Abstract
Jasmonic acid and its precursors are potent regulatory molecules in plants. We devised a method for the simultaneous extraction of these compounds from plant leaves to quantitate changes in the levels of jasmonate family members during health and on wounding. During our study, we identified a novel 16-carbon cyclopentenoic acid in leaf extracts from Arabidopsis and potato. The new compound, a member of the jasmonate family of signals, was named dinor-oxo-phytodienoic acid. Dinor-oxo-phytodienoic acid was not detected in the Arabidopsis mutant fad5, which is incapable of synthesizing 7Z,10Z,13Z-hexadecatrienoic acid (16:3), suggesting that the metabolite is derived directly from plastid 16:3 rather than by β-oxidation of the 18-carbon 12-oxo-phytodienoic acid. Simultaneous quantitation of jasmonate family members in healthy leaves of Arabidopsis and potato suggest that different plant species have different relative levels of jasmonic acid, oxo-phytodienoic acid, and dinor-oxo-phytodienoic acid. We term these profiles “oxylipin signatures.” Dinor-oxo-phytodienoic acid levels increased dramatically in Arabidopsis and potato leaves on wounding, suggesting roles in wound signaling. Treatment of Arabidopsis with micromolar levels of dinor-oxo-phytodienoic acid increased the ability of leaf extracts to transform linoleic acid into the α-ketol 13-hydroxy-12-oxo-9(Z) octadecenoic acid indicating that the compound can regulate part of its own biosynthetic pathway. Tightly regulated changes in the relative levels of biologically active jasmonates may permit sensitive control over metabolic, developmental, and defensive processes in plants.
Keywords: fatty acids, jasmonic acid, phytodienoic acid, wounding
Jasmonates are potent biological regulators in the plant kingdom, playing diverse roles in healthy and wounded tissues (1–4). In particular, the jasmonate family of regulators is known to stimulate the expression of many wound-activated and defense-related genes (5–8). A role for jasmonic acid (JA) in defense against insects has been established (9, 10). The situation concerning potential roles for jasmonates in pathogenesis is less clear, but strong evidence for the roles of jasmonates in some pathosystems is emerging. For example, in Arabidopsis, jasmonates are involved in defense gene expression in response to the fungal pathogen Alternaria brassicola (11). Jasmonates are known to be involved in several developmental phenomena. For example, jasmonate synthesis and perception also appears to be essential for normal pollen development in Arabidopsis (12, 13). Additionally, JA and its precursor 12-oxo-phytodienoic acid (OPDA) stimulate the coiling response of Bryonia tendrils and are thought to be endogenous regulators of this phenomenon (14).
All known jasmonates (and almost all other fatty acid signals in plants) have been shown to derive from unsaturated octadecanoic acids. It is not known whether other unsaturated fatty acids can act as precursors for the biosynthesis of other regulators in plants. With the possibility that as yet undiscovered fatty acid signals exist in plants (4), we decided to develop a quantitative method for the simultaneous extraction of jasmonate family regulators. Using this method, we quantitated the levels of jasmonate family members in healthy and wounded leaves and by this means observed a novel wound-inducible fatty acid derivative. Its characterization led to the detection of a powerful hexadecanoid signal compound in plants and raised the possibility that other C16-derived signals exist in these organisms.
MATERIALS AND METHODS
Growth and Treatment of Plants.
Arabidopsis thaliana ecotype Columbia were grown under short day conditions [9 h light (150 μE m−2 s−1) at 23°C, 70% relative humidity]. Twelve-week-old plants were used for most experiments; however, 9-week-old plants were used in some experiments to compare wild-type with the fad5 Arabidopsis mutant (previously named fadB) (15). The apical halves of rosette leaves from 4-week-old plants were wounded with forceps and kept under light (150 μE m−2 s−1) for 90 min. Solanum tuberosum cv. Matilda was grown for 3 weeks from tubers under a 17 h light regime (200–300 μE m−2 s−1) at 20°C/75% relative humidity (light) and 16°C/65% relative humidity (dark). The leaflets of fully expanded leaves were wounded several times across the midvein using forceps (equivalent to wounding about 40% of the leaf surface) and then kept under light for 4 h.
Analytical Methods.
