Abstract
The aa3 type cytochrome c oxidase consisting of the core subunits I and II only was isolated from the soil bacterium Paracoccus denitrificans and crystallized as complex with a monoclonal antibody Fv fragment. Crystals could be grown in the presence of a number of different nonionic detergents. However, only undecyl-β-d-maltoside and cyclohexyl-hexyl-β-d-maltoside yielded well-ordered crystals suitable for high resolution x-ray crystallographic studies. The crystals belong to space group P212121 and diffract x-rays to at least 2.5 Å (1 Å = 0.1 nm) resolution using synchrotron radiation. The structure was determined to a resolution of 2.7 Å using molecular replacement and refined to a crystallographic R-factor of 20.5% (Rfree = 25.9%). The refined model includes subunits I and II and the 2 chains of the Fv fragment, 2 heme A molecules, 3 copper atoms, and 1 Mg/Mn atom, a new metal (Ca) binding site, 52 tentatively identified water molecules, and 9 detergent molecules. Only four of the water molecules are located in the cytoplasmic half of cytochrome c oxidase. Most of them are near the interface of subunits I and II. Several waters form a hydrogen-bonded cluster, including the heme propionates and the Mg/Mn binding site. The Fv fragment binds to the periplasmic polar domain of subunit II and is critically involved in the formation of the crystal lattice. The crystallization procedure is well reproducible and will allow for the analysis of the structures of mechanistically interesting mutant cytochrome c oxidases.
Cytochrome c oxidase (ferrocytochrome c:oxygen oxidoreductase, EC 1.9.3.1, or complex IV) is the terminal enzyme in the respiratory chains of mitochondria and many aerobic bacteria (see refs. 1–6 for reviews). Being located in the inner mitochondrial or bacterial membranes it uses electrons from cytochrome c to reduce molecular oxygen. The product of the reaction is water. Because the protons needed for water formation originate from the mitochondrial matrix or the bacterial cytoplasm, and cytochrome c donates electrons from the opposite side of the membrane, an electric field and a pH difference are generated across the membrane. In addition, cytochrome c oxidases translocate (“pump”) up to four protons per oxygen molecule across the inner membrane, thereby enhancing the electric field and the proton gradient. Both drive protons back across the membrane through ATP synthases, a process that results in the formation of ATP from ADP and inorganic phosphate.
The cytochrome c oxidases are members of a large superfamily of heme and copper containing terminal oxidases (3, 5). The number of subunits varies between 3 and 5 in bacteria and up to 13 in mammalian mitochondria. However, only subunits I and II are essential for the function of the enzyme. This functional core catalyzes both oxygen reduction and proton pumping (7). Subunit III, which is also conserved, may play a role in assembly or oxygen delivery to the core of the enzyme (8, 9).
Subunits I and II of various terminal oxidases show a high degree of sequence conservation (1, 3). Subunit II of cytochrome c oxidases contains the binuclear CuA center, which is the primary acceptor of electrons from reduced cytochrome c. Subunit I contains one low-spin heme A (“heme a”) and a binuclear metal center formed by a high-spin heme A (“heme a3”) and CuB. Within the binuclear center oxygen is bound to the heme a3 iron, then reduced, and two molecules of water are formed. How these processes are coupled to proton pumping across the membrane is unknown.
Prerequisite to understanding the mechanism of action of cytochrome c oxidases is the knowledge of their structure. In 1995, the structure of the cytochrome c oxidase from Paracoccus denitrificans (10) and the metal center structure of the mitochondrial cytochrome c oxidase from bovine heart (11) were published, followed by the presentation of the structures of the 13 protein subunits (12). Both structures were determined by x-ray crystallography to a nominal resolution of 2.8 Å (1 Å = 0.1 nm). However, the Paracoccus cytochrome c oxidase crystals were very difficult to grow and showed a high degree of anisotropy in diffraction (13).
Getting well-ordered crystals of membrane proteins is still extremely difficult. For the crystallization of the bacterial enzyme a new strategy was developed, namely enforcement of crystallization by using a monoclonal antibody Fv fragment. Membrane protein crystals are hold together mainly by interactions of the polar parts of the membrane protein surface, whereas detergents cover the hydrophobic parts. The effect of the Fv fragment is to enlarge the polar surface of the membrane protein, thereby increasing the chances of getting highly ordered crystals (13). Only very mild detergents can be tried for crystallization of the four subunits containing cytochrome c oxidase from P. denitrificans, since otherwise subunits III and IV are removed from the complex. So far, crystals of the bacterial four-subunit cytochrome c oxidase complex could only be grown with dodecyl-β-d-maltoside as detergent. The cytochrome c oxidase from P. denitrificans, however, was first isolated as a two-subunit enzyme using Triton X-100 as detergent (14). Later, the gene for subunit III was discovered (15) and a “three-subunit enzyme” was isolated subsequently with dodecyl-β-d-maltoside as detergent (16). Recently, it was recognized that an additional small subunit, subunit IV, was present in such preparations (17, 18).
