Abstract
Continuous growth and development in plants are accomplished by meristems, groups of undifferentiated cells that persist as stem cells and initiate organs. While the structures of the apical and floral meristems in dicotyledonous plants have been well described, little is known about the underlying molecular mechanisms controlling cell proliferation and differentiation in these structures. We have shown previously that the CLAVATA1 (CLV1) gene in Arabidopsis encodes a receptor kinase-like protein that controls the size of the apical and floral meristems. Here, we show that KAPP, a gene encoding a kinase-associated protein phosphatase, is expressed in apical and young floral meristems, along with CLV1. Overexpression of KAPP mimics the clv1 mutant phenotype. Furthermore, CLV1 has kinase activity: it phosphorylates both itself and KAPP. Finally, KAPP binds and dephosphorylates CLV1. We present a model where KAPP functions as a negative regulator of the CLAVATA1 signal transduction pathway.
Keywords: meristem development, protein kinase, signal transduction, negative regulator
Flowering plant development is characterized by the establishment and maintenance of shoot apical meristems (SAMs). These are structures, usually at the growing tips of the plants, that contain slowly dividing stem cells in the center that are surrounded by more mitotically active cells that produce organ primordia (1). In Arabidopsis, the SAM is initiated during embryonic growth and then lies dormant until germination. After germination, the SAM initiates leaves, branches, and floral meristems in succession. The floral meristems then produce sepal, petal, stamen, and carpel primordia. The SAM and floral meristems have a similar structure, but differ in the types of organs initiated, in the phyllotactic pattern of organ initiation, and in ultimate meristem fate. SAMs initiate primordia in a spiral pattern and retain indeterminate growth and meristematic activity. In contrast, floral meristems produce organs in a whorled pattern and are determinate—ceasing growth after the initiation of carpels. Therefore, the control of cell proliferation is developmentally regulated, and in addition, cell division patterns result from communication between cells (2).
Mutations have been isolated that disrupt meristem structure and/or function in Arabidopsis. Among these, mutations in the CLAVATA1 (CLV1) and CLAVATA3 (CLV3) loci result in plants with enlarged apical and floral meristems, indicating a defect in the control of cell proliferation versus differentiation in these meristems (3–7). On the basis of double mutant phenotypes and interactions of clv1 and clv3 alleles, the CLV1 and clv3 gene products are thought to function in the same pathway (7). A third gene, WUSCHEL (WUS; ref. 8), has mutant phenotypes opposite those of and epistatic to clv1, suggesting that WUS may encode a downstream component in the CLV1 pathway.
CLV1 encodes a putative receptor kinase similar in organization and sequence to the Arabidopsis RLK5 protein, which was isolated by its sequence similarity to protein kinases but is of unknown function (9, 10). Both proteins contain an N-terminal signal sequence followed by 21 tandem leucine-rich repeats, a membrane-spanning region, and a putative cytoplasmic serine/threonine kinase domain. The CLV1 gene is specifically expressed in the interior layers of the apical and floral meristems but its RNA is not detected in organ primordia (9). Therefore, CLV1 may receive a signal that regulates cell proliferation in the meristematic regions.
The RLK5 kinase domain has been shown to autophosphorylate (11) and interact with a type 2C protein phosphatase, called kinase-associated protein phosphatase (KAPP; ref. 12). Because CLV1 and RLK5 are closely related members of a larger gene family (9), we tested if KAPP may act in the CLV1 pathway. Here we show that the KAPP RNA expression domain includes the region of CLV1 RNA expression and that constitutive KAPP expression mimics clv1 mutant phenotypes. Furthermore, KAPP interacts with the phosphorylated form of the CLV1 kinase domain, and KAPP can dephosphorylate CLV1 in vitro. These data suggest that KAPP may function as a negative regulator of the CLV1 signal transduction pathway, as well as in other signaling pathways.
MATERIALS AND METHODS
In Situ Hybridization.
The entire KAPP ORF was amplified by PCR using KAPP-1 (ATGGCGATGATAGGGATGAAC) and KAPP-6 (TTACAGGGAAGTATCGAAATC) as primers and ligated into a TA cloning vector (Invitrogen). A single plasmid, called pKAPP16, was sequenced to determine the orientation of the insert. Antisense KAPP RNA probe was synthesized using Sp6 RNA polymerase on an EcoRV-linearized template. Synthesis of the CLV1 probe was as described in Clark et al. (9). Tissue was fixed, sectioned, hybridized, washed, and exposed following the procedure of Drews et al. (13) with modifications by Sakai et al. (14).
