The structure of AKR1C21 holoenzyme was determined at 1.8 Å resolution. A model describing the interaction between AKR1C21 and steroid substrates is proposed that explains the bifunctionality of the enzyme.
Keywords: 3(17)α-hydroxysteroid dehydrogenase
Abstract
Mouse 3(17)α-hydroxysteroid dehydrogenase (AKR1C21) is a bifunctional enzyme that catalyses the oxidoreduction of the 3- and 17-hydroxy/keto groups of steroid substrates such as oestrogens, androgens and neurosteroids. The structure of the AKR1C21–NADPH binary complex was determined from an orthorhombic crystal belonging to space group P212121 at a resolution of 1.8 Å. In order to identify the factors responsible for the bifunctionality of AKR1C21, three steroid substrates including a 17-keto steroid, a 3-keto steroid and a 3α-hydroxysteroid were docked into the substrate-binding cavity. Models of the enzyme–coenzyme–substrate complexes suggest that Lys31, Gly225 and Gly226 are important for ligand recognition and orientation in the active site.
1. Introduction
Hydroxysteroid dehydrogenases (HSDs) play a pivotal role in regulating the activity of potent steroid hormones at the pre-receptor level. HSDs stereospecifically catalyse the interconversion of active and inactive steroid substrates through the oxidoreduction of hydroxyl/keto groups at specific positions on the steroid molecules. The reductase and oxidase activities of HSDs permit them to function as molecular switches that regulate the occupancy of steroid hormone receptors (Bauman et al., 2004 ▶; Penning, 2003 ▶). HSDs are divided into two different protein superfamilies: the short-chain dehydrogenase/reductase (SDR) and aldo-keto reductase (AKR) superfamilies (Jez & Penning, 2001 ▶; Oppermann et al., 2003 ▶). The members of the AKR superfamily are cytosolic NADPH-dependent oxidoreductases that metabolize a variety of substrates ranging from aldehydes and monosaccharides to steroids and prostaglandins (Jez et al., 1997 ▶) and possess an (α/β)8-barrel motif characteristic of triose phosphate isomerase (TIM) (El-Kabbani et al., 1995 ▶; Hoog et al., 1994 ▶; Rondeau et al., 1992 ▶; Wilson et al., 1995 ▶; Hyndman et al., 2003 ▶).
The mammalian HSDs in the AKR superfamily share more than 60% sequence identity and are grouped into the AKR1C subfamily. In spite of their high sequence identity, these enzymes display varying ratios of 3-, 17- and 20-keto steroid reductase activity and varying ratios of 3α-, 17β- and 20α-hydroxysteroid dehydrogenase activity (Penning et al., 2000 ▶). The functional plasticity of these enzymes highlights their ability to regulate steroid hormone action by controlling receptor occupancy in target tissues. Mouse 3(17)α-hydroxysteroid dehydrogenase was originally found in the kidney as one of the multiple forms of dihydrodiol dehydrogenase (Nakagawa et al., 1989 ▶) and was recently identified as a new member (AKR1C21) of the AKR1C subfamily by cloning its cDNA (Ishikura et al., 2004 ▶; Bellemare et al., 2005 ▶). Like other members of the AKR1C subfamily, AKR1C21 exhibits functional plasticity and can oxidize/reduce steroid substrates at positions C3 and C17. However, AKR1C21 is unique in its ability to produce 17α-hydroxyl derivatives from the respective 17-keto steroids, as opposed to other HSDs, which can only reduce the 17-keto steroids to the corresponding 17β-hydroxysteroids (Penning et al., 2000 ▶; Ishikura et al., 2004 ▶).