For analysis and quantitation of fatty acids, leaf material was harvested, weighed, and immediately frozen in liquid nitrogen. One gram of frozen leaves was ground in a mortar to a fine powder and added to 3 ml of ice-cold methanol containing 100 ng tetrahydro-12-oxo-PDA and 100 ng dihydrojasmonic acid (H2JA) as internal standards (below). The solution was immediately homogenized with a Polytron (Kinematica, Lucerne, Switzerland) for 1 min on ice. Extraction of the fatty acids was continued by rotating the samples in tubes for 2 h at 4°C. Ice-cold water (1.5 ml) was added and, after centrifugation, the supernatant was passed through a C18-SepPak classic cartridge (Waters), which had been prewashed with methanol followed by methanol/water (70:30, vol/vol). The column was washed with an additional 7 ml of methanol/water (70:30, vol/vol) and both the eluates were combined and concentrated to less than 1 ml under reduced pressure at 40°C in a rotary evaporator. The samples were adjusted to a volume of 5 ml with water, acidified with 0.3 ml of concentrated HCl, and extracted twice with 5 ml CHCl3. The CHCl3 phase was dried over anhydrous MgSO4 and evaporated under a stream of N2 at 35°C. Fatty acids were methylated with 450 μl of diazomethane in ether, dried under N2, and redissolved in 1 ml of hexane. The samples were loaded onto a silica-SepPak classic cartridge (Waters), which had been prewashed first with diethyl ether (10 ml) and then hexane (10 ml). The loaded column was washed with 5 ml of hexane, and the fatty acids were eluted with 7 ml of hexane/diethyl ether (2:1, vol/vol) (7). After evaporation of the solvent under N2, the samples were taken up in 10 μl of hexane and stored at −80°C. Fatty acids were analyzed by gas chromatography–mass spectrometry [Hewlett–Packard 5890 gas chromatograph linked to a Hewlett–Packard 5927 mass spectrometer, electron ionization mode, 70 eV electron potential, 11 psi column head pressure, HP-5MS column 30 m × 0.25 mm]. The temperature gradient was 100°C for 1 min, 100°C–160°C at 20°C/min, 160°C–238°C at 3°C/min, and 238°C–300°C at 30°C/min. Retention times were as follows: 3R,7R-jasmonic acid methyl ester, 8.98 min; methyl-3R,7R dihydrojasmonate, 9.05 min; 3R,7S-jasmonic acid methyl ester, 9.52 min; methyl cis-dnOPDA (dinor-oxo-phytodienoic acid), 20.6 min; trans-tetrahydro-OPDA (H4OPDA) methyl ester, 24.63 min; 9S,13S-OPDA methyl ester, 26.28 min. Quantitation was done using selective ion monitoring measuring ions m/z = 224 for methyl jasmonate isomers, m/z = 226 for methyl-dihydrojasmonate, m/z = 210 for methyl-dnOPDA, m/z = 240 for methyl-H4OPDA, and m/z = 238 for methyl-OPDA. H2JA was used as the internal standard for quantitation of jasmonic acid isomers. Because this compound was detected in broad bean (16), we first checked potato and Arabidopsis tissues to confirm its absence. H4OPDA was used for quantitation of dnOPDA and OPDA. 3R,7R-jasmonic acid and 9S,13S-OPDA standards were purchased from Cayman Chemicals (Ann Arbor, MI). H4OPDA was synthesized by hydrogenation of 9S,13R-OPDA. 9S,13R-OPDA (250 μg) was dissolved in 4.75 ml ethanol and 10 mg PtO2 was added. Reduction was performed in an H2 atmosphere for 3 h at room temperature. After removal of the catalyst by filtration, the solution was concentrated by rotary evaporation. H2JA was a generous gift from Patrick Schweizer (University of Fribourg). Response factors were established for the two internal standards as well as for the methyl esters of 3R,7S and 3R,7R-JA, 9S,13S-OPDA, and cis-dnOPDA. Recoveries of internal standards were as follows: 50% for H2JA and 65% for H4OPDA in potato and 52% for H2JA and 27% for H4OPDA in Arabidopsis. Reproducibility of recovery was (coefficients of variation, n = 15) 18.7% for H2JA and 23.9% for H4OPDA (potato) and 25.6% for H2JA and 33.3% for H4OPDA (Arabidopsis). Note that the extraction and measurement procedure does not allow us to distinguish between the free acid or methyl ester form of endogenous jasmonates although, from separate experiments, levels of endogenous methyl esters appear to be very low in comparison to free acids. 1H-NMR spectra were recorded on a Bruker AM-400. The sample (≈30 μg) was dissolved in CDCl3 (20 μl), sealed in a capillary tube, and placed in a 5-mm sample tube for NMR analysis. Two- or 3-hr collections were necessary to gain adequate signal-to-noise ratios. Chemical shifts are reported in relation to tetramethylsilane (δ 0.0), with the shift for residual CHCl3 observed at δ 7.26.
Purification of dnOPDA from Plant Tissues.
For the isolation of dnOPDA from potato, 67 g of leaf material was homogenized in a Waring blender with 120 ml of ice-cold buffer (10 mM Hepes, pH 7.0/150 mM NaCl). The homogenate was filtered through four layers of gauze and centrifuged at 15,000 × g for 15 min at 4°C. The supernatant was extracted with organic solvents according to the method of Clancy and Hugli (17). From the resulting chloroform phase the solvent was evaporated with N2 and the residue was taken up in ethanol. For purification of dnOPDA, the fatty acid extract was chromatographed by normal-phase HPLC on a Whatman Partisil 10 Magnum 9 column (9.4 × 250 mm) at a flow rate of 3 ml/min. The mobile phase consisted of a gradient formed from solvent A (hexane/acetic acid, 100:0.1) and solvent B (hexane/isopropanol/acetic acid, 70:30:0.1) as follows: 3% B from 0 to 10 min; 3% to 17% B from 10 to 30 min; 17% to 50% B from 30 to 50 min; 50% B from 50 to 55 min; 50% to 100% B from 55 to 60 min; 100% B from 60 to 70 min. Fractions were collected in 2 min intervals during the entire run. After evaporation of the solvents under nitrogen, each fraction was taken up in 400 μl of ethanol and aliquots analyzed by GCMS after derivatization with diazomethane. The fraction containing dnOPDA (fraction from 30 to 32 min) was further purified by normal-phase HPLC on a Whatman Partisil 5 column (4.6 x 250 mm) at a flow rate of 1 ml/min. The mobile phase consisted of a gradient formed from solvent A (hexane/acetic acid, 100:0.1) and solvent B (hexane/isopropanol/acetic acid, 70:30:0.1) as follows: 3% B from 0 to 10 min; 3% to 16.7% B from 10 to 70 min. The run was monitored at 240 nm and peaks were manually collected. After removing the solvents under a stream of N2, each fraction was redissolved in 50 μl of ethanol and an aliquot derivatized with diazomethane and subsequently analyzed by GCMS. Elution of dnOPDA occurred at 31.6 min.