In light of the limited quality of the crystals of the four subunits containing cytochrome c oxidase from P. denitrificans, we have tried to crystallize the two subunits containing core cytochrome c oxidase, which allows a more extensive detergent screening. We were successful again using the strategy of cocrystallization with an Fv fragment of a monoclonal antibody. Here we describe the crystallization, structure determination, and new features revealed by the crystallographic analysis.
MATERIALS AND METHODS
Preparation of the Two-Subunit Cytochrome c Oxidase–Fv Complex.
P. denitrificans membranes and the Escherichia coli periplasmic fractions containing the recombinant monoclonal antibody Fv fragment 7E2C50S were prepared as described (7, 13) at 4°C. P. denitrificans (20–30 ml) membranes (50–80 mg protein per ml) were solubilized at pH 8.0 by adding 7 ml of a 30% (wt/vol) lauryl-N,N-dimethylamine-N-oxide solution. After 5 min of stirring on ice, 50 ml of E. coli periplasmic fraction was added. After centrifugation at 100,000 × g for 70 min the supernatant was loaded onto a 30 ml pre-equilibrated (20 mM Tris⋅HCl, pH 8.0/1 mM EDTA/0.1% lauryl-N,N-dimethylamine-N-oxide) streptavidin Sepharose column (18). A specifically engineered streptavidin (a kind gift of Arne Skerra, Institute of Biochemistry, Technical University, Darmstadt, Germany) was used for the isolation, because it has a higher affinity to the strep tag affinity tag (19) and shows a much better binding and elution behavior than the wild-type streptavidin used previously. After washing the column with 75 ml of equilibration buffer, the Fv–cytochrome c oxidase complex was eluted with 2.5 mM desthiobiotin (Sigma) in the same buffer. A sharp elution peak (≈15 ml) was obtained. This single-step purification yielded very pure material and subsequent steps were performed only to remove the excess of the Fv fragment and to exchange detergents. After adding 30 mg of solid undecyl-β-d-maltoside (Biomol, Hamburg, Germany) or of another detergent, the fraction was loaded onto a 10-ml Q-Sepharose fast flow column (Pharmacia) and washed with 120 ml buffer (0.2% undecyl-β-d-maltoside or another detergent/10 mM Tris⋅HCl, pH 8.0) to replace the lauryl-N,N-dimethylamine-N-oxide by undecyl-β-d-maltoside. The complex was then eluted using 600 mM NaCl/0.2% undecyl-β-d-maltoside (or another detergent)/10 mM Tris⋅HCl, pH 8.0. The green fractions were concentrated to about 1 ml using centriprep concentrators (Amicon). Gel filtration was performed on a TSK 3000 column (60 × 0.7 cm, Tosohaas, Montgomeryville, PA) in the presence of 10 mM Tris⋅HCl (pH 7), 20 mM NaCl, 0.06% undecyl-β-d-maltoside (or the 2-fold critical micellar concentration of another detergent). The cytochrome c oxidase-containing fractions were concentrated using centriprep concentrators with a cutoff of 50 kDa and finally adjusted to 10–20 mg/ml. A typical preparation took 6 hr and yielded more than 10 mg of pure protein.
Crystallization, Data Collection, and Structure Determination.
Crystallization trials were performed using the vapor diffusion technique with a hanging drop setup at 293K. Protein droplets (2–3 μl) were combined with the same volume of bottom solution (usually 100 mM sodium acetate, pH 5.5/7–8.5% polyethylene glycol 2000 monomethylether). Crystals appeared within 2 days and reached a final size of 0.3 × 0.3 × 1.0 mm after 1–2 weeks. The cocomplex crystals were initially examined with a MAR research imaging plate detector connected to a Rigaku RU-200 rotating anode (CuKα) operated at 40 kV and 100 mA. The diffraction patterns showed systematic absences characteristic for space group P212121. The unit cell dimensions were: a = 93.5 Å, b = 151.0 Å, c = 156.7 Å. The asymmetric unit contains one cocomplex with a molecular weight of 118 kDa resulting in an apparent Vm of 4.8 Å3/Da and a solvent content of about 72%. With the rotating anode x-ray generator a data set with a completeness of 80% up to 3.5 Å was collected from a single crystal at 293K. Data were processed with the HKL programs denzo and scalepack (20). Molecular replacement using the data between 10 and 4 Å resolution was performed with the CCP4 (21) program amore (22) and the coordinates of SU I, II and the Fv fragment of the structure published previously (10) as search model. For refinement, high resolution data were collected at beamline BM14 of the European Synchroton Radiation Facility in Grenoble, France, at 277K. Data were collected by 0.5° rotations with a crystal-to-detector distance of 150 mm at a wavelength of 1.078 Å using a CCD camera as detector. The images showed diffraction up to 2.5 Å. After rotation and translation searches that both yielded clear, unique solutions, the initial crystallographic R-factor for 95% of the data was 42.3%. The initial model was refined using slow cool, energy minimization and the B-factor refinement protocols of xplor (23), version 3.851, with the Engh and Huber set of stereochemical parameters (24). A bulk solvent was included in the later cycles of refinement. The crystallographic model was adjusted with the program O (25). Solvent (water) molecules were added when the Fo–Fc electron density was above 3σ and the geometry was suited. They were maintained when the 2 Fo–Fc electron density was greater than 1 σ after subsequent refinement. The current model comprises a total of 8,300 nonhydrogen atoms. During several rounds of interactive and automatic refinement procedures, the crystallographic R-factor and the free R-factor (26) dropped to 20.5% and 25.9%, respectively.