Previous studies have shown that AKR1C21 efficiently reduces androstenedione to epitestosterone (epi-T) through its 17α-HSD activity (Ishikura et al., 2004 ▶; Bellemare et al., 2005 ▶). Epi-T, an epimer of testosterone, has been found to accumulate in breast cyst fluid and the prostate (Bellemare et al., 2005 ▶). Although the exact pathway leading to the formation of epi-T in the human body is unknown, administration of androstenedione has been reported to increase urinary excretion of epi-T, which suggests that androstenedione may be the source of epi-T (Catlin et al., 2002 ▶). AKR1C21 is expressed in many mouse tissues (Ishikura et al., 2004 ▶) and also exhibits high 3α-HSD activity towards various 3-keto steroids, including 21-hydroxy-5β-pregnane-3,20-dione (HPD), which is a precursor of the potent positive modulators of the γ-aminobutyric acid type A (GABAA) receptor (Rupprecht & Holsboer, 1999 ▶; Morris et al., 1999 ▶). The stereospecificity and functional plasticity of AKR1C21 raises questions as to how the enzyme selectively recognizes the two positions on the steroid molecules and whether these substrates differ in the type of interactions that are formed with residues in the substrate-binding pocket.
In an attempt to elucidate the molecular determinants of substrate selectivity for AKR1C21, we studied the crystal structure of the AKR1C21–NADPH binary complex and modelled physiological substrates in the active site. In this report, we present a detailed account of molecular-docking simulations of three steroidal substrates (5β-pregnane-3α,20α-diol, HPD and androstenedione) docked into the substrate-binding cavity of AKR1C21 (Fig. 1 ▶). Our models suggest that the 3-keto/3α-hydroxysteroids and the 17-keto steroids bind differently within the steroid-binding pocket and are orientated by hydrogen-bonding interactions with residues Lys31, Gly225 and Gly226 of the steroid-binding pocket.
Figure 1.
Chemical structures of the three steroid molecules that were docked into the active site of AKR1C21. The C13 and C17 atoms are labelled. H atoms and substituted groups that are oriented towards the α-side of the steroid molecule are indicated by broken bonds and those oriented towards the β-side are indicated by solid bonds.
2. Materials and methods
2.1. Crystallization
Recombinant AKR1C21 was expressed and purified as described previously (Ishikura et al., 2004 ▶; Nakagawa et al., 1989 ▶). Crystals of the AKR1C21–NADPH binary complex were grown using the hanging-drop vapour-diffusion method (McPherson, 1985 ▶) at 295 K in crystallization buffer containing 0.1 M HEPES pH 7.5, 10% polyethylene glycol 6000 and 5% 2-methyl-2,4-pentanediol (MPD) as described previously (El-Kabbani et al., 2005 ▶). The final concentration of protein in the binary complex solution was 18 mg ml−1. Droplets were prepared by mixing 2 µl of the binary complex solution (comprising AKR1C21 and NADPH in a 1:3 molar ratio) with an equal volume of the crystallization buffer. The mixture was placed on siliconized cover slips and allowed to equilibrate at 295 K against 1 ml reservoir solution. Crystals with a longest dimension of approximately 0.3 mm were obtained within a week and were used for X-ray diffraction analysis.
2.2. Data collection and analysis
The crystals used for X-ray diffraction analysis were directly soaked in a cryoprotective solution (20% ethylene glycol added to the mother liquor) and flash-cooled at 100 K. Diffraction data were collected using a MAR CCD detector on the synchrotron beamline X06SA at the Swiss Light Source (λ = 1.00002 Å). Each frame was recorded with 3 s exposure and 0.4° oscillation around ϕ. The detector was set at a distance of 165 mm so that the spots were clearly resolved. The data were processed using XDS and scaled using the XSCALE software package (Kabsch, 1993 ▶). There were two monomers of AKR1C21 per asymmetric unit, with a calculated solvent content of 47.38% and a Matthews coefficient of 2.34 Å3 Da−1.