Isolation of 7Z,10Z,13Z-Hexadecatrienoic Acid (16:3) from Potato Leaves.
Total lipids from fresh potato leaves were extracted (18). Monogalactosyldiacylglycerol (MGDG), the lipid class enriched in 16:3, was isolated by preparative thin-layer chromatography (TLC) as follows: about 2.5 mg of total lipids were separated on a silica–TLC (Kieselgel 60, Merck, 20 × 20 cm, 0.25 mm) with a mobile phase consisting of CHCl3/MeOH/ammonia/H2O, 65:35:2:3 (vol/vol). After development, a small portion of the plate was stained with iodine to visualize the MGDG band. The MGDG was scraped from the plate and eluted with CHCl3/MeOH, 1:1 (vol/vol). The solvent was evaporated under N2 and the residue was saponified with 2 ml of 1 M KOH in 95% (vol/vol) ethanol at room temperature. After acidification with HCl the fatty acids were extracted with diethyl ether, the solvent dried with anhydrous MgSO4 and evaporated under N2. Purification of 16:3 was by reversed-phase HPLC on a Whatman Partisil 10 ODS-3 Magnum 9 column (9.4 × 250 mm). The flow rate was 5 ml/min and the mobile phase consisted of a gradient formed from solvent A (H2O/trifluoroacetic acid, 100:0.1, vol/vol) and solvent B (acetonitrile/trifluoroacetic acid, 100:0.1, vol/vol) as follows: 60% B from 0 to 3 min, 60%–88% B from 3 to 17 min, 88%–100% B from 17 to 19 min, 100% B from 19 to 24 min. The column eluate was monitored at 210 nm; 16:3 eluted at 11.6 min.
Direct Enzymatic Preparation of dnOPDA from 16:3.
A flaxseed acetone powder extract (19) was prepared by the incubation of 1 g of the powder with 10 ml of 0.1 M sodium phosphate buffer (pH 6.3) at 4°C for 30 min. Insoluble material was removed by centrifugation. The supernatant was used as the enzyme source for the conversion of 16:3 to dnOPDA. Approximately 1.8 mg of 16:3 dissolved in 0.4 ml of ethanol was added to 40 ml of 50 mM sodium phosphate buffer (pH 6.3), followed by the addition of 1 ml of the flaxseed acetone powder extract. After 60 min, the reaction mixture was adjusted to pH 4 with 1 N HCl, passed through a C18-SepPak column (Waters), which had been prewashed with 5 ml each of methanol, diethyl ether, methanol, and water, respectively. The fatty acids were eluted from the column with 3 ml of diethyl ether, which was then dried with anhydrous MgSO4. The extract was concentrated under N2, then applied to 10 cm × 10 cm Kieselgel-60 F254 HP-TLC plates (Merck). The plates were developed once in hexane/diethyl ether/acetic acid solvent (70:30:1, vol/vol), and a second time in chloroform/acetic acid (100:1, vol/vol). The cyclopentenone product dnOPDA could be visualized as a dark band under fluorescent light. Other products were visualized by exposure of a small portion of the plate to iodine vapor. For recovery, the dnOPDA product was scraped from the plate and eluted with diethyl ether.
Effects of dnOPDA on Fatty Acid Metabolism in Arabidopsis.
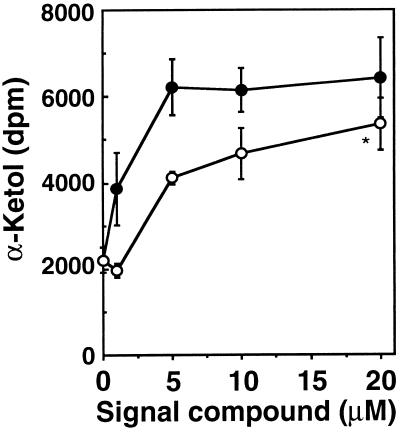
Arabidopsis (Columbia) seeds were surface sterilized and germinated and grown in liquid medium (pH 5.7) containing 1 g liter−1 Murashige and Skoog salts (Sigma) and 5 g liter−1 sucrose with 12 h light (150 μE m−2 s−1) at 24°C. Fourteen days after germination JA or dnOPDA were added in culture medium in varying concentrations up to 20 μM (see Fig. 3) and the plants incubated an additional 48 h. Leaves were crushed and leaf juice (1 μl) added directly to the assay, which contained [1-14C]linoleic acid (2.1 GBq mmol−1, 50,000 dpm per assay) in 40 mM 4-morpholinepropanesulfonic acid buffer (pH 7.0) containing 10% (vol/vol) glycerol. After a 15 min incubation at 25°C, the reaction was extracted and chromatographed as described (20). Radioactive bands were quantitated with a phosphoimager (Bio-Rad GS250). Production of the α-ketol [13-hydroxy-12-oxo-9(Z)-octadecenoic acid] was confirmed by gas chromatography mass spectroscopy (positive ion mode, 70 eV) after derivatization of the molecule with methoxyamine and diazomethane. A characteristic mass spectrum was obtained with a parent ion of m/z = 427 and fragments of 357 (20%), 173 (60%), and 73 (100%).
