We have previously shown that BTBR islets are hyperresponsive to KIC-induced insulin secretion and that alpha-KG can directly stimulate insulin release from isolated pancreatic islets (Rabaglia et al. 2005).
The magenta islet module, also enriched with transcripts highly correlated with plasma glucose (Supplemental Fig. S4), contains transcripts from a number of genes previously highlighted for their potential role in glucose homeostasis, including: gamma-subunit of the gamma-aminobutyric acid A-receptor, Gabrd (plasma glucose correlation = 0.93); peroxisome proliferator-activated receptor-alpha, Ppara (0.92); and cocaine- and amphetamine-regulated transcript, Cartpt (0.86). Glucose suppresses glucagon release by activation of the GABA-A receptors on alpha-cells, mediated by GABA released from neighboring beta-cells (Rorsman et al. 1989; Wendt et al. 2004). More recent evidence suggests that glucose may directly suppress glucagon release from alpha-cells (Vieira et al. 2007). Chronic treatment of Ins-1 cells or primary rat islets with a high glucose medium has been shown to decrease Ppara expression (Roduit et al. 2000). Finally, Cartpt expression is up-regulated in the beta-cells of type 2 diabetic models in rat (Wierup et al. 2006), and Cartpt knockout mice have impaired GSIS and are glucose intolerant (Wierup et al. 2005). Taken together, our results indicate that modules highly correlated with plasma glucose may identify compensatory changes in gene expression elicited by hyperglycemia.
Only islet, liver, and adipose contained modules correlated with plasma insulin. Adipose contained the greatest number of modules correlated with insulin (11 of 19), whereas liver contained the module with the greatest absolute correlation with plasma insulin (Supplemental Fig. S4). Obesity is a well-known driver of changes in plasma insulin, due to an obesity-induced increase in insulin resistance. The lack of correlation for insulin in the muscle tissues and hypothalamus is consistent with their “age-responsive” expression patterns, with few obesity-related transcripts (Fig. 2). For islets, liver, and adipose, modules with a high correlation with insulin were generally not correlated with glucose.
We identified a module of correlated transcripts in adipose tissue enriched with genes involved in inflammation (the brown module, Supplemental Fig. S3). These transcripts show increased expression in the obese animals, and older animals, of both strains. These data are consistent with the previous finding that adipose tissue from the B6 ob/ob mouse is enriched in transcripts found in macrophages (Weisberg et al. 2003; Xu et al. 2003). These cells are believed to secrete cytokines that affect peripheral insulin resistance. Up-regulated transcripts in our module include: Emr1, the macrophage-specific surface marker; Cd68, a macrophage transmembrane protein; Itgam, an integrin found primarily on macrophages and other inflammatory cells; and Adam8, a monocyte metalloproteinase. Transcripts in this module showed a high correlation with plasma glucose (Supplemental Fig. S4).
An inter-tissue gene expression network model
A major challenge in diabetes research is to better understand the potential relationships between gene expression changes among various tissues. For example, there are dramatic changes in gene expression in liver, muscle, and adipose tissue associated with insulin resistance and consequent changes in islets. Insulin-resistant tissues may communicate with islets through the production and secretion of blood-borne factors.
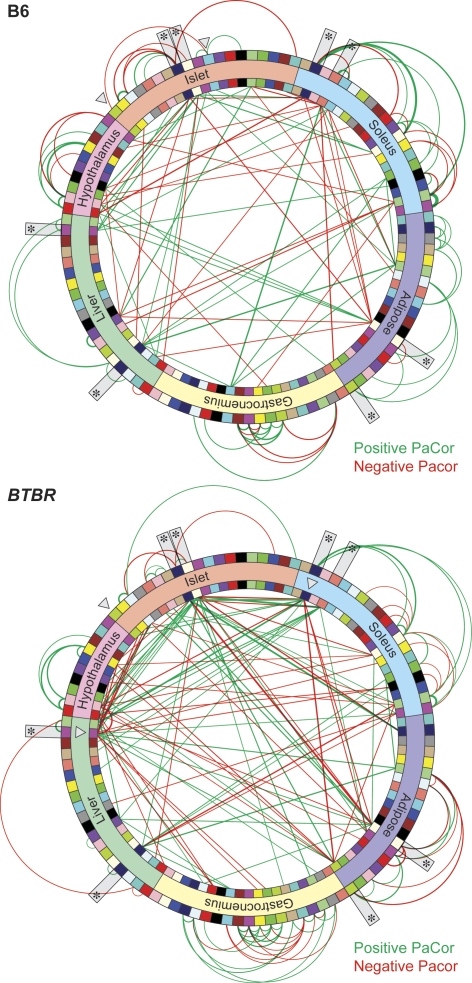
One way to search for molecules that mediate inter-organ communication is to identify relationships among the 105 modules across all six tissues. For each strain we represented each module with its strain-specific first principal component (PC1strain), which is highly correlated with all the module-specific transcripts (Supplemental Fig. S5). Each module corresponds to a node within the network (Fig. 4, see also http://diabetes.wisc.edu/kelleretal2008/fig4.php for hyperlinked data page). Strain-specific edges are shown when two modules have significant partial correlation (PaCor). PaCor is distinct from an ordinary correlation in that PaCor reveals the “direct” correlation between two PC1s after adjusting for the effects of all other module PC1s, as well as plasma glucose and insulin (see Methods). Adjusting for these two physiological traits has allowed us to more clearly establish direct gene-to-gene networks in B6 and BTBR. The strain-specific networks contained only 66 and 62 of 105 module nodes, forming just 125 and 161 significant edges out of potentially 5460 edges, in B6 and BTBR, respectively (Fig. 4). Few connections were formed out of the total possible number (∼2%), which is consistent with the assumptions made in the network construction algorithm (Schafer and Strimmer 2005b). Thus, in the diabetic strain, fewer network nodes were associated with a greater number of inter-tissue connections.
Figure 4.
A gene-gene network model is distinct between B6 and BTBR mice. A gene-gene network was constructed based on the PaCor between the strain-specific PC1 calculated between all modules identified in the six tissues profiled. Modules are illustrated as colored bricks along the inside and outside of the network wheels and preserve the color scheme illustrated in Figure 2. Inter-tissue edges within the network are shown as lines connecting inside modules; intra-tissue edges are depicted as arcs connecting the outside modules. The cell cycle regulatory module in islet and those modules that form a direct connection to the cell cycle islet module are highlighted with open arrowheads. Network hot spots are indicated with asterisks. Line thickness is proportional to the magnitude of the PaCor, which ranged from 0.487 to 0.093 in B6 and from 0.303 to 0.086 in BTBR, for maximum and minimum, respectively. Positive predictive values for edge accuracy, obtained from simulations (see Methods), were on average 78% in B6 and 77% in BTBR. Red, negative PaCor; green, positive PaCor. Significance is set to control FDR at 0.5%. This figure is hyperlinked to our microarray gene expression database at http://diabetes.wisc.edu/kelleretal2008/fig4.php.