2.3. Structure solution and refinement
The initial phases of the binary complex were determined by molecular replacement using AKR1C1 (73% sequence identity) as a search model with the MOLREP software from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶; Storoni et al., 2004 ▶). Prior to molecular-replacement calculations, residues that were non-identical to those in AKR1C21, the ligand (NADP+) and solvent molecules were omitted from the AKR1C1 (PDB code 1mrq) search model (Couture et al., 2003 ▶). Refinement was performed using REFMAC5 and the difference Fourier maps (2F o − F c and F o − F c) were visualized in Coot (Murshudov et al., 1997 ▶; Collaborative Computational Project, Number 4, 1994 ▶; Emsley & Cowtan, 2004 ▶). The model was refined by simple energy minimization followed by refinement of isotropic B factors and was corrected by manually fitting the amino-acid side chains into the 2F o − F c and F o − F c electron-density maps. After several rounds of manual rebuilding, NADPH, MPD and water molecules were added to the model and it was further refined to a resolution of 1.8 Å. Structure validation was conducted using PROCHECK (Laskowski et al., 1993 ▶). A summary of the data-collection and refinement statistics are presented in Table 1 ▶.
Table 1. Summary of the data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell (2.0–1.8 Å).
| Data collection and processing | |
| X-ray source | SLS beamline X06SA |
| Detector | MAR CCD |
| Wavelength (Å) | 1.00002 |
| Unit-cell parameters (Å, °) | a = 84.91, b = 84.90, c = 95.83, α = β = γ = 90.0 |
| Space group | P212121 |
| Diffraction data | |
| Resolution range (Å) | 50–1.8 |
| Unique reflections | 59131 |
| Completeness (%) | 91.1 (84.8) |
| Data redundancy | 3.2 (3.2) |
| Rmerge | 6.8 (47.5) |
| Refinement (12–1.8 Å) | |
| Rfree (%) | 26.8 |
| Rcryst (%) | 19.9 |
| Size of Rfree set (%) | 5.1 |
| Protein residues (dimer) | 646 |
| NADPH molecules | 2 |
| MPD molecules | 2 |
| Water molecules | 569 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.018 |
| Bond angles (°) | 1.873 |
| Ramachandran plot | |
| Residues in favoured regions (%) | 92 |
| Residues in disallowed regions (%) | 0.3 |
| Estimated coordinate error | |
| Luzzati plot | 0.248 |
| B factors (Å2) | |
| Average for protein (chain A + B) | 27.093 |
| NADPH | 23.815 |
| MPD | 50.499 |
| Waters | 34.590 |
2.4. Molecular docking and energy minimization
In order to determine the mode of binding for different steroid substrates, such as 5β-pregnane-3α,20α-diol, HPD and androstenedione, the substrates were manually docked into the active-site pocket of AKR1C21. Based on the optimal substrate-positioning criteria reported in previous studies, all three substrates were modelled into the active site with the β-face exposed to the coenzyme and the C3/C17 carbonyl/hydroxyl group at an appropriate distance from the C4 atom of the nicotinamide ring for hydride transfer (Bennett et al., 1997 ▶; Nahoum et al., 2001 ▶; Couture et al., 2003 ▶). The reactive hydroxyl/keto groups on the substrates were positioned within hydrogen-bonding distance of the catalytic residues Tyr55 and His117. The torsion angles of the residues lining the substrate-binding cavity, such as Trp227, Tyr224, Phe219, Tyr118 and Tyr55, were adjusted in order to avoid close contacts with the docked substrates. NADPH was used as the coenzyme for the reduction of ketosteroids, whereas NADP+ was used for the oxidation of hydroxysteroids. H atoms, partial charges, atomic potentials and bond orders were assigned to the substrate–protein complexes using automatic procedures within the InsightII 2.1 package (Biosym Technologies Inc., San Diego, CA, USA). The protein–substrate complexes were energy-minimized to reduce steric hindrance using the Discover 2.7 package (Biosym Technologies Inc., San Diego, CA, USA) on a Linux workstation. Energy-minimization calculations were performed using the steepest-descent and conjugate-gradient algorithms down to a maximum root-mean-square derivative of 41.8 and 0.04 kJ Å−1, respectively. Energy minimization was carried out using previously established protocols that had been found to be effective for visualizing the protein–ligand complex in a conformation close to its lowest energy structure (Darmanin & El-Kabbani, 2000 ▶, 2001 ▶). Figures were generated using PyMOL (DeLano Scientific, San Carlos, CA, USA).