RESULTS AND DISCUSSION
Crystallization.
The aim of this study was to obtain better diffracting crystals of the cytochrome c oxidase from P. denitrificans and thus to improve the accuracy of the x-ray structure. For this purpose we have concentrated on the functional core of cytochrome c oxidase, consisting of subunits I and II only, in the presence of the Fv fragment 7E2 (13). This two-subunit cytochrome c oxidase is more stable, thus allowing a wider range of detergents to be tried, and probably more homogeneous due to a lower content of lipids. As a first step, a pure two-subunit cytochrome c oxidase complexed with the antibody Fv fragment, not contaminated by subunits III or IV, was isolated as described in Materials and Methods. The use of an improved streptavidin engineered to bind the strep tag at the Fv fragment much tighter (19) facilitated the isolation substantially. Subunits III and IV were lost when the rather harsh detergent lauryl-N,N-dimethylamine-N-oxide was used. This detergent was found to be the best one for removing subunits III and IV. However, because the long-term stability of the Paracoccus two-subunit cytochrome c oxidase in this detergent was low, it could not be used for crystallization trials. Exchange to other detergents was achieved by binding the complex of the two-subunit cytochrome c oxidase and the antibody fragment to an ion exchange column, washing it in the presence of the new detergent, and eluting it in a buffer containing the new detergent. A subsequent broad screening (with respect to pH, precipitating agent, and buffer) yielded crystals under rather restricted conditions at a comparatively low pH value (5.5). Table 1 provides an overview of the detergent effects on the crystallization behavior of the complex. Crystals were obtained with detergents of the maltoside type and with octaethylene glycol–monododecylether. However, crystals grown in the presence of the latter detergent were found to be insufficient for an x-ray structure determination. The quality of the crystals observed with the alkyl-maltoside detergents depended strongly on the length of the alkyl chains. Use of undecyl-β-d-maltoside yielded the best crystals, with a large difference in diffraction quality compared with the dodecyl homologue. Similarly, with cyclohexyl-hexyl-β-d-maltoside good crystals were observed, but no crystals with cyclohexyl-pentyl-β-d-maltoside. Cyclohexyl-heptyl-β-d-maltoside is not available. Such a strong dependence of the crystallization behavior and crystal quality on the length of the alkyl chain is commonly observed in membrane protein crystallization (see ref. 27 for review).
Table 1.
Summary of the results of the crystallization trials using different detergents
| Detergent | Supplier | CMC, mM | Crystals |
|---|---|---|---|
| C12-β-d-maltoside | B | 0.15 | + (8 Å) |
| C11-β-d-maltoside | B | 0.59 | + (2.5 Å) |
| C10-β-d-maltoside | B | 1.8 | — |
| CYMAL-6 | A | 0.56 | + (2.6 Å) |
| CYMAL-5 | A | 2.4 | — |
| Dodecylsucrose | C | 0.3 | — |
| Decylsucrose | C | 2.5 | — |
| C9-β-d-glucoside | C | 6.5 | — |
| C12E8 | F | 0.07 | + (>8 Å) |
| C12E6 | F | 0.06 | — |
| C12E5 | F | 0.06 | — |
| C10E6 | F | 0.9 | — |
| C10E5 | F | 0.8 | — |
The resolution of the crystals obtained is given in parentheses. CMC, critical micellar concentration (as provided by the vendor); C12, dodecyl; C11, undecyl; C10, decyl; C9, nonyl; CYMAL-6, cyclohexyl-hexyl-β-d-maltoside; CYMAL-5, cyclohexyl-pentyl-β-d-maltoside; Ex, x stands for the number of oxyethylen units in the alkyl polyoxyethylen detergents. The detergents were purchased from Anatrace (Maumee, OH) (A), Biomol (B), Calbiochem (C), or Fluka (F).
The stability of the crystals in the x-ray beam turned out to be reasonable so that a structure determination became possible.
Structure Determination and Quality of the Model.
The present x-ray diffraction data have been collected to a resolution of 2.69 Å. The completeness and quality of the data were excellent (Table 2). The initial structure determination (see Materials and Methods) using molecular replacement with subunits I and II and the bound Fv fragment of the four-subunit cytochrome c oxidase from P. denitrificans (10) was straightforward. As a result of the high solvent contents the ratio of observables to parameters was very good for a 2.7 Å resolution structure. Therefore, 52 tentatively identified solvent (water) molecules could be included during refinement. This has not been possible during the refinement of the four-subunit Paracoccus cytochrome c oxidase. Elongated electron densities were observed and attributed to nine alkyl chains of detergent molecules or lipids. These alkyl chains were included into the model. On the grounds of the clear electron density we have identified the Mg/Mn site, as well as a new metal binding site (see below). The refined structural model contains 1,007 residues (subunit I, 17–545; subunit II, 1–252; VL, 1–108; and VH, 1–118), 3 copper atoms, 2 heme A molecules, 1 Mg/Mn and another metal atom, 52 water molecules, and 9 alkyl chains. The current crystallographic R-factor is 20.5% (Rfree = 25.9%) with very good stereochemistry of the model (Table 2).