Construction of Transgenic Plants.
The KAPP gene was amplified from a cDNA library using the following oligonucleotides with BamHI sites at the ends: SKAPP1, CGGGATCCAGAGAAGCGAAGACGATGGCG; and SKAPP2, CGGGATCCCCTAACATTACAGGGAAGTATC. The resulting PCR product was digested with BamHI and cloned into pGEM-7zf (Promega). The sequenced insert was transferred, using BamHI, to the cauliflower mosaic virus 35S promoter-containing vector pCGN18 constructed by Leslie Sieburth (15). A sense construct was identified, transformed into Agrobacterium strain ASE, and transformed into Arabidopsis thaliana ecotype Landsberg-erecta using the vacuum infiltration method (16). Transgenic seedlings were selected on Murashige/Skoog (Sigma)+kanamycin plates and transferred to soil. The resulting line was called 35S::KAPP.
Construction and Purification of Fusion Proteins.
Various regions of CLV1 and KAPP were expressed as glutathione S-transferase (GST) fusion proteins. The kinase domain of CLV1 (corresponding to amino acids 657–980; GenBank accession number U96879) was used to make the CKD construct. The kinase interaction domain (KI; amino acids 50–337 of KAPP; see ref. 12) and the KI domain plus the protein phosphatase domain (KAPP; amino acids 50–582) of the KAPP cDNA were amplified by PCR using the following oligonucleotides with BamHI sites engineered at each 5′ end: KD1, CGGGATCCGTAGCGATTCGTCAGATGAA; KD4, CGGGATCCAATAGAACGAACCCAGTCTTG; KID1, CGGGATCCGTGGGGGATTTGCAGAGACCA; KID2, CGGGATCCTTAAGCCCCAGGAAGCGGCCAC; and SKAPP2, as above.
The PCR products were digested with BamHI and ligated into the BamHI site of pGEX-2TK, which encodes GST, a thrombin cleavage site, and a heart muscle kinase site upstream and in frame with the BamHI site (Pharmacia). The clv1-1 mutation, which results in a glycine to aspartic acid change at position 856 in the CLV1 amino acid sequence (9), was introduced into GST-CKD to make the GST-CKD1-1 construct and the conserved lysine (at position 720 of the CLV1 amino acid sequence) was altered to a glutamic acid to make the GST-CKDK720E construct. All mutant constructs were made using the Chameleon Double Stranded Site Directed Mutagenesis kit following the manufacturer’s instructions (Stratagene). All plasmids were verified by sequencing, and transformed into Escherichia coli strain ADA 94 (Novagen) for protein expression.
GST-fusion proteins were isolated using a modified protocol from Chang et al. (17). E. coli cells were grown in 200 ml of Luria–Bertani medium to approximately an OD600 0.8 and then induced with 0.1 mM isopropyl β-d-thiogalactopyranoside for 2–3 hr at 37°C. The induced cells were harvested by centrifugation, resuspended in 10 ml of lysis buffer containing 10 mM Tris (pH 7.5), 150 mM NaCl, 1 mM DTT, 0.2% Tween 20 with 10 μg/ml aprotinin, 10 μg/ml leupeptin, 0.50 mg/ml Pefabloc SC (Boehringer Mannheim), and 10 μg/ml pepstatin as protease inhibitors, lysed by sonication, and separated into soluble and insoluble fractions by centrifugation. The soluble GST-fusion proteins were purified from the supernatant using Glutathione Sepharose 4B (Pharmacia) following the manufacturer’s instructions.
CLV1 Autophosphorylation and Interactions with KAPP.
The GST-CKD, GST-CKD1-1, and GST-CKDK720E fusion proteins were assayed for autophosphorylation by incubation in 30 μl of 20 mM Tris (pH7.5), 100 mM NaCl, 12 mM MgCl2 with 10 μCi of [γ-32P]ATP for 1 hr at room temperature. Samples were boiled in SDS loading buffer and analyzed by SDS/PAGE. Coomassie blue-stained gels were dried and exposed to film. CLV1 transphosphorylation was assessed by mixing CKDK720E protein, removed from GST by thrombin cleavage, with GST-CKD in an autophosphorylation reaction. CKDK720E-dependent inhibition of GST-CKD autophosphorylation was demonstrated by removing the CKDK720E protein from GST by thrombin cleavage, then adding increasing amounts of CKDK720E to GST-CKD during autophosphorylation reactions. Phosphoamino acid analysis was performed as described by Boyle et al. (18).