3. Results and discussion
3.1. Overall structure description of AKR1C21
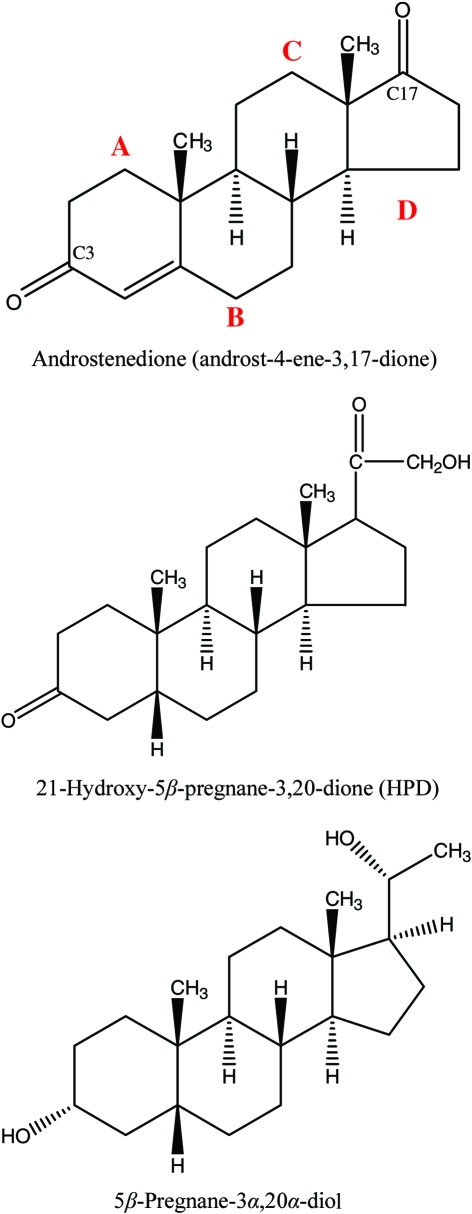
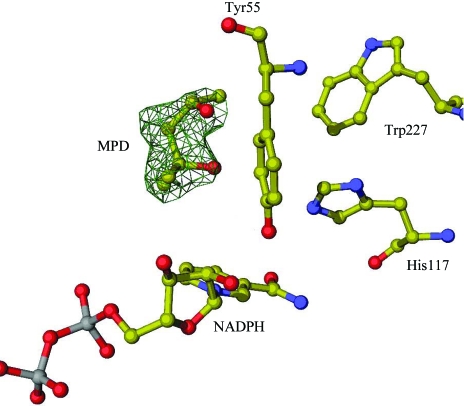
Fig. 2 ▶ shows a representative electron-density map for MPD, which was bound in the active site of AKR1C21. The backbone dihedral angles of 92% of the residues are in the favoured regions of the Ramachandran plot. Only Thr221, which forms a hydrogen bond with the coenzyme through its main chain, is in the disallowed regions. Like most other members of the family, AKR1C21 adopts an (α/β)8 TIM-barrel motif comprising eight parallel β-sheets each alternating with an α-helix running antiparallel to the β-sheet (Fig. 3 ▶).
Figure 2.
Difference electron-density map (F o − F c) corresponding to MPD in the active site of AKR1C21 calculated at 1.8 Å resolution and contoured at the 2.5σ level. The residues surrounding the MPD molecule and the coenzyme NADPH are labelled.
Figure 3.
A ribbon representation of the AKR1C21 monomer coloured according to secondary structure (α-helices are shown in cyan, loop regions in orange and β-sheets in purple). The ligands MPD and NADPH are shown as sticks.
A comparison of the AKR1C21–NADPH interactions in the orthorhombic and the recently reported monoclinic (PDB code 2hej) crystal forms revealed minor differences in the orientations of the side chains of Asn167 and Asn280 (Faucher et al., 2006 ▶). In the monoclinic structure Asn167 OD1 is within hydrogen-bonding distance of the carbonyl O atom of the nicotinamide ring of the coenzyme, whereas in the orthorhombic structure Asn167 ND2 forms a hydrogen bond with the carbonyl O atom of the nicotinamide ring. Furthermore, in the monoclinic model Asn280 OD1 and ND2 are within hydrogen-bonding distance of the N7 and amino group on the adenine ring of NADPH, respectively. In this model, Asn280 ND2 and OD1 form hydrogen bonds with N7 and the amino group of the adenine ring, respectively.