Table 2.
Data collection, structural, and refinement statistics for the two subunit cytochrome c oxidase from P. denitrificans at 2.7 Å resolution
| Measurements | Values |
|---|---|
| Unit cell dimensions | |
| a, b, c, Å | 93.5, 151.0, 156.7 |
| α, β, γ, ° | 90 |
| Space group | P212121 |
| Number of reflections | 198,394 |
| Unique data | 57,945 |
| Resolution, Å | 30.0 – 2.7 |
| Completeness, % | 93.8 |
| Rmerge,* % | 6.2 |
| Rcryst,▴ % | 20.5 |
| Rfree,▴ % | 25.9 |
| Rms deviations from ideal geometry | |
| Bond lengths, Å | 0.010 |
| Bond angles, ° | 1.6 |
| Improper angles, ° | 2.4 |
| Dihedral angles, ° | 23.4 |
| Number of nonhydrogen atoms | |
| All atoms | 8248 |
| Waters | 52 |
Rmerge (I) = ΣΣI(I(i) − 〈I(hkl)〉I)/Σ(i), summed over all reflections and all observations, where I(i) is the ith observation of the intensity of the hkl reflection and 〈I(hkl)〉 is the mean intensity of the hkl reflection.
▴ Rcryst (F) = ΣhIIFobs(h) − Fcalc(h)/ΣhIFobs(h)I, where Fobs and Fcalc are the observed and calculated structure–factor amplitudes for the reflection with Miller indices h = (h, k, l). The free R-factor (Rfree) is calculated for a “test” set of reflections (5%), which were not included in atomic refinement.
Structure of the Complex of Two-Subunit Cytochrome c Oxidase and the Fv Fragment.
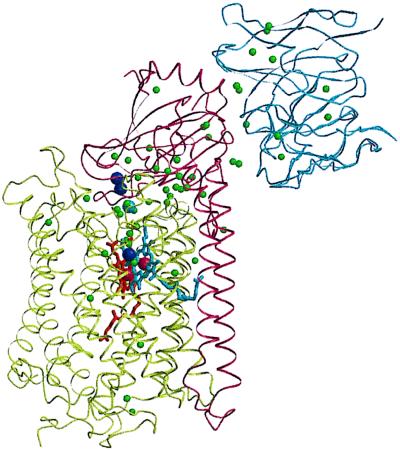
As expected from the result of molecular replacement the structure of the core cytochrome c oxidase complexed with the antibody Fv fragment is very similar to that published for the four-subunit enzyme complexed with the same antibody Fv fragment. The higher resolution and the better quality of the data resulted in some revisions of structural details during refinement. The improved assignment of regular secondary structure being dominated by 12 regular transmembrane helices of subunit I and 2 of subunit II is presented in Table 3. The structure of the complex, including the tentatively identified water molecules is shown in Fig. 1. It is remarkable that only four of these water molecules are located in the periplasmic half—this means below the heme groups in Fig. 1—of the core cytochrome c oxidase. The significance of this finding is unknown. Many of the water molecules are found at the interface between subunits I and II.
Table 3.
Elements of regular secondary structure in subunits I and II of the cytochrome c oxidase from P. denitrificans
| Subunit I
|
Subunit II
|
||
|---|---|---|---|
| Elements | Residues | Elements | Residues |
| TM I | 27–59 | TM I | 25–60 |
| Tm II | 83–120 | TM II | 75–105 |
| Tm III | 127–151 | β-1 | 112–119 |
| 3/10-helix | 170–174 | β-2 | 122–127 |
| TM IV | 177–206 | β-3 | 132–136 |
| 3/10-helix | 213–217 | 3/10-helix | 144–157 |
| TM V | 218–251 | β-4 | 162–166 |
| 3/10-helix | 257–261 | β-5 | 169–177 |
| TM VI | 263–299 | β-6 | 181–185 |
| TM VII | 304–319 | β-7 | 190–194 |
| 3/10-helix | 320–331 | β-8 | 200–204 |
| TM VIII | 333–364 | β-9 | 210–214 |
| TM IX | 370–395 | β-10 | 229–234 |
| α-helix | 397–403 | α-helix | 235–250 |
| TM X | 405–437 | ||
| TM XI | 441–469 | ||
| TM XII | 479–514 | ||
| 3/10-helix | 529–533 | ||
The secondary structure elements were assigned using the program dssp (28). All transmembrane helices (TM) are regular α-helices, with the exception of transmembrane helix VII which becomes a 3/10-helix (residues 320–331) near the CuB binding site.
Figure 1.
Ribbon representation of the structure of the two-subunit cytochrome c oxidase from P. denitrificans complexed with the antibody Fv fragment 7E2. Subunit I, olive green; subunit II, dark red; Fv fragment, blue; heme a, red; heme a3, blue; copper atoms, dark blue spheres; water molecules, green spheres. The programs molscript (29) and raster 3d (30) were used to prepare the figure.