Strain-specific PaCor among the 105 PC1strain variables, plasma glucose and insulin, revealed remarkable strain differences (Supplemental Fig. S6). In B6, there were 14 connections between glucose and gene modules, whereas in BTBR plasma, glucose made no connections with any module after adjusting for insulin and other module nodes. For insulin, there were significant PaCor connections formed with six and seven modules in B6 and BTBR mice, respectively, with only one in common. Many modules contain transcripts highly correlated with plasma glucose or insulin (Supplemental Fig. S4).
There were substantial differences between tissues in the degree of intra-tissue connectivity in the co-expression network (Fig. 4). The two muscle tissues had the most intra-tissue connections, whereas liver and hypothalamus had the fewest, in both strains. Gastrocnemius showed a twofold increase in the number of intra-tissue connections in BTBR vs. B6 mice. The number of connections present in the other tissues was similar between the strains. There was two- to eightfold more inter-tissue than intra-tissue connections in all tissues, except gastrocnemius, where there were ∼60% fewer inter-tissue connections. All tissues showed an increase in inter-tissue connectivity in BTBR vs. B6. Soleus and liver had the greatest number of inter-tissue connections, whereas gastrocnemius had the fewest. Overall, connection strength was greater for intra-tissue connections than for inter-tissue connections throughout the network. Taken together, our results suggest that obesity-dependent diabetes results in dramatic changes in the intra-tissue correlation structure of the co-expression network.
There were several nodes in each of the strain-specific networks that appear to be “hot spots”, or nodes that form the greatest number of connections within the network (Fig. 4, asterisks). These hot spots were found in both strains and in all tissues, excluding gastrocnemius, consistent with the relatively low inter-tissue connectivity of this tissue. Several of the hot spot nodes form as many as 10 or more connections with other nodes. It is interesting to note that these hot spots are highly inter-connected among themselves, as well as with other nodes.
The islet cell cycle module, Isletgreenyellow (Fig. 4, arrowhead), showed dramatic strain-dependent changes in its interconnectivity within the network. In the B6 network there is one connection between this node and a node derived from another islet module, Isletcyan, with a negative association (Fig. 4, arrowhead). Interestingly, the Isletcyan module contains transcripts with expression patterns highly correlated with plasma glucose (Supplemental Fig. S4), suggesting that genes positively correlated with glucose have negative association with the genes within the cell cycle module in B6 islets.
In the BTBR network, the intra-islet connection was lost, being replaced with two new connections: a negative association with the livermagenta node and a positive association with the soleusmidnightblue node (Fig. 4, arrowheads). Both of these nodes are highly interconnected within the network, forming 16 and 10 connections, respectively. The soleus node contains transcripts with expression patterns highly correlated with plasma glucose (Supplemental Fig. S4), suggesting glucose-regulated genes in soleus may positively associate with cell cycle regulatory genes in BTBR islets.
The islet cell cycle module is highly correlated with individual transcripts in insulin target tissues
The correlations between modules do not allow the identification of instances where there are strong correlations between a module and a transcript not belonging to a module. To identify transcripts highly correlated with the islet cell cycle module, we calculated the correlation between the module’s PC1 and all transcripts in each of the five non-islet tissues. We identified a few transcripts with high correlations to the islet cell cycle PC1, including: Gdf10 (0.85, gastrocnemius), Bmp1 (0.84, soleus), Igf2 (0.86, soleus), Igf2bp1 (0.85, adipose), and Ngf (0.8, soleus). These genes could potentially mediate a signal that promotes islet cellular proliferation. These results offer a hypothesis that one or more of these proteins mediate beta-cell proliferation in insulin-resistant mice.
A searchable type 2 diabetes gene expression database
We have created a searchable resource (http://diabetes.wisc.edu) of the gene expression data that was used to generate the network model described herein. This search tool allows the user to enter one to multiple genes and will display the gene expression profiles of our eight experimental groups (lean and obese B6 and BTBR mice at 4 and 10 wk of age) in any of six tissues (islets, adipose, liver, gastrocnemius, soleus, and hypothalamus). In addition, we have incorporated a transcript-to-transcript correlation tool that can be used to identify groups of genes with highly correlated expression profiles. For example, searching the database for the expression profiles for Ccna2 (cyclin A2) shows that this transcript was included in four co-expression gene modules (Gastrocnemiuscyan, Isletgreenyellow, Livermidnightblue, and Soluesgreen), all of which were significantly enriched with GO terms associated with cell cycle regulation (see Supplemental Table S4). Plots of the expression profiles reveal, in general, that Ccna2 decreases with age in all tissues and increases with obesity in adipose for both mouse strains and in islet for B6 mice only. Searching the mlratio database in islets for transcripts that correlate with Ccna2 yields a list of 100 transcripts with a Pearson’s correlation, r > 0.94. Many of these highly correlated transcripts themselves are involved in cell cycle regulation, including Foxm1 (r = 0.99), Ccnb1 (r = 0.98), Ccnb2 (r = 0.98), Brca1 (r = 0.97), and Aurka (r = 0.96), and are included in the cell cycle regulatory modules. In addition to positively correlated transcripts, the most negatively correlated transcripts are displayed, which, in the case of Ccna2, included Ccnk (r = −0.77), a cyclin that has been shown previously to suppress cellular proliferation when overexpressed in human glioblastoma cells (Mori et al. 2002). This brief example has illustrated the utility of our searchable database. It can be used to survey gene expression in six key tissues as a function of obesity, strain, and age in a mouse model of type II diabetes. We believe it will be a powerful resource tool that will benefit many in the diabetes research community.
Discussion
During the transition to diabetes, tissues undergo coordinated changes in gene expression and arrive at a new highly regulated, although pathological, steady state. These changes are highly correlated and thus enable us to identify modules of coordinately regulated genes. In this study, we exploited three primary variables that converge to cause type 2 diabetes in our model—obesity, age, and mouse strain—to study the correlation structure of the changes in gene expression in six tissues. We obtained a gene expression network model containing 105 modules and establish the modules’ inter- and intra-tissue relationships in the non-diabetic and diabetic state.
A major finding in our study is that, in five of the six tissues profiled, at least one of the tissue-specific modules was significantly enriched with cell cycle regulatory genes. Of direct relevance to diabetes pathogenesis, the islet cell cycle module exhibited a striking strain difference in expression pattern; B6, but not BTBR, showed an obesity-dependent increase in expression at both 4 and 10 wk of age.
These results predict a strain difference in beta-cell proliferation during the onset of obesity that would correlate with resistance or susceptibility to diabetes. This prediction was tested with a direct measure of cellular proliferation. We found that B6 islets have a robust, obesity-dependent increase in islet cell replication, consistent with previous reports. In contrast, BTBR islets failed to increase proliferation in response to obesity. Thus, the islet cell cycle module predicted diabetes.