Figure 3.
Extracts from Arabidopsis plants treated with dnOPDA show an increased ability to convert linoleic acid to oxygenated metabolites including 13-hydroxy-12-oxo-9(Z)-octadecenoic acid. 3R,7R JA (○) or dnOPDA (•) were added at concentrations of 1, 5, 10, and 20 μM to Arabidopsis seedlings grown in liquid culture. Forty-eight hours after addition of dnOPDA or JA, leaf extracts were assayed for enzymatic conversion of [1-14C]linoleic acid to the α-ketol 13-hydroxy-12-oxo-9(Z)-octadecenoic acid. Values represent the mean ± SE of four independent experiments except for ∗, where n = 3.
RESULTS
A 16-Carbon Homologue of Oxo-Phytodienoic Acid.
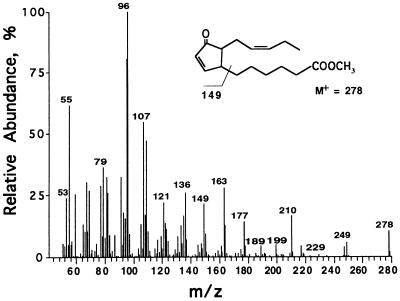
An extraction method allowing the simultaneous quantification of jasmonate family members was developed. The development of the procedure was influenced by a previously published method (7). We first compared gas chromatograph profiles of methylated fatty acid derivatives from wounded and unwounded Arabidopsis and potato leaves. Most wound-inducible peaks were identified by their mass spectra and/or by comparison with known compounds. However, an unidentified peak with a retention time between that of methyl jasmonate and methyl-OPDA was observed in extracts from both wounded Arabidopsis and wounded potato leaves. Fig. 1 shows the mass spectrum of the methylated compound. The compound showed a parent ion of m/z = 278 and principle fragments of m/z = 210 (due to McLafferty rearrangement), 163, 149, 107, 96, and 95. The similarity of the spectrum to that of methyl-OPDA (not shown) led us to speculate that the compound might be a 16-carbon homologue of OPDA.
Figure 1.
Identification of dnOPDA. Mass spectrum of methyl-dnOPDA isolated from potato leaves.
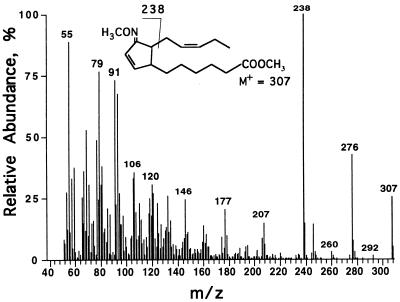
The 18-carbon compound OPDA can be produced in vitro from linolenic acid or its 13-hydroperoxide using a flaxseed acetone powder extract (19). To test whether the 16-carbon compound could be synthesized in vitro, we first purified the putative precursor 7Z,10Z,13Z-hexadecatrienoic acid (16:3). We used MGDG purified from potato leaves as a source of 16:3. The yield was estimated to be 0.28 mol of 16:3 per mol of MGDG. Products of incubating 16:3 with the flaxseed extract were separated by TLC. Three major products were observed, which migrated in positions corresponding to the homologous γ-ketol, OPDA, and α-ketol products of linolenic acid (18:3) metabolism. The compound that migrated similarly to OPDA had an absorbance maximum at 221 nm in ethanol (not shown), the same as that of OPDA. After methylation, the molecule showed the same mass spectrum as the molecule discovered in leaf extracts (Fig. 1). To further examine its structure, the molecule was methylated and derivatized with methoxyamine prior to mass spectrometry. This derivative (Fig. 2) showed a parent ion of m/z = 307 (26%) and fragments of m/z = 238 (M-C5H9, 100%) and 276 (M-OCH3, 43%). The fragmentation pattern was very similar to that of the methyloxime derivative of OPDA (21).
Figure 2.
Mass spectrum of the methyloxime derivative of methyl-dnOPDA.
An initial NMR spectrum of the compound synthesized in the flaxseed system showed a mixture of two isomers with the predominant isomer (≈60%) assigned as the cis orientation of the side chains. Subsequent NMR spectra showed increasing amounts of the trans isomer. The NMR spectra were essentially identical to the spectra previously reported for OPDA (22, 23) and these observations together with the mass spectra indicated that the compound is an OPDA homologue lacking two carbons in the alkanoic acid side chain. Based on the previous assignments for cis- and trans-OPDA and the observation of signal intensity changes due to epimerization during the course of our NMR analysis, the following chemical shift assignments have been made for the compound: δ 7.73 (H8, dd, J8,9 = 6.1 Hz, J7,8 = 2.4 Hz), 6.19 (H9, dd, J8,9 = 6.1 Hz, J7,9 = 2.4 Hz), 5.35–5.43 (H13, m, and H14, m), 2.98 (H7, m), 2.53 (H12a, m), 2.49 (H11, m), 2.37 (H2, t, J2,3 = 7.4 Hz), 2.14 (H12b, m), 1.75 (H6a, m), 1.18 (H6b, m), 0.97 (H16, t, J15,16 = 7.5 Hz). Several resonances were not resolved from interfering signals thought to derive from a trans epimer(s) of the compound and are tentatively assigned at: δ 2.04 (H15, m), 1.66 (H3, m), 1.25–1.50 (H4, m and H5, m). Overall the mass spectral and NMR data imply that the molecule is a 16-carbon homologue of OPDA. We chose the trivial name dinor-oxo-phytodienoic acid (which can be abbreviated dnOPDA). The IUPAC name is 6-[2′-(pent-2"-enyl)-3′-oxocyclopent-4′-en-1′-yl]hexanoic acid.