Structure of and Around Bound Cofactors.
The refinement also led to minor changes in the environment of the bound cofactors and metals. In our present model the distance between both copper atoms of the CuA center is 2.5 Å, the Cu-Sγ (Cys), Cu-Sδ (Met-227), Cu-Nδ1 (His), and Cu-O (Glu-218 backbone carbonyl oxygen) distances are 2.2, 2.6, 2.1, and 2.8 Å, respectively, the distance from the CuA2 atom to the heme a-iron is 19.5 Å, to the heme a3-iron 22.2 Å. The distance between both heme iron atoms is 13.1 Å. The heme a3-iron is 0.36 Å out of plane. The distance between the heme a3-iron and CuB is 4.5 Å, compared with 5.2 Å in the published structure of the four-subunit cytochrome c oxidase. This difference appears to be real and not to be due to the better quality of the two-subunit cytochrome c oxidase structure, since importing the two-subunit structure back into the four-subunit enzyme leads again to an increase in the distance upon further refinement (A.H., unpublished work). The difference could be caused by the removal of subunits III and IV, due to crystal packing effects, or more likely, due to the different pH (5.5 versus 8).
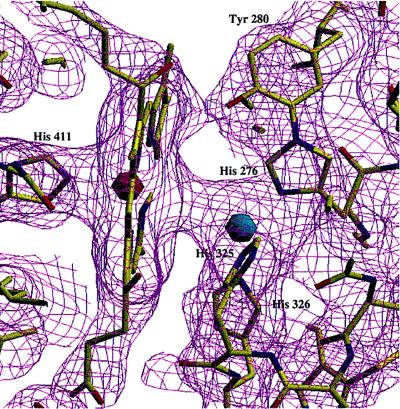
Fig. 2 shows the electron density around the binuclear CuB-heme a3 center, including the model. In contrast to the published structure of the four-subunit cytochrome c oxidase, which had been crystallized in the presence of azide electron density for all three histidine ligands to CuB is seen. There is a continuous electron density between CuB and the heme a3-iron, which might be indicative of bridging ligands. However, due to the heavy CuB and Fe atoms, a precise interpretation is difficult, and attempts to include bridging ligands and to refine their positions have not been undertaken. We favor the possibility of a water molecule as a heme a3-iron ligand, and—in hydrogen bonding distance—a hydroxy ion as an additional CuB ligand. Such an interpretation is in agreement with recent spectroscopic data (33). Recently, the electron density of the CuB ligand His-276 (His-240 in the bovine sequence) and of Tyr-280 (Tyr-244 in the bovine sequence) has been interpreted in terms of a covalent linkage between these two side chains in the crystal structure of the bovine heart mitochondrial enzyme by Tsukihara, Yoshikawa and coworkers (S. Yoshikawa, personal communication). Such a covalent linkage is in better agreement also with an unbiased electron density map (see Fig. 2) of our two-subunit cytochrome c oxidase than the published hydrogen bond between Tyr-280 (244) and His-276 (240) (10, 12). It will be of great importance to confirm this crosslink by chemical analyses, and to elucidate its function and biosynthesis. Having in mind the discussions about the structure of the “peroxy intermediate” (see refs. 5 and 34 for reviews), one is tempted to speculate that such a Tyr-His dimer might donate an electron and become a radical in the “peroxy intermediate” if the O-O bond were already broken in this state.
Figure 2.
“Unbiased” electron density map at the 1σ level of the binuclear heme a3-CuB site and the atomic model. A simulated annealing omit map (31), omitting all residues (including heme a3) with an atom closer than 4.5 Å to CuB, was calculated. The heme a3 iron atom is shown in red, CuB in blue, nitrogen atoms in blue, oxygen atoms in red, carbon atoms in yellow, and the electron density in purple. The figure was prepared using the program setor (32).
A New Metal Binding Site.
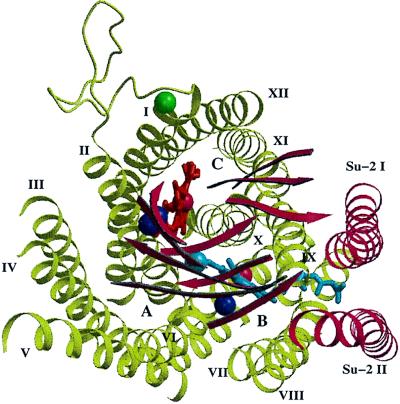
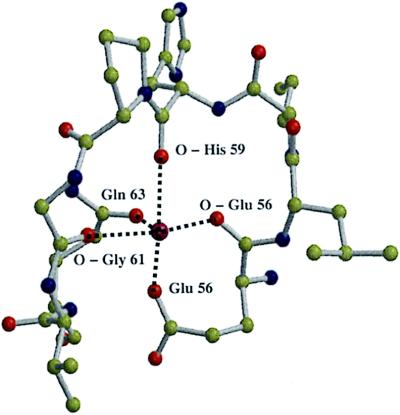
During the refinement of the two-subunit cytochrome c oxidase structure it also became apparent that there is another metal binding site. It is located close to the periplasmic end of the first membrane-spanning helix of subunit I. Its position can nicely be seen in Fig. 3. As indicated in Fig. 4, the backbone carbonyl oxygens of Glu-56, His-59, and Gly-61, as well as the side chain oxygens of Glu-56 and Gln-63 are involved in binding the metal. Neither the chemical identity of the metal nor its function is known. Our model contains a calcium ion, because the coordination is typical of a calcium binding site. Mutagenesis data around this new metal binding site are not available.