Given the dramatic islet growth phenotypes that have been reported for the Cdk4 knockout (Rane et al. 1999; Tsutsui et al. 1999) and constitutively active transgenic (Rane et al. 1999) mouse, we asked whether Cdk4 or any of the D-type cyclins were present in the islet and other tissue-specific cell cycle modules. These genes were not present in the cell cycle modules, suggesting that age, obesity, and strain were not factors that alter their expression in our experimental model. However, cyclins A, B, and E were identified in several cell cycle modules, suggesting these molecules play a critical role in obesity and age-related changes in cell cycle progression. Ccna2 was present in the cell cycle modules in all tissues except adipose, whereas Ccne2 was unique to adipose.
There are a number of cyclin-dependent kinase inhibitors, but only two that were identified in the cell cycle modules: Cdkn2c (also known as p18) in the liver module and Cdkn3 in both liver and islet modules. Similar to the partnership that is formed between Cdk4 or Cdk6 and the D cyclins, Cdk2 partners with either the E or A cyclins to mediate phosphorylation of retinoblastoma tumor suppressor protein (Rb1) at sites distinct from those phosphorylated by Cdk4 and Cdk6 (Cozar-Castellano et al. 2006). Remarkably, Cdk2 was uniquely included in the cell cycle module of the islets, suggesting that Cdk2-dependent phosphorylation of Rb1 may be a key regulatory step that mediates the strain difference in islet cell proliferation between B6 and BTBR mice. Rb1 is widely regarded as the molecular “brake” that controls transition from G1 into S phase of cellular growth (Cozar-Castellano et al. 2006). Once relieved of Rb1-dependent inhibition, a family of E2F transcription factors mediates coordinate regulation of gene expression that is required for cellular replication. However, it is important to note that beta-cell specific ablation of Rb1 does not lead to pancreatic beta-cell mass or glucose homeostatic phenotypes, suggesting that other factors (e.g., the additional pocket proteins, Rbl1 or Rbl2) can compensate to achieve growth arrest in the absence of Rb1 (Vasavada et al. 2007).
E2f1 and E2f2 were found in soleus and liver, whereas E2f8 was in adipose and islet cell cycle modules. E2f7 was found exclusively in the islet cell cycle module. Our results reveal key molecular components of the cell cycle regulatory machinery that form expression pathways in most of the tissues profiled. Some of these components are uniquely contained in islet pathways and may play a critical role in islet cell proliferation during aging or under the stimulus evoked by obesity-induced insulin resistance.
The cell cycle module results also predicted an obesity-dependent increase in adipose cell proliferation in both mouse strains. The prediction was tested in vivo and, again, the results are entirely consistent with the cell cycle module data; both mouse strains showed an obesity-dependent increase in adipose cell proliferation.
What is the link between obesity and an increase in islet cellular proliferation? One possibility is expression and secretion of mitogenic factors from peripheral insulin-resistant tissues that circulate in the blood and stimulate beta-cells to proliferate, a mechanism supported by transplantation studies (Flier et al. 2001). Perhaps B6 mice express a beta-cell mitogenic signal in peripheral tissues and this signal is missing or not functional in BTBR mice.
To search for possible candidates for these factors, we identified genes in insulin target tissues with expression profiles highly correlated to the islet cell cycle regulatory module and whose products were putative secreted peptides. Several candidates were identified, including Ngf, Igf2, Igf2bp1, Gdf10, and Bmp1.
Several studies have provided compelling evidence that these molecules play a critical role in regulating beta-cell mass. Widespread (Petrik et al. 1999) or local (Devedjian et al. 2000; Okamoto et al. 2006) overexpression of Igf2 has been shown to promote islet cell hyperplasia, and application of Igf2 to beta-cells in culture induces proliferation (Milo-Landesman and Efrat 2002). Recent genome-wide association studies have identified genetic variants in IGF2BP2 that are associated with type 2 diabetes in human patients (Saxena et al. 2007; Scott et al. 2007; Zeggini et al. 2007). While distinct from Igf2bp2, Igf2bp1 functions similarly to Igf2bp2 to stabilize Igf2 mRNA, resulting in increased synthesis and secretion of Igf2 protein. Removal of Ngf from isolated islets in culture induces apoptosis (Pierucci et al. 2001). Finally, recent work has shown that Bmp3, a member of the bone morphogenetic protein ligand super-family that includes Gdf10 and Bmp1, may play a critical role in regulating beta-cell mass. Bmp3 knockout mice have decreased islet Mki67-positive immunoreactivity, reduced beta-cell mass, and increased random-fed blood glucose, suggesting a role for Bmp3 in regulating beta-cell proliferation (Lee et al. 2007).
Recent evidence suggests that beta-cell replication is the primary means by which animals increase beta-cell mass during adulthood and under conditions of islet regeneration (Dor et al. 2004). All beta-cells have equal capacity to replicate (Brennand et al. 2007) and this replicative capacity requires functional Ccnd2 (Georgia and Bhushan 2004). Mice harboring beta-cell specific ablations of the transcript factor Foxo1 (Okamoto et al. 2006) or Insr (Okada et al. 2007) fail to show islet cell hyperplasia in response to severe insulin resistance. Taken together, these data strongly support a model whereby the expansion of beta-cell mass in response to insulin resistance is due to replication of preexisting beta-cells. Our finding of obesity-dependent differences in the expression of genes critical for cell cycle regulation in isolated islets, coupled with our direct in vivo measure of islet cellular proliferation, corroborates this model.
These studies provide the scientific community with the first gene expression network model of type 2 diabetes across multiple tissues. In addition, it provides a large database inferring intra- and inter-tissue connections between gene expression modules across a wide array of cellular functions. The modules can be broadly used to make predictions about the regulation of numerous pathways as we did with the cell cycle module.
Methods
Animals
All mice used in this study were male, bred from our in-house colonies at the University of Wisconsin Biochemistry Department, and housed in an environmentally controlled facility on a 12-h light/dark cycle (6 am–6 pm, respectively). Mice were provided free access to water at all times and to a standard rodent chow (Purina no. 5008) ab libitum, except during a fasting period (8 am–noon) in order to obtain plasma at 4 or 10 wk of age, after which they were sacrificed by decapitation. For each animal, the following tissues were collected in order: left lateral lobe of the liver, hypothalamus, right gonadal fat pad (adipose), pancreas, soleus, and gastrocnemius. For the two muscle tissues, both the right and left were combined for gene expression profiling. All tissues, except the pancreas, were flash-frozen in liquid nitrogen. All procedures were approved by the University of Wisconsin Animal Care and Use Committee.
Materials
Collagenase Type XI, RIA-grade BSA, dextrose, and Ficoll Type 400-DL were purchased from Sigma. Hanks’ balanced salt solution was from GIBCO. RNeasy Mini Kit was from Qiagen. DEPC-treated water was from Ambion.