GST-KI was phosphorylated with heart muscle kinase (Sigma) in 20 mM Tris (pH 7.5), 100 mM NaCl, 12 mM MgCl2 with 10 μCi (1 Ci = 37 GBq) of [γ-32P]ATP, and then eluted from the matrix by washing with 20 mM Tris (pH 7.5), 100 mM NaCl, 12 mM MgCl2, and 20 mM glutathione. The resulting supernatant was used to probe various GST fusion proteins bound to nitrocellulose filters as described by Stone et al. (12). Phosphorylation dependence of the CLV1/KAPP interactions was tested by dephosphorylating immobilized GST-CKD with 20 units of alkaline phosphatase (AP) with and without 100 mM PO4− as an inhibitor of AP. GST-CKD was washed after phosphatase treatment and incubated with cold ATP in a standard autophosphorylation reaction. CLV1-dependent KAPP phosphorylation was assessed by adding GST-KAPP to GST-CKD, GST-CKD1-1, and GST-CKDK720E during a standard autophosphorylation reaction.
GST-KAPP-dependent dephosphorylation of GST-CKD was tested using a modified protocol from Stone et al. (12). The GST-CKD protein, still bound to the Glutathione Sepharose 4B matrix, was allowed to autophosphorylate in the presence of [γ-32P]ATP, then washed three times in 20 mM Tris (pH7.5), 0.1% 2-mercaptoethanol. Labeled GST-CKD, still immobilized on the matrix, was incubated with and without GST-KAPP in 20 mM Tris (pH 7.5), 0.1% 2-mercaptoethanol with either 10 mM EDTA or 10 mM MgCl2/10 mM MnCl2 for 2 hr at room temperature.
RESULTS
KAPP Expression Patterns.
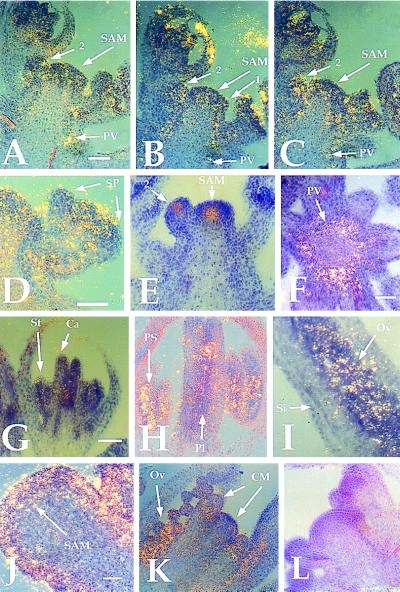
We determined the pattern of KAPP mRNA expression by in situ hybridization. Because KAPP does not appear to cross-hybridize to any other Arabidopsis DNA sequences at low stringency (R.W.W. and E.M.M., data not shown), the entire KAPP ORF was used to make an antisense RNA probe that was hybridized to 8 μm sections of wild-type and clv1-4 inflorescence tissue. In wild-type tissue, KAPP was expressed throughout the SAM and stage 1–2 flowers (19) but appeared to be excluded from the sepal primordia of stage 3 flowers (Fig. 1 A–D). CLV1 is expressed in a much more restricted domain, the central cells of the SAM and young floral meristems (ref. 9; Fig. 1E). Therefore, the CLV1 expression domain is encompassed within that of KAPP. Below the SAM, KAPP RNA was detected in the provascular tissue, but not differentiated vascular or pith tissue (Fig. 1 A–C, F). After stage 5 of flower development, KAPP RNA levels appeared to decrease with only weak signal detected in the stamen and carpel primordia (Fig. 1G). Later in flower development, KAPP was expressed in the developing pollen sacs and ovules (Fig. 1H). KAPP expression was maintained during ovule development (Fig. 1I).
Figure 1.