To establish that the in vitro synthesized compound was identical to the plant-derived compound, we purified the latter from potato leaves by HPLC. This compound and our in vitro synthesized compound displayed identical retention times upon gas chromatography and had identical mass spectra. OPDA is probably synthesized in vivo as the 9S,13S diastereomer (24), but in the flaxseed system the compound is produced as a racemic mixture of 9S,13S and 9R,13R isomers (22). Epimerization of cis-OPDA to trans-OPDA is easily achieved by heating the dried cis-epimer(s) (25) and these stereoisomers have different retention times during gas chromatography. All our samples of dnOPDA (those isolated from potato and Arabidopsis and those synthesized in the flaxseed system) showed one dominant peak on gas chromatography and we reasoned that the compound might represent one or both cis enantiomers of dnOPDA. We synthesized dnOPDA in the flaxseed system and subjected 2 μg to identical heat treatment. Analysis of the compound by gas chromatography revealed quantitative conversion to a peak with a retention time 1 min earlier than the parent compound and we interpret this to indicate epimerization of cis- to trans-dnOPDA. By analogy to OPDA, our data are consistent with the generation of 7S,11R-dnOPDA from a 7S,11S parent form, which we suppose is the epimer produced in vivo, although this has to be experimentally confirmed.
To investigate possible biosynthetic origins for dnOPDA we measured the levels of this molecule in Arabidopsis wild-type and the fad5 mutant, which does not accumulate 16:3 (15, 26, 27), the putative precursor for dnOPDA. Table 1 shows that dnOPDA was undetectable in both healthy and wounded fad5 plants, unlike in the wild type, where the compound was easily detected. Levels of JA stereoisomers in wild-type and mutant plants were similar, but the fad5 plants had OPDA levels of 23.12 ng g−1 fresh weight (fwt) in unwounded leaves, whereas the OPDA level in unwounded wild-type leaves was 343.4 ng g−1 fwt.
Table 1.
Levels of jasmonate family members in unwounded and wounded leaves of wild-type and fad5 Arabidopsis plants
| Compound | Wild type
|
fad5 mutant
|
||
|---|---|---|---|---|
| Unwounded | Wounded | Unwounded | Wounded | |
| dnOPDA | 18.81 ± 8.50 | 86.98 ± 17.37 | 0 | 0 |
| 9S,13S-OPDA | 343.41 ± 65.98 | 820.84 ± 203.95 | 23.12 ± 6.04 | 836.3 ± 231.5 |
| 3R,7S and 3R,7R JA | 36.65 ± 5.72 | 1,426 ± 228.1 | 25.52 ± 1.94 | 1,022 ± 72.76 |
The apical half of leaves were wounded 90 min prior to harvest and extraction of the entire leaf. The content of dnOPDA, OPDA, and JA was measured. Values are in ng⋅g−1 fwt ± SE (n = 4 for all measurements except those with unwounded fad5 plants, where n = 5). The detection limit was approximately 1 ng⋅g−1 fwt for all compounds.
Extracts from dnOPDA-Treated Arabidopsis Show an Increased Ability to Oxidize Linoleic Acid.
The in vitro production of dnOPDA allowed the generation of sufficient quantities of this substance to permit a preliminary study of its biological effects. Because methyl jasmonate is known to enhance the level of mRNA encoding lipoxygenase 2 in Arabidopsis (28) we reasoned that it should be possible to measure an increase in AtLOX2 activity, directly or indirectly, in JA-treated Arabidopsis plants. We incubated leaf extracts from this plant with the lipoxygenase substrate [1-14C]linoleic acid and were able to detect increased production of its metabolite, 12,13 α-ketol, generated by the action of lipoxygenase and allene oxide synthase (3). Fig. 3 shows that low micromolar levels of JA and dnOPDA increase the ability of Arabidopsis leaf extracts to generate α-ketol from linoleic acid. However, dnOPDA appeared to be more active than JA. The biological activity of OPDA was not tested in this system.
Quantitation of dnOPDA in Healthy and Wounded Leaves.