Plasma measurements
Glucose was measured by the glucose oxidase method using commercially available kits (Sigma-Aldrich). For lean mice, insulin was measured by radioimmunoassay (RIA; RI-13K, Linco Research). For ob mice, insulin was measured by an in-house developed ELISA using a pair of anti-insulin/proinsulin antibodies (clones D6C4 and D3E7-BT) purchased from Research Diagnostics. Briefly, 96-well high-binding plates (Corning) were coated (50 μL/well) overnight with 3 μg/mL of D6C4. After removal of D6C4, plates were blocked with PBS containing 4% RIA-grade BSA (Sigma) for 1 h (100 μL/well) and then incubated for 1 h with insulin standards (Linco Research, 0.1–10 ng/mL), whole plasma or whole pancreas extract (25 μL/well). D3E7-BT (25 μL/well) and 1 μg/mL in PBS/1% BSA were added, gently mixed, and incubated for an additional hour. After washing each well three times (50 mM Tris, 0.2% Tween-20, pH 8.0), 1 μg/mL of streptavidin-HRP (Pierce) in PBS/0.1% BSA was added (50 μL/well) and incubated for 30 min. Following an additional three washes, 16 μmol/mL of o-phenylenediamine (Sigma), dissolved in citrate buffer (0.1 M citrate-phosphate, 0.03% H2O2 at pH 5.0), was added (50 μL/well) and incubated for 30 min, followed by an equal volume of 0.18 M sulfuric acid to quench the reaction. Absorbance at 492 nm was determined by a plate reader (Ultra 384 TECAN). Insulin contents in plasma were calculated by comparison to known standards. Adiponectin, PAI-1, and resistin were determined by commercial ELISA services at Linco Research.
Islet isolation and RNA purification
Intact pancreatic islets were isolated from mice using a collagenase digestion procedure as previously described (Rabaglia et al. 2005). Islets were carefully hand-picked under a stereo microscope to remove contaminating acinar tissue, after which the islets were washed twice with phosphate buffered saline (PBS) and centrifuged at 2500 rpm, 5 min, 4°C. The PBS supernatant was removed and 200 μL RLT buffer (Qiagen) was added. Islets were homogenized by hand for 1 min with a plastic micro-pestel (USA Scientific) and stored at −80°C until RNA purification. RNA was purified using the Qiagen RNeasy Mini Kit, according to manufacturer directions. An Agilent Bioanalyzer 2100 was used to assess RNA quality for all islet samples, which typically showed a 28/18S ratio of ∼1.5 or greater.
RNA isolation from non-islet tissues, gene expression profiling, data normalization, and GO term enrichment analysis
RNA preparations (liver, muscles, adipose, and hypothalamus) and all array hybridizations were performed at Rosetta Inpharmatics (Merck & Co.). The custom ink-jet microarrays used in this study were manufactured by Agilent Technologies and consisted of 4732 control probes and 39,558 noncontrol oligonucleotides extracted from mouse Unigene clusters and combined with RefSeq sequences and RIKEN full-length cDNA clones. Mouse tissues were homogenized and total RNA extracted using Trizol reagent (Invitrogen) according to manufacturer's protocol. Total RNA was reverse-transcribed and labeled with either Cy3 or Cy5 flurochrome. For a given strain, labeled complementary RNA (cRNA) from each animal of that strain was hybridized against a pool of labeled cRNAs constructed from equal aliquots of RNA from all of the animals for that strain (over both time points). All hybridizations were performed in fluor-reversal for 48 h in a hybridization chamber, washed, and scanned using a confocal laser scanner. Arrays were quantified on the basis of spot intensity relative to background, adjusted for experimental variation between arrays using average intensity over multiple channels, and fitted to a previously described error model to determine significance (type I error) (He et al. 2003). Gene expression measures are reported as the ratio of the mean log10 intensity (mlratio). Gene expression data that were used for the trait-gene correlations were generated using the ratio splitter pairwise ratio builder function in Resolver 6.0 (Rosetta Biosoftware) to account for the strain-specific reference pools. This pipeline allows the creation of new experiments based on comparisons of intensity channels from existing ratio hybridizations without having to prepare new hybridizations. The ratio-splitting operation which generates intensity profiles includes error modeling of the channels of the ratio scan, group normalization, forward transformation of intensities, group de-trending, and inverse transformations. Experiments are then rebuilt by ratioing each sample to a new baseline value, represented here as a super-pool (average of all array hybridizations in the experiment). The statistical significance of the overlap between input sets from the co-expression networks and GO biological process gene sets was assessed using the hypergeometric distribution and a multiple test correction (Bonferroni).
In vivo islet proliferation measurement
The proliferation rate of islet cells was measured using a recently developed heavy water (2H2O) labeling technique (Busch et al. 2004; Shankaran et al. 2006, 2007). Briefly, the incorporation of deuterium (2H) from 2H2O into the deoxyribose moiety of deoxyribonucleotides in cells replicating their DNA is measured by gas chromatography/mass spectrometry (GC/MS). To rapidly attain stable 2H2O enrichment in body water, mice were given an IP injection of 2H2O in 0.9% NaCl at 6 wk of age. The volume, V (mL), of the IP injection was calculated for each animal according to the formula for lean and ob animals, respectively, Vlean = 0.03 × body weight (gm) and Vob = 0.015 × body weight (gm). On the same day of the IP injection, mice were placed on drinking water containing 8% 2H2O for a period of 2 wk ad libitum. Mice were sacrificed at 8 wk of age, at which time plasma was collected and islets and adipose tissue isolated as described above. This procedure yielded average 2H2O enrichment in plasma of 5.7% ± 0.5% in all mice except BTBR ob where 7.8% ± 0.5% enrichment was achieved, owing to polyuria, in turn due to hyperglycemia, in these diabetic mice. Hind legs were collected in order to determine the 2H2O enrichment in the DNA of bone marrow, a cellular population considered to have completely, or nearly completely, turned over during the 2-wk labeling period (Neese et al. 2002). DNA was extracted from islets, whole adipose tissue, or bone marrow using Qiagen DNeasy tissue kits (Qiagen Inc.) and hydrolyzed to deoxyribonucleosides. The deoxyribose moiety of purine deoxyribonucleosides was then converted to the pentafluorobenzyl triacetate derivative by reaction with excess pentafluorobenzyl hydroxylamine under acidic conditions, followed by acetylation with acetic anhydride. GC/MS analysis was performed in negative chemical ionization mode using an Agilent model 5973 mass spectrometer and a 6890 gas chromatograph fitted with a db-225 column. Selected ion monitoring was performed on ions with mass-to-charge ratios (m/z) 435 and 436. Incorporation of 2H into purine deoxyribose was quantified as the molar excess fraction M1 (EM1), correcting for injected amount of material as described (Neese et al. 2002).The fraction of newly replicated cells in islet or adipose was calculated as the ratio of the 2H-enrichment for each tissue to that observed in bone marrow.
In vivo adipose proliferation measurement
Measurement of adipose cell proliferation was performed as described above for islets. The whole epididymal adipose tissue was removed and frozen. Genomic DNA was isolated and 2H enrichment was determined by GC/MS as above.
