The crystallization of peanut allergen Ara h 3 is reported.
Keywords: food safety, allergy, cupin, glycinin
Abstract
The peanut is a significant food source, but is responsible for many cases of anaphylaxis. The peanut 11S legumin-like seed storage protein Ara h 3 is one of the best characterized allergens. In this study, Ara h 3 was extracted from peanut kernels and purified by sequential anion-exchange, hydrophobic interaction and gel-filtration chromatography to very high purity to facilitate crystallization and structural studies. Well diffracting single crystals were obtained by the vapor-diffusion method. A molecular-replacement structural solution has been obtained and refinement of the structure is currently under way.
1. Introduction
Peanut (Arachis hypogaea) allergy is a serious and potentially life-threatening condition (Palmer & Burks, 2006 ▶) that accounts for the majority of severe anaphylactic reactions to foods (Al-Muhsen et al., 2003 ▶). Several proteins including Ara h 1, Ara h 2, Ara h 3/4, Ara h 5, Ara h 6, Ara h 7, Ara h 8 and oleosin have been identified as peanut allergens (Palmer & Burks, 2006 ▶) that can be recognized by patients’ sera. Of these, Ara h 1, Ara h 2 and Ara h 3/4 are categorized as major allergens (Palmer & Burks, 2006 ▶); each reacts with more than 45% of patients’ sera (Scurlock & Burks, 2004 ▶).
Ara h 3, also known as glycinin, is an 11S legumin-type seed storage protein. It has a theoretical pI of 5.68 and a molecular weight of 58 349.76 Da as calculated from the published sequence of one of its isoforms (Entrez Protein Database accession No. AAC63045). It has been reported that in mature seeds of a number of species, the 11S storage proteins exist as hexamers that are post-translationally cleaved by an endopeptidase, leaving an inter-chain disulfide bond as the only covalent bond between the N-terminal acidic chain of ∼40 kDa and the C-terminal basic chain of ∼20 kDa (Gruis et al., 2004 ▶; Kitamura et al., 1976 ▶; Scott et al., 1992 ▶; Vonder Haar et al., 1988 ▶).
Structural characterization of allergens provides information that is indispensable to understanding the molecular basis of protein allergenicity and allergic cross-reactivity. The only reported structures of 11S seed storage proteins are those of a soybean (Glycin max) glycinin (Adachi et al., 2003 ▶) and a soybean proglycinin (Adachi et al., 2001 ▶). The PDB coordinates (2evx) of the 11S protein from winter squash (Cucurbita maxima) were recently released. Additional three-dimensional structures of 11S seed storage proteins are required in order to further characterize the stability and allergenicity of the 11S allergens and Ara h 3 is one of the most important allergens to study because of the high prevalence of peanut allergies. Here, we report the crystallization, X-ray data collection and initial solution of the structure of Ara h 3.
2. Methods
2.1. Protein purification
20 g of dry peanut kernels were ground and extracted in 200 ml buffer (1 M NaCl, 20 mM Tris–HCl pH 7.9) and subjected to centrifugation at 20 000g for 10 min. The supernatant was collected as a crude extract for further use. The crude extract was dialyzed against distilled water twice for 12 h. The dialyzed sample was filtered with 0.45 µm pore-size syringe filters and loaded onto an 8 ml Source Q15 column (GE Healthcare, Piscataway, NJ, USA) pre-equilibrated with 10 mM Tris buffer (10 mM Tris–HCl pH 7.9) and eluted with Tris buffer and a linear NaCl gradient of 0–0.5 M in 75 ml. Fractions containing Ara h 3 were pooled and ammonium sulfate powder was added to the pooled sample to a final concentration of 1.5 M. The sample was then filtered with 0.45 µm pore-size syringe filters prior to loading onto a 10 ml phenyl Sepharose hydrophobic interaction column (GE Healthcare) pre-equilibrated with buffer A (1.5 M ammonium sulfate, 50 mM Tris–HCl pH 7.9). The bound proteins were eluted with a 95 ml linear gradient of ammonium sulfate by mixing 0–100% buffer B (10 mM Tris–HCl pH 7.9) with buffer A. Ara h 3 was eluted as the major peak and the fractions making up the peak were pooled and loaded onto a 320 ml Superdex 200 column (GE Healthcare) pre-equilibrated and eluted with buffer B. The gel-filtration column was calibrated with thyroglobulin (669 kDa), ferritin (440 kDa), catalase (242 kDa), aldolase (158 kDa) and bovine serum albumin (67 kDa).