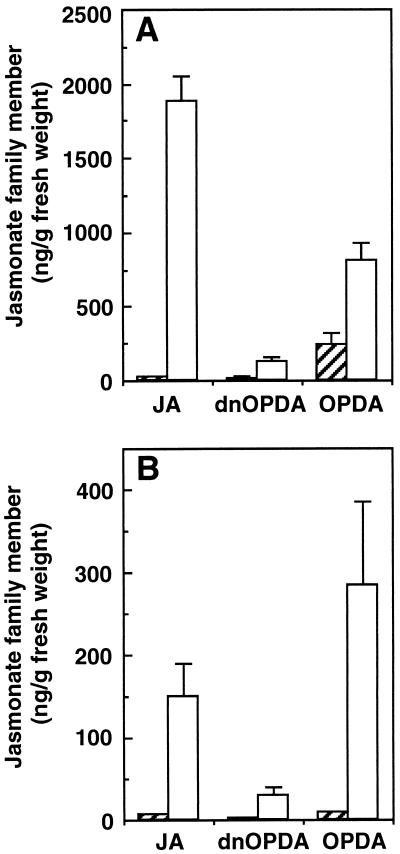
Using our procedure for the quantitative extraction of jasmonate family members we estimated levels of the compounds JA, OPDA, and dnOPDA in unwounded and wounded Arabidopsis and potato leaves (Fig. 4). In Arabidopsis, cis-OPDA (presumably in the 9S,13S conformation) was the dominant measurable signal in unwounded leaves. On wounding, the dominant measurable compound was JA. In Arabidopsis, wounding increased dnOPDA levels approximately 9-fold from a resting level of 14.40 ± 6.24 ng g−1 fwt to a level of 128.10 ± 27.69 ng g−1 fwt in wounded leaves. In potato, OPDA and JA were the most abundant jasmonate family signals in healthy leaves, but after wounding potato leaves cis-OPDA became the dominant signal. Wounding potato leaves caused a 14-fold increase in dnOPDA levels from 2.01 ± 0.014 ng g−1 fwt to 29.03 ± 10.68 ng g−1 fwt.
Figure 4.
Levels of jasmonate family members in unwounded and wounded leaves of potato and Arabidopsis. Arabidopsis (A) leaves were extracted 90 min after wounding, potato (B) leaves were extracted 4 h after wounding, and tissues were analyzed for their content of JA, OPDA, and dnOPDA. Open bars, wounded leaves; hatched bars, unwounded control plants. For JA, amounts of both 3R,7S and the 3R,7R isoforms were cointegrated. Values represent the mean ± SE of three plants, except for unwounded Arabidopsis, where n = 5.
DISCUSSION
dnOPDA: A Hexadecanoid Signal.
Using a simple and rapid extraction procedure designed to recover jasmonate family members, we identified a novel, wound-inducible compound in Arabidopsis and potato leaf tissues. Structural analysis of the compound revealed that it was a 16-carbon cyclopentenone of the jasmonate family, which we named dinor-oxo-phytodienoic acid (abbreviated here as dnOPDA, Fig. 1). We were able to synthesize the compound in vitro from 16:3 in the flaxseed acetone powder system and this allowed further structural studies on the compound as well as suggesting a route to its biosynthesis. The in vitro synthesis of dnOPDA from 16:3 would be consistent with its direct synthesis in planta from plastid-derived 16:3. Our results suggest that plants have evolved lipoxygenase and allene oxide synthase enzymes with chain length specificities broad enough to use either 16:3 or 18:3 as a precursor of fatty acid signals. In addition to their activity with 16- and 18-carbon substrates, these enzymes are known to be active with 20-carbon substrates (25). It was necessary to test whether or not dnOPDA was likely to be derived from 16:3, rather than by β-oxidation of OPDA. We tried to detect the compound in the fad5 Arabidopsis mutant, a plant with massively reduced capacity to desaturate 16-carbon fatty acids (15). No dnOPDA could be detected in the fad5 mutant (Table 1) suggesting that, in vivo, dnOPDA is derived directly from 16:3, via its 11-hydroperoxide. This result could have implications in the study of plant fatty acid signaling mutants because, for jasmonate family signal generation, the 16:3 pool could serve as a replacement for the 18:3 pool and vice versa. It is not yet known whether dnOPDA is further metabolized, but it might be a substrate for the enzyme OPDA reductase. The presence of dnOPDA in leaf tissue from both Arabidopsis and potato (see Table 1 and Fig. 4) hints to a broad distribution of this molecule in the plant kingdom, because these two plants belong to widely separate subclasses of the angiospermae. Over 30 plant families are known to contain 7Z,10Z,13Z-hexadecatrienoic acid (29), the proposed dnOPDA precursor.
The plastid was originally proposed to be the site for the initial steps in jasmonate synthesis (30). Recent evidence supports this view and suggest that galactolipids such as MGDG might be the source of jasmonates (31, 32). In “16:3” plants such as Arabidopsis and potato, 16:3 is found almost exclusively in MGDG, implying that this molecule is the in vivo source for dnOPDA synthesis. MGDG is made via two metabolic pathways, the plastid localized “prokaryotic” pathway (MGDG rich in 16:3 in the sn-2 position in the case of Arabidopsis and potato) and the “eukaryotic” pathway, localized in the endoplasmic reticulum (MGDG rich in 18:3 in the sn-2 position, ref. 26). It is thus probable that both eukaryotic and prokaryotic MGDG pools contribute to the biosynthesis of jasmonates in Arabidopsis and potato. Additionally, our data on dnOPDA strongly suggest that the molecule is derived from the sn-2 position of MGDG and would thus be liberated by a phospholipase A2.
In plants, octadecanoid-derived signal compounds are well known, but few hexadecanoid-derived signal molecules have been characterized. The topical spray application of several cutin monomers, including hexadecanoids, to barley induced resistance to Erysiphe (33). How many other 16-carbon fatty acid derivatives with potential signaling functions exist in plants? Apart from the dnOPDA described herein, other candidates might include some previously characterized 16-carbon fatty acids of unknown function, for example the 16:3 isomer 8Z,11Z,14Z-hexadecatrienoic acid and a hydroxyl-substituted 16-carbon cyclopentenone (34).
Oxylipin Signatures.