Figure 3.
View down onto the two-subunit cytochrome c oxidase from the periplasmic side showing the localization of the new metal binding site. Only the transmembrane helices, the loop connecting transmembrane helices I and II of subunit I, the β-strands of subunit II, the prosthetic group, and the metals are shown. The metal of the new metal binding site is shown in green. Subunit I, olive green; subunit II, red; heme a, red; heme a3, blue; copper atoms, dark blue spheres; iron atoms, red spheres. The transmembrane helices are indicated by roman numerals, the positions of the three “pores” are indicated by A, B, and C. The figure was created using the programs molscript (29) and raster 3d (30).
Figure 4.
Ball and stick model of the new metal binding site. Oxygen atoms are shown in red, nitrogen atoms in blue, carbon atoms in yellow, and the new metal in purple. The programs molscript (29) and raster 3d (30) were used to prepare the figure.
Putative Proton Transfer Pathways.
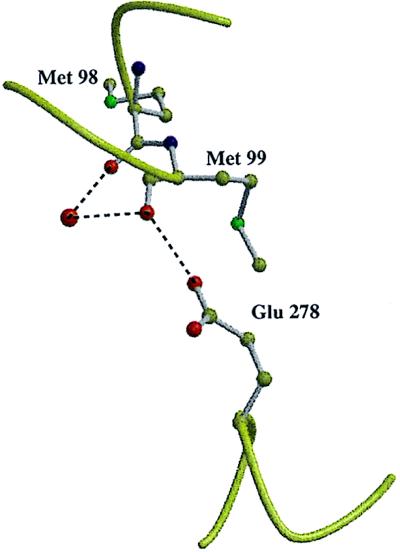
As mentioned above the protons needed for water formation originate from the cytoplasm and have to reach the oxygen binding site through the protein. In addition, the same number of protons is pumped across the membrane. Based on the results of site-directed mutagenesis, two potential proton transfer pathways have been identified (10), one involving Lys-354, leading straight to the active site in pore B (see Fig. 3), and another, leading up to the center of the membrane via Asp-124 in pore A (see Fig. 3) and then crossing over toward Glu-278 and the active site through a presumably solvent filled cavity. The former is called the K pathway due to Lys-354, the latter the D pathway due to Asp-124. The original assignment of the K pathway being used by protons, which are consumed in water formation, and the D pathway by pumped protons, which was based on mutagenesis data, is no longer in agreement with the results of the further characterization of site-directed mutants corresponding to Lys-354 → Met, Glu-278 → Gln. These results suggest that an intact K pathway is needed for the reduction of heme a3 and CuB (35), whereas Glu-278 (or an Asp) is necessary for the reactions leading from the peroxy-intermediates onwards (36). The existence of cavities in both pathways suggests that water in these cavities contributes to proton transfer. However, at the present resolution we could tentatively identify only two solvent molecules, one in the K pathway connecting Ser-291 and Lys-354, and one in the D pathway, being in hydrogen bonding distance to the backbone carbonyl oxygen of Met-99, which is also a hydrogen bond acceptor of Glu-278, which is most likely protonated. The solvent molecule is too far away to form a hydrogen bond to Glu-278 also (Fig. 5). The tentative identification of only two water molecules in both pathways is surprising bearing in mind that a complete uninterrupted water chain presumably involved in proton transfer from the cytoplasm to the binding site of the secondary quinone acceptor QB was found in the photosynthetic reaction center from Rhodobacter sphaeroides at a comparable resolution (37).
Figure 5.
Model of Glu-278 (subunit I) and its hydrogen bond to Met-99. A water molecule (larger red sphere) connects the backbone oxygen atoms of Met-98 and Met-99. The figure was prepared using the programs molscript (29) and raster 3d (30).
In the cytochrome c oxidase from bovine mitochondria the same K pathway was identified (12). However, it was suggested that the D pathway does not lead to Glu-278 and the binuclear site, but continues straight above Ser-193 to the periplasm via polar residues instead. These residues are not conserved in P. denitrificans. They are either alanines or glycines in the bacterial enzyme, leaving the possibility that water replaces the side chains, thereby enabling proton transfer. Another pathway between transmembrane helices XI and XII was also suggested. It is found also in the cytochrome c oxidase from P. denitrificans with the exception of one residue. Mutagenesis data supporting proton transfer along these pathways do not exist.