Expression of KAPP in wild-type and clv1-4 tissue. Labeled KAPP and CLV1-specific RNA probes were used for hybridization to sections of inflorescence meristem and floral tissue. (A–C) Serial sections of wild type tissue showing KAPP RNA is detected throughout the apical and young floral meristems and down the provascular tissue. (D) Section of a wild-type stage 3 flower where KAPP expression is reduced in sepal primordia. (E) CLV1 mRNA expression in the center of the apical and young floral meristems only. CLV1 is not expressed in stage 1 flowers. (F) Transverse section of the stem, below the SAM, showing the punctate ring of KAPP expression in the provascular tissue. (G) KAPP weak expression in the stamen and carpel primordia of a stage 7 flower; (H) KAPP expression in the pollen sacs and carpels of a stage 9 flower; (I) KAPP expression in the developing ovules. (J) KAPP is expressed throughout the enlarged SAM of a clv1-4 mutant. (K) KAPP is expressed in the central meristematic regions in mature clv1-4 flowers. (L) A KAPP sense probe hybridized to a wild-type inflorescence section. All photos are combined bright-field and dark-field exposures, with signal indicated by yellow grains. PV, provascular; SP, sepal primordia; St, stamen, Ca, carpel; PS, pollen sac; Pl, placenta; Ov, ovule; Si, silique wall; CM, central meristematic regions. Numbers indicate stages of floral development (all stages according to ref. 19). Bars = 40 μm.
KAPP has a similar expression pattern in clv1-4 tissue as it does in wild-type tissue. KAPP is expressed throughout the enlarged apical and floral meristems (Fig. 1J) as well as in the pollen sacs, ovules, and central proliferating dome of the mature flowers (Fig. 1K). This result indicates that clv1 mutations do not dramatically affect the level or the spatial regulation of KAPP expression. In situ hybridization of KAPP to clv3-2 tissue results in an RNA expression pattern similar to that in clv1-4 tissue (data not shown). All expression patterns are specific for KAPP and not due to background, as a KAPP sense probe gave very little signal (Fig. 1L).
KAPP Transgenic Studies.
To identify possible roles of KAPP in meristem and flower development, we constructed transgenic lines that express the full-length KAPP cDNA under control of the strong, constitutive 35S-cauliflower mosaic virus promoter (20). Two independent lines exhibited a dominant, heritable floral phenotype that cosegregated with the transferred DNA. Both lines appear similar, so only one line was analyzed in detail.
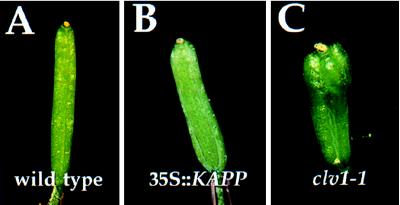
The 35S::KAPP line has a floral phenotype reminiscent of that caused by weak loss-of-function clv1 alleles (4). Flowers of the transgenic plant have a small but consistent increase in the number of carpels compared with wild type (Fig. 2 A–C). Wild type Landsberg-erecta plants have 2.0 ± 0.0 carpels per flower. On average, 35S::KAPP expressing plants have 0.30 ± 0.105 extra carpels per flower, and plants homozygous for the weak clv1-7 allele have 0.75 ± 0.143 extra carpels relative to wild type. The only other observed phenotypic alteration in the 35S::KAPP line was the presence of occasional petalloid stamens in the third whorl of the flowers (data not shown). Flowering time, apical dominance, and male and female fertility appear normal.
Figure 2.
35S::KAPP phenotype. A comparison of (A) wild-type, (B) 35S::KAPP, and (C) clv1-1 siliques. (A) A typical silique from a wild-type Landsberg-erecta flower is composed of two fused carpels. The 35S::KAPP line has on average 2.3 carpels per flower; panel (B) shows a silique comprised of three fused carpels. (C) clv1 mutants have extra carpels that fuse into a functional silique. This representative clv1-1 flower contains four carpels.
CLV1 Autophosphorylation.
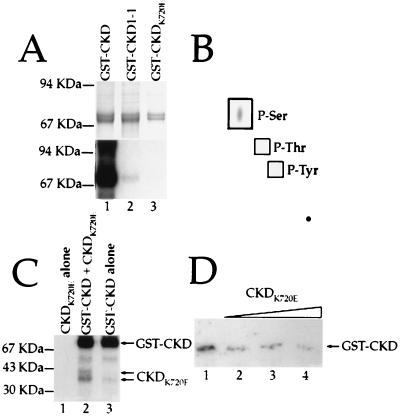
To test for CLV1 protein kinase activity, we expressed three different versions of the CLV1 kinase domain in E. coli. The kinase domain from CLV1 was used to make a GST fusion protein, called GST-CKD. Fusion protein was purified from E. coli and shown to autophosphorylate on serine residues (Fig. 3 A and B). This CLV1 kinase activity requires Mn2+ or Mg2+ (data not shown). The clv1-1 mutation results in a protein with an aspartic acid substituted for the first glycine in box VIII of the kinase domain (9, 21). We made the CLV1-1 form of the kinase domain fused to GST, GST-CKD1-1, and found that it is able to autophosphorylate but at greatly reduced levels (Fig. 3A). When the catalytic lysine residue was changed to glutamic acid (GST-CKDK720E), the resulting protein was unable to autophosphorylate (Fig. 3A).