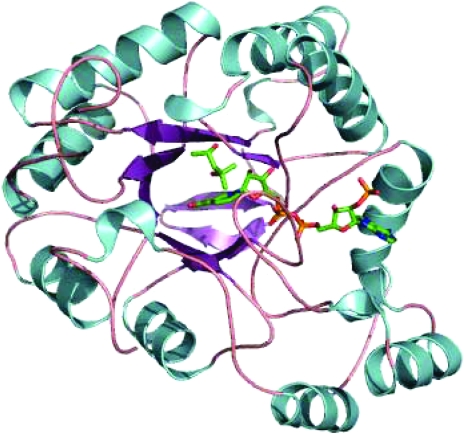
Interesting observations can be drawn upon comparison of the orientations of the MPD molecules bound to the monoclinic (Faucher et al., 2006 ▶) and orthorhombic AKR1C21 structures and of the type of interactions they form with the surrounding residues. Both structures contain a water molecule present in close proximity to His117, Tyr118 and the coenzyme NADPH. The presence of a water molecule in the active site is thought to mimic the carbonyl group of the steroid substrate and has also been observed in the crystal structures of other members of the AKR family (Jez et al., 1997 ▶). In the orthorhombic structure, the water molecule is within hydrogen-bonding distance of residues His117, Tyr118 and MPD. In the monoclinic structure no such interaction is present between MPD, His117 and the water molecule; instead, the MPD molecule is shown to form a hydrogen bond to a byproduct of MPD, 1,2-ethanediol (Fig. 4 ▶). In addition to MPD, the monoclinic model also contains five molecules of 1,2-ethanediol and two molecules of 2-mercaptoethanol forming disulfide bonds with Cys29 from each monomer that are absent in this model. A disulfide bond between the N-terminal residues Cys5 and Cys7 present in this model is missing in the monoclinic structure, together with the five N-terminal residues (Faucher et al., 2006 ▶).
Figure 4.
Superimposed MPD molecules from the orthorhombic model (green) and the monoclinic model (orange) within the binding cavity of AKR1C21. The active-site water molecule is shown in blue, MPD, His117, Tyr118 and NADPH from the orthorhombic model are shown in green and MPD and 1,2-ethanediol from the monoclinic model are shown in orange. Hydrogen bonds and the distance from C4 of the nicotinamide are represented by dotted lines, with the respective distances shown.
3.2. Modelled steroid-binding interactions
AKR1C21 is a bifunctional 3(17)α-HSD. In order to gain an understanding of the mode of binding of 3-keto-, 3α-hydroxy- and 17-keto steroid substrates, we manually docked HPD, 5β-pregnane-3α,20α-diol and androstenedione into the active site of AKR1C21. The enzyme–substrate complexes were energy-minimized to reduce steric hindrance with other amino-acid residues in the active site. Our model of the AKR1C21–substrate complex revealed that the steroid molecule is held in the substrate-binding cavity through contacts between residues in this cavity and atoms at both ends (the A and D rings) of the steroid molecule. Details of the individual interactions are discussed below.