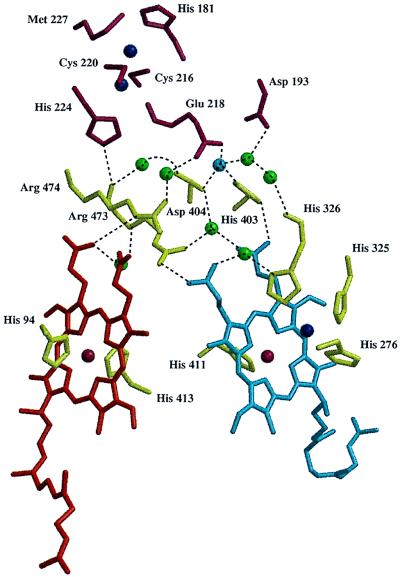
A negative charge cluster has been discussed to be near or part of the exit pathway for pumped protons (10). We now find that this charge cluster is part of an extended hydrogen bonded network, including the heme propionates and the Mg/Mn binding site. This hydrogen bonded network, including seven water molecules, is shown in Fig. 6. Interestingly, the propionate side chains of heme a and those of heme a3 are each connected through a hydrogen-bonding water molecule. In the case of the heme a3 propionates, this water molecule forms an additional hydrogen bond with the CuB ligand His-326. Glu-218 of subunit II, Asp-404 and His-403, and one water molecule, which is stabilized by a hydrogen bond to Asp-193 of subunit II, are ligands of the Mg/Mn atom. There is space for two more water molecules as ligands resulting in an octahedral ligation. These are, however, not resolved in our present structure. The negative charge cluster may play a role in accepting part of those protons (A. Kannt, C. R. D. Lancaster, and H.M., unpublished work), which are taken up upon reduction of the enzyme (38).
Figure 6.
The hydrogen bonded network between the heme groups and CuA. Residues of subunit I are shown in olive green, those of subunit II in dark red, copper atoms are dark blue spheres, iron atoms dark red spheres, the Mg/Mn atom is a blue sphere, water molecules are green spheres, heme a is red, and heme a3 is blue. The figure was produced using the programs molscript (29) and raster 3d (30).
Role of the Fv Fragment and Crystal Packing.
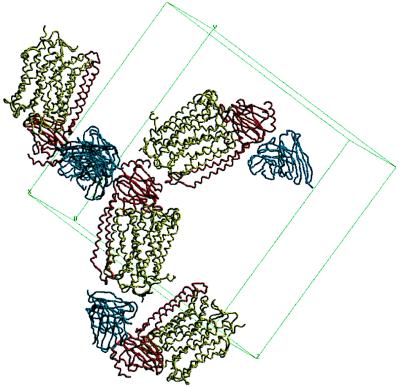
The same antibody Fv fragment was already used to crystallize the four-subunit cytochrome c oxidase from P. denitrificans (13). In that crystal form all protein–protein contacts between cytochrome c oxidases were made via the Fv fragment. However, a problem with these crystals was the lack of direct protein–protein interactions along the c-axis, explaining the anisotropy of the diffraction, especially the low resolution in the c* direction. In the new crystal form a completely different crystal packing is found (Fig. 7). Direct interactions between the cytoplasmic and the periplasmic surfaces of cytochrome c oxidase are seen. Residues near the C terminus of subunit I and the cytoplasmic loop connecting the transmembrane helices of subunit II are in contact with the C-terminal helix and N-terminal residues in the polar periplasmic domain of subunit II. All further contacts are mediated exclusively by the Fv fragment, involving parts of the Fv fragment opposite to the antigen binding site, as well as side-by-side interactions of the Fv fragment. The Fv fragment has also been crystallized in the absence of the antigen (39), and its structure was determined at 1.28 Å resolution (40). Comparison of the structure of the Fv fragment in the presence and absence of the antigen indicates that there are no significant structural changes upon binding of the antigen. At least eight Tyr residues of the Fv fragment contribute to antigen binding.
Figure 7.
The crystal packing of the two-subunit cytochrome c oxidase complexed with the antibody Fv fragment (backbone wire models). Subunit I is shown in olive green, subunit II in red, and the Fv fragment in dark blue. The figure was prepared with the program setor (32).
CONCLUSIONS
Using the more stable two-subunit core cytochrome c oxidase from P. denitrificans we have obtained crystals that are better than those of the four-subunit cytochrome c oxidase. The crystal structure analysis provided a much more detailed insight into the structure of this fundamental enzymes and revealed new features. The results will form the basis for a combined approach using site-directed mutagenesis, x-ray crystallography, and spectroscopy to elucidate the mechanism of this molecular machine. The strategy of isolating and crystallizing membrane proteins with the help of engineered antibody fragments should allow one to crystallize and to determine the structure of other membrane proteins that have refused to crystallize.
Acknowledgments
We thank D. Vinzenz, W. Schiweck, and W. Grabarse for help with the initial x-ray work, Hanne Müller for excellent technical assistance, and A. Skerra for generously providing the engineered streptavidin. We are grateful to A. Thompson (European Molecular Biology Laboratory outstation) and M. Merckel for their help during the high resolution data collection at beamline BM 14 of the European Synchroton Radiation Facility (Grenoble, France), to B. Ludwig for providing Paracoccus cells and for general discussion, to I. Sinning and M. Wikström for useful hints, and to J. Behr, A. Kannt, and W. Kühlbrandt for reading the manuscript. This work was supported by the Fonds der Chemischen Industrie, the Sonderforschungsbereich 472 of the Deutsche Forschungsgemeinschaft, and the Max-Planck-Gesellschaft.