Identification of differentially expressed (DE) genes
To classify genes into differential expression patterns, we used an empirical Bayes hierarchical modeling approach called EBarrays (Newton et al. 2001; Kendziorski et al. 2006; Yuan and Kendziorski 2006), which is implemented in R, a publicly available statistical analysis environment (R Development Core Team 2005) and available through Bioconductor (www.bioconductor.org). EBarrays describes the probability distribution of a set of expression measurements. It accounts generally for differences among genes in their true underlying expression levels, measurement fluctuations, and distinct expression patterns for a given gene among cell types or conditions. An expression pattern is an arrangement of a gene’s true underlying intensities (μ) in each condition. The number of patterns possible depends on the number of conditions from which the expression measurements were obtained. For example, when measurements are taken from two conditions, two patterns of expression are possible: equivalent expression (μ1 = μ2) and differential expression (μ1≠μ2). Given the four conditions within each strain (4 or 10 wk; lean or obese), 15 expression patterns are possible (Supplemental Table S1). Since we do not know a priori which genes are in which patterns, the marginal distribution of the data is a mixture over the possible patterns with model parameters determined by the full set of array data. In this way, the approach utilizes information across a set of arrays to optimize model fit and is thus more efficient than a number of methods that make gene inferences one gene at a time. Posterior probabilities for each of the 15 patterns are calculated for every transcript and used for transcript classification. For each tissue, a transcript is assigned to the expression pattern with maximum posterior probability (MPP). Differentially expressed (DE) transcripts are defined as those with MPP > 0.7 (MPP > 0.5 for hypothalamus) in at least one mouse strain. For a given threshold, FDR is estimated by averaging the posterior probabilities of equivalent expression for each transcript on the list (Newton et al. 2006).
Identification of co-expression modules
We used a previously developed method to identify transcript co-expression modules (Zhang and Horvath 2005). For transcripts identified as DE by EBarrays, an adjacency matrix was constructed. Each entry in the matrix is the absolute value of Pearson’s correlation, adjusted so that the overall network is approximately scale-free. Connection strength between two genes (xi and xj) in the network was determined according to the adjacency function, aij = |cor(xi,xj)|β, using the estimated power parameter β, resulting in a weighted network (Zhang and Horvath 2005). We note that this allows for all correlations to be used, unlike approaches that invoke arbitrary thresholds. For a discussion of the advantages of weighted vs. unweighted networks, see Zhang and Horvath (2005) and references therein. The 8000 most connected transcripts were used in the topological overlap matrix (TOM) calculation, and 1 – TOM was used as a distance matrix in the hierarchical clustering of the transcripts for module identification. When there were fewer than 8000 DE transcripts in a particular tissue (adipose, soleus, and hypothalamus), all were used for module identification. We found that clusters were robust to more stringent thresholds of 0.8 or 0.9 (MPP) for DE transcript identification (Supplemental Fig. S7).
Partial correlation-based networks
A Gaussian graphical modeling framework was used for gene-gene network construction (Schafer and Strimmer 2005a, b). Briefly, the method assumes a linear relationship among variables that can be described by a multivariate normal distribution. In this setting, the partial correlation (PaCor) matrix completely prescribes dependence relationships among variables since a nonzero PaCor between two variables indicates conditional dependence given all other variables; and a zero PaCor indicates that the variables are conditionally independent. More precisely, given (X1, X2, . . . , Xn), the partial correlation between X1 and X2 is defined as the correlation of X1r and X2r where Xir denotes the residuals obtained after regressing Xi upon (X3, . . . , Xn) (i = 1,2). In contrast to Pearson’s correlation coefficient between two variables, which can be high if those two variables are both related to a third variable, the PaCor quantifies the “direct” correlation between two variables since effects from all other variables are “adjusted” for, or more specifically regressed away. Significant PaCor’s were identified as previously described (Schafer and Strimmer 2005a), with FDR controlled at 0.005. For each mouse strain, 1000 simulations of multivariate normal data were generated for 20 mice and 107 nodes, using the strain-specific empirical covariance matrix, to verify that FDR was well-controlled and to evaluate power and the positive predictive value (percent of correctly detected edges) for each network.
Acknowledgments
We thank Karl Broman, Roger Davis, Wes Pike, and Victoria Browning for their critical comments on earlier versions of the manuscript; Yuerong Zhu for construction of the hyperlinked figures and database management; and personnel at the Gene Expression Laboratory at Rosetta for carrying out all of the gene expression profiling. This work was supported by the National Institute of Diabetes and Digestive Kidney Diseases grants 58037 and 66369 (A.D.A.), the National Institute of General Medical Sciences grants 76274 (C.K.), 69430, and PA-02-110 (B.S.Y.), Brazil CNPq (E.C.N.), a Hatch Grant from the College of Natural Resources, University of California at Berkeley and grant Bio 04-10445 from the UC Discovery-BioStar program (M.K.H.) and from in-kind support from Rosetta Inpharmatics and KineMed, Inc.
Footnotes
[Supplemental material is available online at www.genome.org. Primary expression data for all arrays used in this study have been submitted to Gene Expression Omnibus under accession no. GSE10785.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.074914.107.
References
- Brennand K., Huangfu D., Melton D., Huangfu D., Melton D., Melton D. All beta cells contribute equally to islet growth and maintenance. PLoS Biol. 2007;5:e163. doi: 10.1371/journal.pbio.0050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcelin R., Mrejen C., Decaux J.F., De Mouzon S.H., Girard J., Charron M.J., Mrejen C., Decaux J.F., De Mouzon S.H., Girard J., Charron M.J., Decaux J.F., De Mouzon S.H., Girard J., Charron M.J., De Mouzon S.H., Girard J., Charron M.J., Girard J., Charron M.J., Charron M.J. In vivo and in vitro regulation of hepatic glucagon receptor mRNA concentration by glucose metabolism. J. Biol. Chem. 1998;273:8088–8093. doi: 10.1074/jbc.273.14.8088. [DOI] [PubMed] [Google Scholar]