We developed a general procedure for the simultaneous extraction and quantification of jasmonate pathway metabolites (and other oxylipins) from different plant species. Rapid freezing and extraction of tissues with methanol was used to avoid the artifactual generation of OPDA and dnOPDA, which can be formed if tissues are extracted in aqueous solutions (H.W. and E.E.F., unpublished work). The procedure we have developed with potato and Arabidopsis is not optimized for any one species of plant or tissue and provides an easily reproduced protocol. More detailed studies restricted to one plant species or tissue could be improved by focusing on internal standard recoveries. We decided to examine the relative levels of jasmonate family members in healthy and wounded Arabidopsis and potato leaves. Previous quantitative studies have clearly established “resting” levels and wound-inducible increases in JA in plant tissues (35–38). For our work with wounded Arabidopsis and potato leaves we chose to harvest tissues at times of published maximum JA accumulation for the two species studied, i.e., 90 min for Arabidopsis (39) and 4 h for potato (40).
Measurements of JA in the leaves of potato and Arabidopsis were consistent with previously published levels (10, 38–40). Quantitation of jasmonate family members in unwounded and wounded leaves revealed differences between Arabidopsis and potato. In healthy Arabidopsis leaves OPDA ≫ JA ≈ dnOPDA, whereas in healthy potato leaves OPDA ≈ JA > dnOPDA (see Fig. 4). In wounded Arabidopsis leaves the levels of JA > OPDA > dnOPDA, whereas in wounded potato leaves OPDA > JA > dnOPDA. Thus, different species and tissues may show different “oxylipin signatures.”
Based on previous work with JA (35, 36, 41, reviewed in ref. 4) we would expect the relative levels of jasmonate family members to be dynamic and to change over time during development and upon stress. Thus, oxylipin signatures would reflect the physiology of the tissue. The concept of oxylipin signatures may aid in viewing the jasmonate family of signals as a complex, or continuum, rather than attributing biological activities to a single species of molecule. Delicate control over cellular processes could thereby be exerted by changing the relative levels of each of a spectrum of regulatory jasmonates. This idea would be consistent with the fact that the JA precursor OPDA, as well as JA itself, is a powerful signal molecule (37, 42). More work is needed to establish whether levels of jasmonate family members differ in different plant tissues, to characterize how development and stress may influence these levels, and to understand how these changes relate to plant physiology.
Biological Roles of Jasmonates.
Arabidopsis lipoxygenase 2 has been (AtLOX2) identified as a lipoxygenase necessary for the expression of wound-inducible vegetative storage proteins in the leaves of this plant (38). The AtLox2 message is itself wound and jasmonate inducible (28). These previously published results led us to speculate that jasmonate family members including dnOPDA might themselves affect the ability of Arabidopsis leaf tissues to carry out the oxidative modification of the lipoxygenase substrate linoleic acid. Our results (Fig. 3) demonstrate that dnOPDA indeed has strong biological activity, increasing the ability of extracts from Arabidopsis leaves to convert linoleic acid into α-ketol, i.e., increasing the quantity or activity of enzymes of the jasmonate biosynthetic pathway.
A surprising observation was that the unwounded leaves of the fad5 mutant displayed a 15-fold reduced level of OPDA relative to unwounded wild-type leaves (see Table 1). Both wild type and mutant, however, accumulated similar levels of OPDA on wounding. These data suggest that levels of OPDA in unwounded and wounded leaves are regulated differently and might also suggest that an unsaturated 16-carbon fatty acid or derivative thereof (perhaps dnOPDA itself) helps to regulate the levels of OPDA in unwounded leaves. Arabidopsis cells might have a sensing mechanism to control the relative proportions of eukaryotic and prokaryotic MGDG and other lipids (15, 26). If so, it is conceivable that dnOPDA and/or OPDA levels in unwounded leaves could act as sensors, controlling the relative contributions of both metabolic pathways to MGDG synthesis. In a mutant like fad5 with reduced levels of “prokaryotic” type MGDG, levels of “sensor” OPDA might be adjusted to accommodate increased flux of eukaryotic 18:3 into MGDG. Taken together, with the results on OPDA levels in the unwounded leaves of wild-type and fad5 mutant Arabidopsis (Table 1), our data would be consistent with autoregulatory cross-talk in the synthesis and/or accumulation of C16 and C18 fatty acid-derived jasmonates. Jasmonates may to some extent control their own synthesis and/or steady-state levels. Further studies will be necessary to extend our knowledge of how jasmonate family members participate together in the regulation of defense, development, and metabolism and, in 16:3 plants, the potentially important contribution of dnOPDA must be taken into account.
Acknowledgments
E.E.F. dedicates this paper to the memory of Hans Grisebach. The NMR characterization of dnOPDA was expertly performed by J. Huwe. We thank D. Caldelari for help with design of the fatty acid oxidation experiment and the identification of the α-ketol, P. A. Siegenthaler for advice on the isolation of 16:3, W. Ruest for virus-free potato tubers, B. Künstner for expert care of plant material, P. Schweizer for dihydrojasmonate, H. Mosimann for help with the synthesis of H4OPDA, and P. Vogel for advice in naming dnOPDA. This work was supported by Swiss Fonds National de la Recherche Scientifique Biotechnology Grant SPP6 5002–039050-1 and by the Etat de Vaud.