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1ar1).
References
- 1.Saraste M. Q Rev Biophys. 1990;23:331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- 2.Babcock G T, Wikström M. Nature (London) 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 3.Calhoun M W, Thomas J W, Gennis R B. Trends Biochem Sci. 1994;19:325–330. doi: 10.1016/0968-0004(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 4.Malatesta F, Antonini G, Sarti P, Brunori M. Biophys Chem. 1995;54:1–33. doi: 10.1016/0301-4622(94)00117-3. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson-Miller S, Babcock G T. Chem Rev. 1996;96:2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 6.Ostermeier C, Iwata S, Michel H. Curr Opin Struct Biol. 1996;6:460–466. doi: 10.1016/s0959-440x(96)80110-2. [DOI] [PubMed] [Google Scholar]
- 7.Hendler R W, Pardhasaradhi K, Reynafarje B, Ludwig B. Biophys J. 1991;60:415–423. doi: 10.1016/S0006-3495(91)82067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haltia T, Finel M, Harms N, Nakari T, Raitio M, Wikström M, Saraste M. EMBO J. 1989;8:3571–3579. doi: 10.1002/j.1460-2075.1989.tb08529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riistama S, Puustinen A, Garcia-Horsman A, Iwata S, Michel H, Wikström M. Biochim Biophys Acta. 1996;1275:1–4. doi: 10.1016/0005-2728(96)00040-0. [DOI] [PubMed] [Google Scholar]
- 10.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 11.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 12.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 13.Ostermeier C, Iwata S, Ludwig B, Michel H. Nat Struct Biol. 1995;2:842–846. doi: 10.1038/nsb1095-842. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig B, Schatz G. Proc Natl Acad Sci USA. 1980;77:196–200. doi: 10.1073/pnas.77.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saraste M, Raitio M, Jalli T, Perõmaa A. FEBS Lett. 1986;206:154–156. doi: 10.1016/0014-5793(86)81359-x. [DOI] [PubMed] [Google Scholar]
- 16.Haltia T, Puustinen A, Finel M. Eur J Biochem. 1988;172:543–546. doi: 10.1111/j.1432-1033.1988.tb13923.x. [DOI] [PubMed] [Google Scholar]
- 17.Haltia T, Sema N, Arrondo J L R, Goñi F M, Freire E. Biochemistry. 1994;33:9731–9740. doi: 10.1021/bi00198a044. [DOI] [PubMed] [Google Scholar]
- 18.Kleymann G, Ostermeier C, Ludwig B, Skerra A, Michel H. Bio/Technology. 1995;13:155–160. doi: 10.1038/nbt0295-155. [DOI] [PubMed] [Google Scholar]
- 19.Voss, S. & Skerra, A. (1997) Protein Eng. 8, in press. [DOI] [PubMed]
- 20.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer N I, Bailey S, editors. Daresbury, U.K.: SERC Daresbury Lab.; 1993. pp. 51–62. [Google Scholar]
- 21.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 22.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 23.Brünger, A. T. (1993) xplor, Version 3.1: A System for X-Ray Crystallography and NMR (Yale Univ. Press, New Haven, CT).
- 24.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 25.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;7:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 27.Ostermeier, C. & Michel, H. (1997) Curr. Opin. Struct. Biol., in press. [DOI] [PubMed]
- 28.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 30.Merrit E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 31.Hodel A, Kim S-H, Brünger A T. Acta Crystallogr A. 1992;48:851–858. [Google Scholar]
- 32.Evan S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 33.Fann Y C, Ahmed I, Blackburn N J, Boswell J S, Verkhovskaya M L, Hoffmann B M, Wikström M. Biochemistry. 1995;32:10245–10255. doi: 10.1021/bi00032a019. [DOI] [PubMed] [Google Scholar]
- 34.Kitagawa T, Ogura T. Prog Inorg Chem. 1997;45:431–479. [Google Scholar]
- 35.Siletzky S A, Kaulen A D, Mitchell D, Gennis R B, Konstantinov A A. Biochim Biophys Acta EBEC Rep. 1996;9:13–27. [Google Scholar]
- 36.Verkhovskaya M L, Garcia-Horsman A, Puustinen A, Rigaud J L, Morgan J E, Verkhovsky M I, Wikström M. Proc Natl Acad Sci USA. 1997;94:10128–10131. doi: 10.1073/pnas.94.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ermler U, Fritzsch G, Buchanan S K, Michel H. Structure. 1994;2:925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell R, Rich P. Biochim Biophys Acta. 1994;1186:19–26. doi: 10.1016/0005-2728(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 39.Ostermeier C, Essen L-O, Michel H. Proteins Struct Funct Genet. 1995;21:74–77. doi: 10.1002/prot.340210110. [DOI] [PubMed] [Google Scholar]
- 40.Essen, L. O. (1995) Thesis (University of Frankfurt, Germany).