- Busch R., Cesar D., Higuera-Alhino D., Gee T., Hellerstein M.K., McCune J.M., Cesar D., Higuera-Alhino D., Gee T., Hellerstein M.K., McCune J.M., Higuera-Alhino D., Gee T., Hellerstein M.K., McCune J.M., Gee T., Hellerstein M.K., McCune J.M., Hellerstein M.K., McCune J.M., McCune J.M. Isolation of peripheral blood CD4+ T cells using RosetteSep and MACS for studies of DNA turnover by deuterium labeling. J. Immunol. Methods. 2004;286:97–109. doi: 10.1016/j.jim.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Carlson M.R., Zhang B., Fang Z., Mischel P.S., Horvath S., Nelson S.F., Zhang B., Fang Z., Mischel P.S., Horvath S., Nelson S.F., Fang Z., Mischel P.S., Horvath S., Nelson S.F., Mischel P.S., Horvath S., Nelson S.F., Horvath S., Nelson S.F., Nelson S.F. Gene connectivity, function, and sequence conservation: Predictions from modular yeast co-expression networks. BMC Genomics. 2006;7:40. doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozar-Castellano I., Fiaschi-Taesch N., Bigatel T.A., Takane K.K., Garcia-Ocana A., Vasavada R., Stewart A.F., Fiaschi-Taesch N., Bigatel T.A., Takane K.K., Garcia-Ocana A., Vasavada R., Stewart A.F., Bigatel T.A., Takane K.K., Garcia-Ocana A., Vasavada R., Stewart A.F., Takane K.K., Garcia-Ocana A., Vasavada R., Stewart A.F., Garcia-Ocana A., Vasavada R., Stewart A.F., Vasavada R., Stewart A.F., Stewart A.F. Molecular control of cell cycle progression in the pancreatic beta-cell. Endocr. Rev. 2006;27:356–370. doi: 10.1210/er.2006-0004. [DOI] [PubMed] [Google Scholar]
- Devedjian J.C., George M., Casellas A., Pujol A., Visa J., Pelegrin M., Gros L., Bosch F., George M., Casellas A., Pujol A., Visa J., Pelegrin M., Gros L., Bosch F., Casellas A., Pujol A., Visa J., Pelegrin M., Gros L., Bosch F., Pujol A., Visa J., Pelegrin M., Gros L., Bosch F., Visa J., Pelegrin M., Gros L., Bosch F., Pelegrin M., Gros L., Bosch F., Gros L., Bosch F., Bosch F. Transgenic mice overexpressing insulin-like growth factor-II in beta cells develop type 2 diabetes. J. Clin. Invest. 2000;105:731–740. doi: 10.1172/JCI5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y., Brown J., Martinez O.I., Melton D.A., Brown J., Martinez O.I., Melton D.A., Martinez O.I., Melton D.A., Melton D.A. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Flier S.N., Kulkarni R.N., Kahn C.R., Kulkarni R.N., Kahn C.R., Kahn C.R. Evidence for a circulating islet cell growth factor in insulin-resistant states. Proc. Natl. Acad. Sci. 2001;98:7475–7480. doi: 10.1073/pnas.131192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargalovic P.S., Imura M., Zhang B., Gharavi N.M., Clark M.J., Pagnon J., Yang W.P., He A., Truong A., Patel S., Imura M., Zhang B., Gharavi N.M., Clark M.J., Pagnon J., Yang W.P., He A., Truong A., Patel S., Zhang B., Gharavi N.M., Clark M.J., Pagnon J., Yang W.P., He A., Truong A., Patel S., Gharavi N.M., Clark M.J., Pagnon J., Yang W.P., He A., Truong A., Patel S., Clark M.J., Pagnon J., Yang W.P., He A., Truong A., Patel S., Pagnon J., Yang W.P., He A., Truong A., Patel S., Yang W.P., He A., Truong A., Patel S., He A., Truong A., Patel S., Truong A., Patel S., Patel S., et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc. Natl. Acad. Sci. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgia S., Bhushan A., Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J. Clin. Invest. 2004;114:963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A., Doss S., Zhang B., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Doss S., Zhang B., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Zhang B., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Wang S., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Plaisier C., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Castellanos R., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Brozell A., Schadt E.E., Drake T.A., Lusis A.J., Schadt E.E., Drake T.A., Lusis A.J., Drake T.A., Lusis A.J., Lusis A.J., et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluzik M., Haluzikova D., Haluzikova D. The role of resistin in obesity-induced insulin resistance. Curr. Opin. Investig. Drugs. 2006;7:306–311. [PubMed] [Google Scholar]
- He Y.D., Dai H., Schadt E.E., Cavet G., Edwards S.W., Stepaniants S.B., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Dai H., Schadt E.E., Cavet G., Edwards S.W., Stepaniants S.B., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Schadt E.E., Cavet G., Edwards S.W., Stepaniants S.B., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Cavet G., Edwards S.W., Stepaniants S.B., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Edwards S.W., Stepaniants S.B., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Stepaniants S.B., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Duenwald S., Kleinhanz R., Jones A.R., Shoemaker D.D., Kleinhanz R., Jones A.R., Shoemaker D.D., Jones A.R., Shoemaker D.D., Shoemaker D.D., et al. Microarray standard data set and figures of merit for comparing data processing methods and experiment designs. Bioinformatics. 2003;19:956–965. doi: 10.1093/bioinformatics/btg126. [DOI] [PubMed] [Google Scholar]
- Horvath S., Zhang B., Carlson M., Lu K.V., Zhu S., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z., Zhang B., Carlson M., Lu K.V., Zhu S., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z., Carlson M., Lu K.V., Zhu S., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z., Lu K.V., Zhu S., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z., Zhu S., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z., Felciano R.M., Laurance M.F., Zhao W., Qi S., Chen Z., Laurance M.F., Zhao W., Qi S., Chen Z., Zhao W., Qi S., Chen Z., Qi S., Chen Z., Chen Z., et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc. Natl. Acad. Sci. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendziorski C.M., Chen M., Yuan M., Lan H., Attie A.D., Chen M., Yuan M., Lan H., Attie A.D., Yuan M., Lan H., Attie A.D., Lan H., Attie A.D., Attie A.D. Statistical methods for expression quantitative trait loci (eQTL) mapping. Biometrics. 2006;62:19–27. doi: 10.1111/j.1541-0420.2005.00437.x. [DOI] [PubMed] [Google Scholar]