Figure 3.
Autophosphorylation of the CLV1 kinase domain. (A) GST fusion-proteins containing the wild-type (lane 1), CLV1-1 (lane 2), and CKDK720E (lane 3) forms of the CLV1 kinase domain were allowed to phosphorylate in the presence of [γ-32P]ATP. (Upper) The Coomassie blue-stained gel showing the relative amounts of fusion protein in each lane. (Lower) The autoradiograph. (B) The wild-type CLV1 kinase domain was allowed to autophosphorylate, then hydrolyzed. The products of the hydrolysis reaction were analyzed by electrophoresis on a TLC plate. Only serine residues were phosphorylated. (C) Autophosphorylation reactions containing CKDK720E alone (lane 1), CKDK720E plus GST-CKD (lane 2), and GST-CKD alone. (D) Increasing amounts of CKDK720E protein were added to a constant amount of GST-CKD during autophosphorylation reactions. The approximate molar ratios of CKDK720E:GST-CKD are 0:1 (lane 1), 1:1 (lane 2), 2:1 (lane 3), and 4:1 (lane 4). Arrow shows the position of the full length GST-CKD protein.
Genetic evidence suggests that the CLV1 protein may function as an oligomer (4, 7). The closely related RLK5 kinase domain can undergo intermolecular phosphorylation (11), and the rate at which GST-CKD autophosphorylates appears to be dependent on the concentration of the fusion protein (data not shown), suggesting a second order reaction. We therefore tested the ability of CLV1 to transphosphorylate. The inactive CKDK720E protein, when removed from the GST-moiety by thrombin cleavage, could be phosphorylated in the presence of the active GST-CKD protein (Fig. 3C). Furthermore, GST-CKD autophosphorylation is reduced when incubated with increasing amounts of CKDK720E (Fig. 3D). This finding indicates that CLV1, like RLK5 (11), can interact intermolecularly and undergo transphosphorylation.
CLV1/KAPP Interactions.
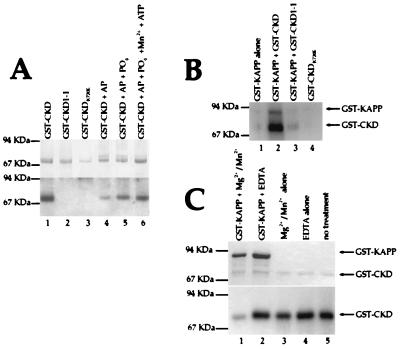
The region of the KAPP protein defined to be the KI domain (12) was also used to make a GST-fusion protein, called GST-KI. The KI protein could be labeled at an engineered heart muscle kinase site. The resulting GST-KI protein interacted with GST-CKD (Fig. 4A) in a filter binding assay. GST-KI did not bind to either GST-CKD1-1 or GST-CKDK720E at detectable levels (Fig. 4A), suggesting that the introduced mutations alter residues or structures that are required for the interaction, or that the CLV1/KAPP interaction is dependent on phosphorylation of the CLV1 kinase domain. To see if this interaction was dependent on the phosphorylation state of the CLV1 protein, we treated GST-CKD with AP alone or with AP plus PO4− (PO4− is an inhibitor of the phosphatase). GST-KI had a reduced interaction with the dephosphorylated GST-CKD as compared with phosphorylated GST-CKD (Fig. 4A). This interaction could be recovered by allowing the dephosphorylated GST-CKD protein to autophosphorylate (Fig. 4A).
Figure 4.