2.2. Crystallization
Purified Ara h 3 was concentrated to 50 mg ml−1 and the buffer was changed to distilled water by repeated dilution and concentration using Ultracel 30k filter devices (Millipore, Bedford, MA, USA). A crystallization screen was set up at room temperature using the Crystal Screen kit (Hampton Research, Aliso Viejo, CA, USA) and the hanging-drop vapor-diffusion method. For each condition, 1 µl protein solution was mixed with 1 µl crystallization solution and sealed against 1 ml crystallization solution in one well of a 24-well Limbro plate. Sizable needle-shaped crystals were obtained in the first screen and crystallization optimization was mainly aimed at screening for a cryoprotectant that could be used during crystallization. Sucrose, glycerol, PEG 400 and ethylene glycol were screened.
2.3. X-ray diffraction experiments and crystal characterization
Single crystals were picked, flash-cooled in liquid nitrogen and stored in a liquid-nitrogen tank. X-ray data collection was performed using a MAR300 CCD detector at the SER-CAT 22ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. The diffraction data were processed using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶) and XGEN (Howard, 2000 ▶). A structural model was derived by molecular-replacement calculations using the program Phaser (McCoy et al., 2005 ▶; Storoni et al., 2004 ▶).
3. Results and discussion
In the gel-filtration step of the purification, Ara h 3 eluted at 162 ml as a single peak with an apparent molecular weight of ∼407 kDa, indicating that it exists as a hexamer in solution. On reducing SDS–PAGE it separated into more than one band, consistent with reports in the literature (Koppelman et al., 2003 ▶), and the only visible bands are those that arise from (one form of) Ara h 3 (Fig. 1 ▶). SDS–PAGE analysis of redissolved crystals showed the same bands as those shown in Fig. 1 ▶. There are 11 entries for peanut 11S storage protein in the Entrez protein-sequence database at NCBI and 52% of the amino acids are identical in all the sequences. Long stretches of deletions in one or two of the sequences contributed to the apparently low sequence identity. The sequence variations may represent differences between peanut cultivars or even different isoforms of the peanut 11S protein. Currently, we are in the process of identifying the cleavage site by N-terminal sequencing and identifying which variation is closest to the 11S protein crystallized by the procedures used in this study.
Figure 1.
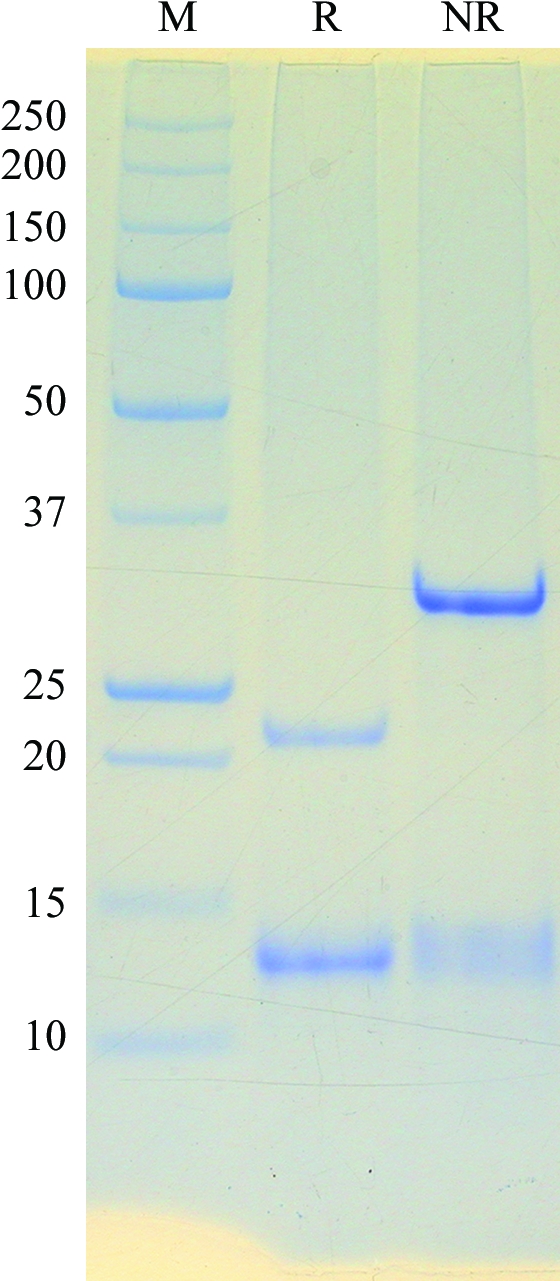
SDS–PAGE analysis of purified Ara h 3. The protein was separated with a 10–20% gel under reducing (lane R) and nonreducing (lane NR) conditions. The molecular weights (in kDa) of the protein standards contained in the marker (lane M) are shown on the left side of the gel image.