- Lee N.K., Sowa H., Hinoi E., Ferron M., Ahn J.D., Confavreux C., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Sowa H., Hinoi E., Ferron M., Ahn J.D., Confavreux C., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Hinoi E., Ferron M., Ahn J.D., Confavreux C., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Ferron M., Ahn J.D., Confavreux C., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Ahn J.D., Confavreux C., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Confavreux C., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Dacquin R., Mee P.J., McKee M.D., Jung D.Y., Mee P.J., McKee M.D., Jung D.Y., McKee M.D., Jung D.Y., Jung D.Y., et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Osborne M.C., Monia B.P., Bhanot S., Gaarde W.A., Reed C., She P., Jetton T.L., Demarest K.T., Osborne M.C., Monia B.P., Bhanot S., Gaarde W.A., Reed C., She P., Jetton T.L., Demarest K.T., Monia B.P., Bhanot S., Gaarde W.A., Reed C., She P., Jetton T.L., Demarest K.T., Bhanot S., Gaarde W.A., Reed C., She P., Jetton T.L., Demarest K.T., Gaarde W.A., Reed C., She P., Jetton T.L., Demarest K.T., Reed C., She P., Jetton T.L., Demarest K.T., She P., Jetton T.L., Demarest K.T., Jetton T.L., Demarest K.T., Demarest K.T. Reduction in glucagon receptor expression by an antisense oligonucleotide ameliorates diabetic syndrome in db/db mice. Diabetes. 2004;53:410–417. doi: 10.2337/diabetes.53.2.410. [DOI] [PubMed] [Google Scholar]
- Milo-Landesman D., Efrat S., Efrat S. Growth factor-dependent proliferation of the pancreatic beta-cell line betaTC-tet: An assay for beta-cell mitogenic factors. Int. J. Exp. Diabetes Res. 2002;3:69–74. doi: 10.1080/15604280212526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Anazawa Y., Matsui K., Fukuda S., Nakamura Y., Arakawa H., Anazawa Y., Matsui K., Fukuda S., Nakamura Y., Arakawa H., Matsui K., Fukuda S., Nakamura Y., Arakawa H., Fukuda S., Nakamura Y., Arakawa H., Nakamura Y., Arakawa H., Arakawa H. Cyclin K as a direct transcriptional target of the p53 tumor suppressor. Neoplasia. 2002;4:268–274. doi: 10.1038/sj.neo.7900235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neese R.A., Misell L.M., Turner S., Chu A., Kim J., Cesar D., Hoh R., Antelo F., Strawford A., McCune J.M., Misell L.M., Turner S., Chu A., Kim J., Cesar D., Hoh R., Antelo F., Strawford A., McCune J.M., Turner S., Chu A., Kim J., Cesar D., Hoh R., Antelo F., Strawford A., McCune J.M., Chu A., Kim J., Cesar D., Hoh R., Antelo F., Strawford A., McCune J.M., Kim J., Cesar D., Hoh R., Antelo F., Strawford A., McCune J.M., Cesar D., Hoh R., Antelo F., Strawford A., McCune J.M., Hoh R., Antelo F., Strawford A., McCune J.M., Antelo F., Strawford A., McCune J.M., Strawford A., McCune J.M., McCune J.M., et al. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc. Natl. Acad. Sci. 2002;99:15345–15350. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M.A., Kendziorski C.M., Richmond C.S., Blattner F.R., Tsui K.W., Kendziorski C.M., Richmond C.S., Blattner F.R., Tsui K.W., Richmond C.S., Blattner F.R., Tsui K.W., Blattner F.R., Tsui K.W., Tsui K.W. On differential variability of expression ratios: Improving statistical inference about gene expression changes from microarray data. J. Comput. Biol. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- Newton M., Wang P., Kendziorski C., Wang P., Kendziorski C., Kendziorski C. Hierarchical mixture models for expression profiles. In: Do K.-A., et al., editors. Bayesian inference for gene expression and proteomics. Cambridge University Press; New York: 2006. pp. 40–52. [Google Scholar]
- Okada T., Liew C.W., Hu J., Hinault C., Michael M.D., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Liew C.W., Hu J., Hinault C., Michael M.D., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Hu J., Hinault C., Michael M.D., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Hinault C., Michael M.D., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Michael M.D., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Krtzfeldt J., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Yin C., Holzenberger M., Stoffel M., Kulkarni R.N., Holzenberger M., Stoffel M., Kulkarni R.N., Stoffel M., Kulkarni R.N., Kulkarni R.N. Insulin receptors in beta-cells are critical for islet compensatory growth response to insulin resistance. Proc. Natl. Acad. Sci. 2007;104:8977–8982. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Hribal M.L., Lin H.V., Bennett W.R., Ward A., Accili D., Hribal M.L., Lin H.V., Bennett W.R., Ward A., Accili D., Lin H.V., Bennett W.R., Ward A., Accili D., Bennett W.R., Ward A., Accili D., Ward A., Accili D., Accili D. Role of the forkhead protein FoxO1 in beta cell compensation to insulin resistance. J. Clin. Invest. 2006;116:775–782. doi: 10.1172/JCI24967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik J., Pell J.M., Arany E., McDonald T.J., Dean W.L., Reik W., Hill D.J., Pell J.M., Arany E., McDonald T.J., Dean W.L., Reik W., Hill D.J., Arany E., McDonald T.J., Dean W.L., Reik W., Hill D.J., McDonald T.J., Dean W.L., Reik W., Hill D.J., Dean W.L., Reik W., Hill D.J., Reik W., Hill D.J., Hill D.J. Overexpression of insulin-like growth factor-II in transgenic mice is associated with pancreatic islet cell hyperplasia. Endocrinology. 1999;140:2353–2363. doi: 10.1210/endo.140.5.6732. [DOI] [PubMed] [Google Scholar]
- Pierucci D., Cicconi S., Bonini P., Ferrelli F., Pastore D., Matteucci C., Marselli L., Marchetti P., Ris F., Halban P., Cicconi S., Bonini P., Ferrelli F., Pastore D., Matteucci C., Marselli L., Marchetti P., Ris F., Halban P., Bonini P., Ferrelli F., Pastore D., Matteucci C., Marselli L., Marchetti P., Ris F., Halban P., Ferrelli F., Pastore D., Matteucci C., Marselli L., Marchetti P., Ris F., Halban P., Pastore D., Matteucci C., Marselli L., Marchetti P., Ris F., Halban P., Matteucci C., Marselli L., Marchetti P., Ris F., Halban P., Marselli L., Marchetti P., Ris F., Halban P., Marchetti P., Ris F., Halban P., Ris F., Halban P., Halban P., et al. NGF-withdrawal induces apoptosis in pancreatic beta cells in vitro. Diabetologia. 2001;44:1281–1295. doi: 10.1007/s001250100650. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2005. [Google Scholar]
- Rabaglia M.E., Gray-Keller M.P., Frey B.L., Shortreed M.R., Smith L.M., Attie A.D., Gray-Keller M.P., Frey B.L., Shortreed M.R., Smith L.M., Attie A.D., Frey B.L., Shortreed M.R., Smith L.M., Attie A.D., Shortreed M.R., Smith L.M., Attie A.D., Smith L.M., Attie A.D., Attie A.D. Alpha-ketoisocaproate-induced hypersecretion of insulin by islets from diabetes-susceptible mice. Am. J. Physiol. Endocrinol. Metab. 2005;289:E218–E224. doi: 10.1152/ajpendo.00573.2004. [DOI] [PubMed] [Google Scholar]