KAPP interactions with the CLV1 kinase domain. (A) GST-KI protein was labeled at the heart muscle kinase site with [γ-32P]ATP and incubated with a filter containing immobilized GST-CKD (lane 1), GST-CKD1-(lane 2), GST-CKDK720E (lane 3), GST-CKD treated with AP (lane 4), GST-CKD treated with AP in the presence of 100 mM PO4− (lane 5), and GST-CKD treated with AP, then allowed to autophosphorylate in the presence of MnCl2 and ATP (lane 6). (Upper) The Coomassie blue-stained gel showing the relative amounts of fusion protein in each lane. (Lower) The autoradiograph. (B) The GST-KAPP protein was incubated alone (lane 1) or with GST-CKD (lane 2), GST-CKD1-(lane 3), or GST-CKDK720E (lane 4) during an autophosphorylation reaction. Equal amounts of GST-CKD, GST-CKD1-1, and GST-CKDK720E were added in each reaction. (C) 32P-labeled GST-CKD was incubated with full-length GST-KAPP protein (lanes 1 and 2), or without GST-KAPP (lanes 3 and 4), and with either MgCl2/MnCl2 (lanes 1 and 3) or EDTA (lanes 2 and 4). Lane 5 is an untreated control. (Upper) Coomassie blue-stained gel showing the approximately equal loading of GST-CKD in each lane. (Lower) The autoradiogram.
The ability of the CLV1 kinase domain to phosphorylate KAPP was also tested. The GST-CKD protein was able to phosphorylate the full-length KAPP protein fused to GST (Fig. 4B). Consistent with their abilities to autophosphorylate, GST-CKD1-1 could phosphorylate GST-KAPP at reduced levels and the GST-CKDK720E protein could not phosphorylate GST-KAPP at any detectable level (Fig. 4B). GST-CKD protein does not phosphorylate GST alone (data not shown).
The KAPP protein had previously been shown to have protein phosphatase activity (12). To further investigate the interaction between the CLV1 kinase domain and KAPP, we tested KAPP’s ability to dephosphorylate GST-CKD in vitro. The phosphorylated GST-CKD protein was incubated with the GST-KAPP fusion protein in the presence and absence of divalent cations. The CKD protein is dephosphorylated by KAPP, and like other type 2C protein phosphatases, KAPP activity is dependent on Mg2+ and/or Mn2+ (Fig. 4C).
DISCUSSION
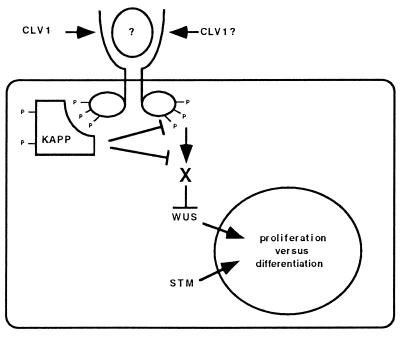
We have started to investigate the molecular mechanisms of CLV1 signal transduction. We have shown that the CLV1 kinase domain can undergo intermolecular autophosphorylation, which suggests that the active CLV1 protein may function as a multimer. The KAPP protein binds the phosphorylated form of CLV1 and exhibits phosphatase activity on the CLV1 protein. CLV1 is expressed in regions that also express KAPP, although the KAPP expression domain is much larger. Finally, transgenic plants overexpressing KAPP mimic the floral defects seen in weak clv1 mutant plants. These data taken together argue that KAPP may function as a negative regulator of the CLV1 signal transduction pathway. A model for the CLV1 signal transduction pathway is given in Fig. 5. Further experimentation will be needed to test the in vivo validity of this model, as the data are suggestive but not conclusive. In particular, KAPP may function in a process parallel to CLV1.
Figure 5.
Model of CLV1 and KAPP Action. The model predicts that CLV1, upon binding a peptide ligand, dimerizes and allows intermolecular phosphorylation of the cytoplasmic kinase domains. The activated receptor then binds and phosphorylates KAPP. The KAPP protein may then modify the CLV1 signal by directly dephosphorylating CLV1. The CLV1 kinase domain probably interacts with other positively acting components (X), which may or may not be dephosphorylated by KAPP. Based on epistasis analysis, WUS may function downstream of CLV1 to promote cell divisions. SHOOTMERISTEMLESS may function independently but competitively with the CLV1 signaling pathway to promote cell division (24).
Mechanisms of CLV1 Action.
The CLV1 biochemical activity may account for the previously observed phenotype seen in the clv1-1 mutant. The clv1-1 allele is predicted to encode a full-length protein with a Gly-856 to aspartic acid missense mutation in a conserved residue of subdomain VIII (9), which is thought to be involved in the recognition of protein substrates (21). clv1-1 homozygous plants have an intermediate phenotype indicating that it is not a null allele. Also, clv1-1/+ heterozygous plants have a weak mutant phenotype (4, 7), whereas the phenotypically stronger clv1-4 allele does not have a heterozygous mutant phenotype, indicating that the CLV1-1 protein may act in a dominant negative manner. This suggests that CLV1 may function as multimer.