ABBREVIATIONS
- JA
jasmonic acid
- OPDA
12-oxo-phytodienoic acid
- H4OPDA
tetrahydro-OPDA
- H2JA
dihydrojasmonic acid
- MGDG
monogalactosyldiacylglycerol
- TLC
thin-layer chromatography
- fwt
fresh weight
References
- 1.Hamberg M, Gardner H W. Biochim Biophys Acta. 1992;1165:1–18. doi: 10.1016/0005-2760(92)90069-8. [DOI] [PubMed] [Google Scholar]
- 2.Sembdner G, Parthier B. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:569–589. [Google Scholar]
- 3.Vick B A. In: Lipid Metabolism in Plants. Moore T S, editor. Boca Raton, FL: CRC; 1993. pp. 167–191. [Google Scholar]
- 4.Farmer E E. Plant Mol Biol. 1994;26:1423–1437. doi: 10.1007/BF00016483. [DOI] [PubMed] [Google Scholar]
- 5.Farmer E E, Ryan C A. Proc Natl Acad Sci USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andresen I, Becker W, Schlüter K, Burges J, Parthier B, Apel K. Plant Mol Biol. 1992;19:193–204. doi: 10.1007/BF00027341. [DOI] [PubMed] [Google Scholar]
- 7.Gundlach H, Müller M J, Kutchan T M, Zenk M H. Proc Natl Acad Sci USA. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constabel C P, Bergey D R, Ryan C A. Proc Natl Acad Sci USA. 1995;92:407–411. doi: 10.1073/pnas.92.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe G A, Lightner J, Browse J, Ryan C A. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McConn M, Creelman R A, Bell E, Mullet J E, Browse J. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penninckx I A M A, Eggermont K, Terras F R G, Thomma B P H J, De Samblanx G W, Buchala A, Métraux J-P, Manners J M, Broakaert W F. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiler W E, Albrecht T, Groth B, Xia Z-Q, Luxem M, Liss H, Andert L, Spengler P. Phytochemistry. 1993;32:591–600. [Google Scholar]
- 15.Kunst L, Browse J, Somerville C. Plant Physiol. 1989;90:943–947. doi: 10.1104/pp.90.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miersch O, Sembdner G, Schreiber K. Phytochemistry. 1989;28:339–340. [Google Scholar]
- 17.Clancy R M, Hugli T E. Anal Biochem. 1983;133:30–39. doi: 10.1016/0003-2697(83)90218-x. [DOI] [PubMed] [Google Scholar]
- 18.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 19.Zimmerman D C, Feng P. Lipids. 1978;13:313–316. [Google Scholar]
- 20.Farmer E E, Caldelari D, Pearce G, Walker-Simmons M-K, Ryan C A. Plant Physiol. 1994;106:337–342. [Google Scholar]
- 21.Vick B A, Zimmerman D C. Plant Physiol. 1982;69:1103–1108. doi: 10.1104/pp.69.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baertschi S W, Ingram C D, Harris T M, Brash A R. Biochemistry. 1988;27:18–24. doi: 10.1021/bi00401a004. [DOI] [PubMed] [Google Scholar]
- 23.Vick B A, Zimmerman D C, Weisleder D. Lipids. 1979;14:734–740. [Google Scholar]
- 24.Hamberg M, Fahlstadius P. Arch Biochem Biophys. 1990;276:518–526. doi: 10.1016/0003-9861(90)90753-l. [DOI] [PubMed] [Google Scholar]
- 25.Vick B A, Zimmerman D C. Plant Physiol. 1986;80:202–205. doi: 10.1104/pp.80.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somerville C, Browse J. Science. 1991;252:80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- 27.Ohlrogge J, Browse J. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell E, Mullet J E. Plant Physiol. 1993;103:1133–1137. doi: 10.1104/pp.103.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamieson G R, Reid E H. Phytochemistry. 1971;10:1837–1843. [Google Scholar]
- 30.Vick B A, Zimmerman D C. Plant Physiol. 1987;85:1073–1078. doi: 10.1104/pp.85.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blée E, Joyard J. Plant Physiol. 1996;110:445–454. doi: 10.1104/pp.110.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conconi A, Miquel M, Browse J A, Ryan C A. Plant Physiol. 1996;111:797–803. doi: 10.1104/pp.111.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer P, Jeanguenat A, Whitacre D, Métraux J-P, Mösinger E. Physiol Mol Plant Pathol. 1996;49:103–120. [Google Scholar]
- 34.Monaco P, Previtera L. Phytochemistry. 1987;26:745–747. [Google Scholar]
- 35.Creelman R A, Tierney M L, Mullet J E. Proc Natl Acad Sci USA. 1992;89:4938–4941. doi: 10.1073/pnas.89.11.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albrecht T, Kehlen A, Stahl K, Knoefel H-D, Sembdner G, Weiler E W. Planta. 1993;191:86–94. [Google Scholar]
- 37.Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan T M, Mueller M J, Xia Z-Q, Zenk M H. Proc Natl Acad Sci USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell E, Creelman R A, Mullet J E. Proc Natl Acad Sci USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laudert D, Pfannschmidt U, Lottspeich F, Hollaender-Czytko H, Weiler E W. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- 40.Harms K, Atzorn R, Brash A, Kühn H, Wasternack C, Willmitzer L, Peña-Cortés H. Plant Cell. 1995;7:1645–1654. doi: 10.1105/tpc.7.10.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller M J, Brodschelm W, Spannagl E, Zenk M H. Proc Natl Acad Sci USA. 1993;90:7490–7494. doi: 10.1073/pnas.90.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiler E W, Kutchan T M, Gorba T, Brodschelm W, Niesel U, Bublitz F. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]