- Rane S.G., Dubus P., Mettus R.V., Galbreath E.J., Boden G., Reddy E.P., Barbacid M., Dubus P., Mettus R.V., Galbreath E.J., Boden G., Reddy E.P., Barbacid M., Mettus R.V., Galbreath E.J., Boden G., Reddy E.P., Barbacid M., Galbreath E.J., Boden G., Reddy E.P., Barbacid M., Boden G., Reddy E.P., Barbacid M., Reddy E.P., Barbacid M., Barbacid M. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat. Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- Roduit R., Morin J., Masse F., Segall L., Roche E., Newgard C.B., Assimacopoulos-Jeannet F., Prentki M., Morin J., Masse F., Segall L., Roche E., Newgard C.B., Assimacopoulos-Jeannet F., Prentki M., Masse F., Segall L., Roche E., Newgard C.B., Assimacopoulos-Jeannet F., Prentki M., Segall L., Roche E., Newgard C.B., Assimacopoulos-Jeannet F., Prentki M., Roche E., Newgard C.B., Assimacopoulos-Jeannet F., Prentki M., Newgard C.B., Assimacopoulos-Jeannet F., Prentki M., Assimacopoulos-Jeannet F., Prentki M., Prentki M. Glucose down-regulates the expression of the peroxisome proliferator-activated receptor-alpha gene in the pancreatic beta-cell. J. Biol. Chem. 2000;275:35799–35806. doi: 10.1074/jbc.M006001200. [DOI] [PubMed] [Google Scholar]
- Rorsman P., Berggren P.O., Bokvist K., Ericson H., Mohler H., Ostenson C.G., Smith P.A., Berggren P.O., Bokvist K., Ericson H., Mohler H., Ostenson C.G., Smith P.A., Bokvist K., Ericson H., Mohler H., Ostenson C.G., Smith P.A., Ericson H., Mohler H., Ostenson C.G., Smith P.A., Mohler H., Ostenson C.G., Smith P.A., Ostenson C.G., Smith P.A., Smith P.A. Glucose-inhibition of glucagon secretion involves activation of GABAA-receptor chloride channels. Nature. 1989;341:233–236. doi: 10.1038/341233a0. [DOI] [PubMed] [Google Scholar]
- Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Hirschhorn J.N., Daly M.J., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Schafer J., Strimmer K., Strimmer K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 2005a;4 doi: 10.2202/1544-6115.1175. Article 32. http://www.bepress.com/sagmb/vol4/iss1/art32. [DOI] [PubMed] [Google Scholar]
- Schafer J., Strimmer K., Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005b;21:754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., Stringham H.M., Chines P.S., Jackson A.U., Chines P.S., Jackson A.U., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran M., King C., Lee J., Busch R., Wolff M., Hellerstein M.K., King C., Lee J., Busch R., Wolff M., Hellerstein M.K., Lee J., Busch R., Wolff M., Hellerstein M.K., Busch R., Wolff M., Hellerstein M.K., Wolff M., Hellerstein M.K., Hellerstein M.K. Discovery of novel hippocampal neurogenic agents by using an in vivo stable isotope labeling technique. J. Pharmacol. Exp. Ther. 2006;319:1172–1181. doi: 10.1124/jpet.106.110510. [DOI] [PubMed] [Google Scholar]
- Shankaran M., Marino M.E., Busch R., Keim C., King C., Lee J., Killion S., Awada M., Hellerstein M.K., Marino M.E., Busch R., Keim C., King C., Lee J., Killion S., Awada M., Hellerstein M.K., Busch R., Keim C., King C., Lee J., Killion S., Awada M., Hellerstein M.K., Keim C., King C., Lee J., Killion S., Awada M., Hellerstein M.K., King C., Lee J., Killion S., Awada M., Hellerstein M.K., Lee J., Killion S., Awada M., Hellerstein M.K., Killion S., Awada M., Hellerstein M.K., Awada M., Hellerstein M.K., Hellerstein M.K. Measurement of brain microglial proliferation rates in vivo in response to neuroinflammatory stimuli: Application to drug discovery. J. Neurosci. Res. 2007;85:2374–2384. doi: 10.1002/jnr.21389. [DOI] [PubMed] [Google Scholar]
- Tsutsui T., Hesabi B., Moons D.S., Pandolfi P.P., Hansel K.S., Koff A., Kiyokawa H., Hesabi B., Moons D.S., Pandolfi P.P., Hansel K.S., Koff A., Kiyokawa H., Moons D.S., Pandolfi P.P., Hansel K.S., Koff A., Kiyokawa H., Pandolfi P.P., Hansel K.S., Koff A., Kiyokawa H., Hansel K.S., Koff A., Kiyokawa H., Koff A., Kiyokawa H., Kiyokawa H. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27Kip1 activity. Mol. Cell. Biol. 1999;19:7011–7019. doi: 10.1128/mcb.19.10.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasavada R.C., Cozar-Castellano I., Sipula D., Stewart A.F., Cozar-Castellano I., Sipula D., Stewart A.F., Sipula D., Stewart A.F., Stewart A.F. Tissue-specific deletion of the retinoblastoma protein in the pancreatic beta-cell has limited effects on beta-cell replication, mass, and function. Diabetes. 2007;56:57–64. doi: 10.2337/db06-0517. [DOI] [PubMed] [Google Scholar]
- Vieira E., Salehi A., Gylfe E., Salehi A., Gylfe E., Gylfe E. Glucose inhibits glucagon secretion by a direct effect on mouse pancreatic alpha cells. Diabetologia. 2007;50:370–379. doi: 10.1007/s00125-006-0511-1. [DOI] [PubMed] [Google Scholar]
- Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Rosenbaum M., Leibel R.L., Ferrante A.W., Leibel R.L., Ferrante A.W., Ferrante A.W. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt A., Birnir B., Buschard K., Gromada J., Salehi A., Sewing S., Rorsman P., Braun M., Birnir B., Buschard K., Gromada J., Salehi A., Sewing S., Rorsman P., Braun M., Buschard K., Gromada J., Salehi A., Sewing S., Rorsman P., Braun M., Gromada J., Salehi A., Sewing S., Rorsman P., Braun M., Salehi A., Sewing S., Rorsman P., Braun M., Sewing S., Rorsman P., Braun M., Rorsman P., Braun M., Braun M. Glucose inhibition of glucagon secretion from rat alpha-cells is mediated by GABA released from neighboring beta-cells. Diabetes. 2004;53:1038–1045. doi: 10.2337/diabetes.53.4.1038. [DOI] [PubMed] [Google Scholar]
- Wierup N., Richards W.G., Bannon A.W., Kuhar M.J., Ahren B., Sundler F., Richards W.G., Bannon A.W., Kuhar M.J., Ahren B., Sundler F., Bannon A.W., Kuhar M.J., Ahren B., Sundler F., Kuhar M.J., Ahren B., Sundler F., Ahren B., Sundler F., Sundler F. CART knock out mice have impaired insulin secretion and glucose intolerance, altered beta cell morphology and increased body weight. Regul. Pept. 2005;129:203–211. doi: 10.1016/j.regpep.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Wierup N., Bjorkqvist M., Kuhar M.J., Mulder H., Sundler F., Bjorkqvist M., Kuhar M.J., Mulder H., Sundler F., Kuhar M.J., Mulder H., Sundler F., Mulder H., Sundler F., Sundler F. CART regulates islet hormone secretion and is expressed in the beta-cells of type 2 diabetic rats. Diabetes. 2006;55:305–311. doi: 10.2337/diabetes.55.02.06.db04-1383. [DOI] [PubMed] [Google Scholar]
- Xu H., Barnes G.T., Yang Q., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Barnes G.T., Yang Q., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Yang Q., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Nichols A., Ross J.S., Tartaglia L.A., Ross J.S., Tartaglia L.A., Tartaglia L.A., et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Kendziorski C., Kendziorski C. A unified approach for simultaneous gene clustering and differential expression identification. Biometrics. 2006;62:1089–1098. doi: 10.1111/j.1541-0420.2006.00611.x. [DOI] [PubMed] [Google Scholar]
- Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., Perry J.R., Rayner N.W., Freathy R.M., Rayner N.W., Freathy R.M., Freathy R.M., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Horvath S., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. http://www.bepress.com/sagmb/vol4/iss1/art17. [DOI] [PubMed] [Google Scholar]