The CLV1-1 form of the kinase domain can autophosphorylate, but only at low levels, consistent with clv1-1 not being a complete loss-of-function allele. We have also shown that the CLV1 kinase domain can transphosphorylate an inactive form of the protein. Furthermore, large amounts of the inactive (CKDK720E) kinase can inhibit the autophosphorylation of the wild-type protein. These data, consistent with the genetic data, argue that the CLV1 protein may function as a multimer. Like receptor-tyrosine kinases in animals (22), the CLV1 extracellular domain may inhibit dimerization, which is overcome by ligand binding. Upon oligomerization, the cytoplasmic domains may autophosphorylate and then recruit specific downstream components.
We have also shown that CLV1 is capable of phosphorylating KAPP, but it remains to be seen if CLV1 phosphorylates other substrates. As will be discussed below, KAPP is likely to be a negative regulator of the CLV1 pathway. No downstream components of the pathway have been identified, but epistasis analysis suggests the product of the WUS (8) gene as a candidate (Fig. 5).
Mechanisms of KAPP Action.
Signaling pathways, like many developmental mechanisms, need both positively acting components and negative regulators. Positively acting components are needed to transduce a signal to the nucleus for changes in gene expression or to the cytoskeleton for changes in cell shape. Negative regulators are needed to attenuate the signaling cascade and to repress the pathway when the signal is no longer present. The KAPP protein may function as a negative regulator of the CLV1 signal transduction pathway. The region of KAPP expression includes the CLV1 expression domain, showing that the two genes are at least expressed in the same cells. Moreover, high levels of KAPP gene expression can mimic clv1 mutants. Mutations in the CLV1 or CLV3 genes do not appear to affect the level of KAPP expression. So the similarity of clv1 and 35S::KAPP phenotypes is probably not the indirect result of increased KAPP expression in clv1 or clv3 plants.
More importantly, the KAPP protein binds to the phosphorylated form of the CLV1 kinase domain. The KAPP protein does not appear to merely bind phosphoserine and/or phosphothreonine, because KAPP does not interact with all kinases tested (23), so there must be other sequence requirements for KAPP function. However, KAPP can bind to denatured proteins and thus is not dependent on a proper structural conformation (Fig. 4A; see also ref. 12), suggesting that KAPP may recognize a short amino acid sequence. Because the KAPP protein exhibits phosphatase activity toward the CLV1 kinase domain, KAPP may function in vivo by directly dephosphorylating CLV1, thus preventing other downstream components from receiving the CLV1 signal. KAPP may dephosphorylate other components of the pathway as well. The KAPP protein is phosphorylated in the presence of CLV1, suggesting that the activity of KAPP may be modulated in response to the activated CLV1 receptor. However, our data suggest that phosphorylation of KAPP is not required for KAPP to dephosphorylate CLV1.
Other Roles for KAPP.
There are several lines of evidence indicating that KAPP may function in a number of different signaling pathways. First, KAPP interacts with RLK5, CLV1, and several other receptor-like kinases (23). RLK5 and CLV1 are members of a large family of leucine-rich repeats containing receptor kinases, of which there are members in Arabidopsis that are closely related to CLV1, and others closely related to RLK5 (ref. 9; R.W.W. and E.M.M., unpublished data). So far, no role has been ascribed to RLK5. Second, based on low stringency hybridization experiments and expressed sequence tag database searches, there does not appear to be another KAPP-like gene in the Arabidopsis genome (R.W.W. and E.M.M., unpublished data). It is therefore possible that KAPP is a promiscuous molecule that regulates a number of signaling pathways.
The expression data for KAPP supports such an idea. Previous work has shown that the KAPP mRNA is found in root and rosette tissue (12). In situ hybridization to inflorescence tissue shows that KAPP is expressed throughout the SAM and young floral primordia and in a punctate ring down the stem. CLV1 is expressed in a much smaller region of the SAM and floral meristems. Later in flower development, KAPP is expressed in the pollen sacs and in the placenta and ovules of the carpel, thus suggesting a role for KAPP in the formation of gametes.