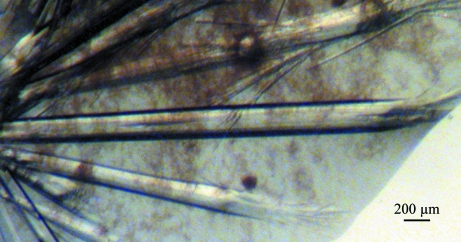
In the crystallization screen, a number of crystals appeared within a week at room temperature in a drop hanging over solution No. 37 of the Crystal Screen kit [8%(w/v) PEG 4000, 0.1 M sodium acetate trihydrate pH 4.6]. After optimization, long single crystals with an average cross-section of ∼100 × 150 µm were obtained using this hit solution with 20%(v/v) glycerol (Fig. 2 ▶).
Figure 2.
Single crystals of Ara h 3 obtained by vapor diffusion using the hanging-drop method.
Noticeable radiation damage to the Ara h 3 crystals was observed during data collection, but we were able to collect a complete 1.74 Å data set. Data processing using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶) and XGEN (Howard, 2000 ▶) revealed a primitive hexagonal crystal system with unit-cell parameters a = b = 144.931, c = 86.924 Å. Based on systematic absences of specific reflections in the diffraction, the space group was determined to be P6322 (Table 1 ▶). Assuming the presence of one Ara h 3 molecule in an asymmetric unit and an average partial specific volume of 0.74 cm3 g−1 for proteins, the Matthews coefficient was 2.44 Å3 Da−1, with a corresponding crystal solvent content of ∼49.6%.
Table 1. X-ray data-collection statistics.
Values in parentheses are for the outer shell.
| Resolution (Å) | 50–1.74 (1.80–1.74) |
| Wavelength (Å) | 1.0 |
| Data-collection temperature (K) | 110 |
| No. of observed reflections | 1018072 |
| No. of unique reflections | 55192 |
| Completeness (%) | 99.1 (93.5) |
| Mean I/σ(I) | 13.9 (2.14) |
| Rsym† (%) | 12.0 (83.1) |
R
sym = 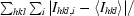
 .
.
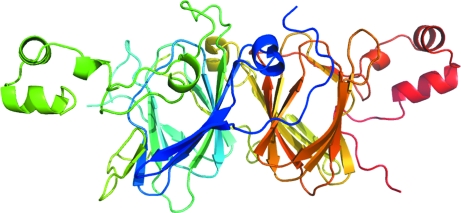
Searching the Protein Data Bank with BLAST showed that peanut glycinin Ara h 3 and soybean proglycinin have 55% sequence identity over a region of 515 amino acids. The two proteins have 57% sequence identity over a region of 510 amino acids (Fig. 3 ▶) when aligned with ClustalW (Thompson et al., 1994 ▶). Thus, we constructed an Ara h 3 structural model (Fig. 4 ▶) with the program SCWRL (Canutescu et al., 2003 ▶) using chain A of the soybean proglycinin structure (Adachi et al., 2001 ▶; PDB code 1fxz) as a template. The monomeric Ara h 3 model was used in a molecular-replacement calculation using the program Phaser (McCoy et al., 2005 ▶; Storoni et al., 2004 ▶). This led to a molecular-replacement solution with one Ara h 3 molecule in the asymmetric unit and a log-likelihood gain of 1225. The R factor of the solution after one round of rigid-body and restrained refinement with the program REFMAC (Murshudov et al., 1997 ▶) was 32%, with an R free of 36% using a 5% set of randomly chosen test reflections.