In the 35S::KAPP lines, we did not observe a defect in female or male fertility. This may be due to higher endogenous levels of KAPP mRNA in these cells and/or a very sensitive requirement for lower levels of KAPP mRNA in the CLV1-expressing cells. Alternatively, the 35S promoter may not be active in these cells. There were also minor floral organ identity defects in the 35S::KAPP line, suggesting that KAPP can function in CLV1 nonexpressing cells. The absence of observable defects outside the flower indicates that KAPP overexpression does not affect all cell types and likely needs other factors to function. It is interesting to speculate that the relatively large, and expanding family of leucine-rich repeat-kinases in plants may play a variety of roles in communicating information needed for proper cell proliferation, elongation, and differentiation, and it is possible that KAPP may be an important negative regulator of a number of these kinases.
Acknowledgments
We thank Hajime Sakai for assistance with the in situ hybridizations, Doris Wagner for advice on protein purification, and Akiko Kumagai for assistance with the phosphoamino acid analysis. We acknowledge Ray Deshaies and Renny Feldman for discussions, comments, and reagents. We also thank Xuemei Chen, Bill Dunphy, Jennifer Fletcher, Jian Hua, Toshiro Ito, Steve Jacobsen, Carolyn Ohno, Jose-Luis Reichmann, Mark Running, Robert Sablowski, Hajime Sakai, Tom Tubman, and Eva Ziegelhoffer for careful review of this manuscript. This work was supported by National Science Foundation Grant MCB-9204839 and a Strategic Research Fund Grant from Zeneca Seeds (to E.M.M). R.W.W. was supported by National Institutes of Health Predoctoral Training Grant GM07616 and the Howard Hughes Medical Institute.
ABBREVIATIONS
- CLV1
CLAVATA1
- SAMs
shoot apical meristems
- WUS
WUSCHEL
- GST
glutathione S-transferase
- KAPP
kinase-associated protein phosphatase
- KI
kinase interaction
- CKD
CLV1 kinase domain
- AP
alkaline phosphatase
References
- 1.Steeves T A, Sussex I M. Patterns in Plant Development. New York: Cambridge Univ. Press; 1989. [Google Scholar]
- 2.Meyerowitz E M. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- 3.Leyser H M O, Furner I J. Development (Cambridge, UK) 1992;116:397–403. [Google Scholar]
- 4.Clark S E, Running M P, Meyerowitz E M. Development (Cambridge, UK) 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- 5.Crone W, Lord E M. Am J Bot. 1993;80:1419–1426. [Google Scholar]
- 6.Alvarez J, Smyth D R. In: Arabidopsis: An Atlas of Morphology and Development. Bowman J L, editor. New York: Springer; 1994. [Google Scholar]
- 7.Clark S E, Running M P, Meyerowitz E M. Development (Cambridge, UK) 1995;121:2057–2067. [Google Scholar]
- 8.Laux T, Mayer K F X, Berger J, Jurgens G. Development (Cambridge, UK) 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 9.Clark S E, Williams R W, Meyerowitz E M. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 10.Walker J C. Plant J. 1993;3:451–456. doi: 10.1111/j.1365-313x.1993.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 11.Horn M A, Walker J C. Biochim Biophys Acta. 1994;1208:65–74. doi: 10.1016/0167-4838(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 12.Stone J M, Collinge M A, Smith R D, Horn M A, Walker J C. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- 13.Drews G N, Bowman J L, Meyerowitz E M. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- 14.Sakai H, Medrano L J, Meyerowitz E M. Nature (London) 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- 15.Jack T, Fox G L, Meyerowitz E M. Cell. 1994;76:703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- 16.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 17.Chang C, Schaller G E, Patterson S E, Kwok S F, Meyerowitz E M, Bleecker A B. Plant Cell. 1992;4:1263–1271. doi: 10.1105/tpc.4.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle W J, van der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 19.Smyth D R, Bowman J L, Meyerowitz E M. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odell J T, Nagy F, Chua N-H. Nature (London) 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- 21.Hardie G, Hanks S. The Protein Kinase Facts Book. London: Academic; 1995. [Google Scholar]
- 22.Carpenter G. Annu Rev Biochem. 1987;56:881–914. doi: 10.1146/annurev.bi.56.070187.004313. [DOI] [PubMed] [Google Scholar]
- 23.Braun D M, Stone J M, Walker J C. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- 24.Clark S E, Jacobsen S E, Levin J, Meyerowitz E M. Development (Cambridge, UK) 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]