Figure 3.
Sequence alignment of soybean glycinin (from PDB entry 1fxz) and Ara h 3. Sequence alignment was carried out with the program ClustalW (Thompson et al., 1994 ▶) using Entrez Protein database accession No. AAC63045 and 1fxz chain A. Identical residues are highlighted in red and similar residues are highlighted in yellow.
Figure 4.
The structural model of Ara h 3 used as a search structure in molecular replacement. The model was built with the program SCWRL (Canutescu et al., 2003 ▶). The model is presented in a rainbow color with the N-terminus blue and the C-terminus red.
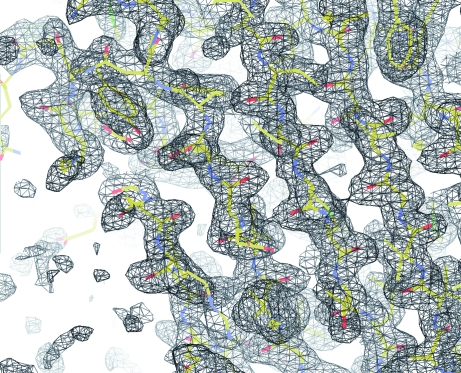
A preliminary inspection of the map calculation and refinement led to an experimental electron-density map at 1.74 Å resolution with a clear protein–solvent boundary. The electron-density map allowed modeling of most of the main-chain atoms of Ara h 3 (Fig. 5 ▶). Currently, model building and refinement of the structure is under way. Structural determination of Ara h 3 will provide information required to understand the allergenicity of this peanut allergen.
Figure 5.
A portion of a 2F o − F c electron-density map. The map was calculated after a rigid-body and restrained refinement cycle starting from a Phaser molecular-replacement solution. The map was contoured at 1.0σ and the structure model is shown in stick representation using the CPK coloring scheme.
Acknowledgments
This work was supported by funding from Illinois Institute of Technology. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the APS, Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38.
References
- Adachi, M., Kanamori, J., Masuda, T., Yagasaki, K., Kitamura, K., Mikami, B. & Utsumi, S. (2003). Proc. Natl Acad. Sci. USA, 100, 7395–7400. [DOI] [PMC free article] [PubMed]
- Adachi, M., Takenaka, Y., Gidamis, A. B., Mikami, B. & Utsumi, S. (2001). J. Mol. Biol.305, 291–305. [DOI] [PubMed]
- Al-Muhsen, S., Clarke, A. E. & Kagan, R. S. (2003). Can. Med. Assoc. J.168, 1279–1285. [PMC free article] [PubMed]
- Canutescu, A. A., Shelenkov, A. A. & Dunbrack, R. L. Jr (2003). Protein Sci.12, 2001–2014. [DOI] [PMC free article] [PubMed]
- Gruis, D., Schulze, J. & Jung, R. (2004). Plant Cell, 16, 270–290. [DOI] [PMC free article] [PubMed]
- Howard, A. J. (2000). In Crystallographic Computing 7, edited by P. E. Bourne & K. D. Watenpaugh. Oxford University Press.
- Kitamura, K., Takagi, T. & Shibasaki, K. (1976). Agric. Biol. Chem.40, 1837–1844.
- Koppelman, S. J., Knol, E. F., Vlooswijk, R. A., Wensing, M., Knulst, A. C., Hefle, S. L., Gruppen, H. & Piersma, S. (2003). Allergy, 58, 1144–1151. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Palmer, K. & Burks, W. (2006). Curr. Opin. Allergy Clin. Immunol.6, 202–206. [DOI] [PubMed]
- Scott, M. P., Jung, R., Muntz, K. & Nielsen, N. C. (1992). Proc. Natl Acad. Sci. USA, 89, 658–662. [DOI] [PMC free article] [PubMed]
- Scurlock, A. M. & Burks, A. W. (2004). Ann. Allergy Asthma Immunol.93, S12–S18. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Vonder Haar, R. A., Allen, R. D., Cohen, E. A., Nessler, C. L. & Thomas, T. L. (1988). Gene, 74, 433–443. [DOI] [PubMed]