3.3. HPD (3-keto steroid)
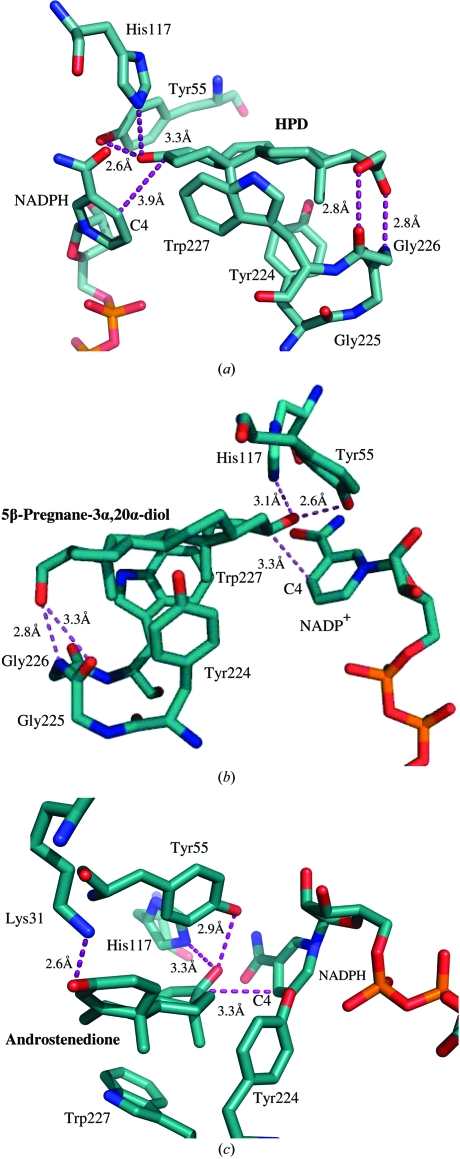
AKR1C21 has been reported to efficiently reduce HPD to 21-hydroxy-5β-pregnane-3α-ol-20-one (Ishikura et al., 2004 ▶). Our model suggests that the carbonyl and hydroxyl groups on the D ring of HPD are within hydrogen-bonding distance of the main-chain imine (2.8 Å) and carbonyl (2.8 Å) groups, respectively, of Gly226 (Fig. 5 ▶ a). In addition, the C3 carbonyl group of the A ring makes contacts with Tyr55 (2.6 Å), His117 (3.3 Å) and the C4 atom on the nicotinamide ring of NADPH (3.9 Å). The methyl groups of the substrate are sandwiched between the aromatic rings of Trp227 and Tyr224 and stabilized via hydrophobic interactions. Interactions between the carbonyl and hydroxyl groups on the D ring of HPD and the main-chain carbonyl and imine groups of residue Gly226 may play an important role in maintaining the stability of the substrate in the binding pocket. This is in agreement with the threefold drop in catalytic efficiency for 5β-pregnane-3,20-dione (Ishikura et al., 2004 ▶), which lacks the carbonyl group on the D ring (Fig. 1 ▶).
Figure 5.
Models of the three substrates (a) HPD, (b) 5β-pregnane-3α,20α-diol and (c) androstenedione docked in the active site of AKR1C21. The cofactor NADP(H) and the active-site residues are labelled. Hydrogen bonds and distances from C4 of the nicotinamide are represented by dotted lines (purple), with the respective distances shown in black.
3.4. β-Pregnane-3α,20α-diol (3α-hydroxysteroid)
The model of 5β-pregnane-3α,20α-diol in complex with AKR1C21 (Fig. 5 ▶ b) revealed a similar mode of binding as that for HPD. The hydroxyl group on the C20 C atom is within hydrogen-bonding distance of the main-chain carbonyl group of Gly225 (3.3 Å) and the imine group of Gly226 (2.8 Å). The 3α-hydroxyl group on the A ring of the steroid is hydrogen bonded to His117 (3.1 Å) and Tyr55 (2.6 Å). The C3 C atom is separated by 3.3 Å from the C4 atom on the nicotinamide ring of the coenzyme. Similar to HPD, the methyl groups of the steroid substrate are sandwiched between the aromatic rings of Tyr224 and Trp227. This observation suggests that substrates that undergo either 3α-dehydrogenase activity or 3-ketosteroid reductase activity bind to the active site in the same manner and are stabilized through similar interactions with residues in the substrate-binding cavity.
3.5. Androstenedione (17-keto steroid)
AKR1C21 is the only enzyme that has been identified to efficiently reduce androstenedione to epi-T, with a k cat/K m of 50 min−1 µm−1 (Ishikura et al., 2004 ▶). Our model of the androstenedione–AKR1C21 suggests that androstenedione is oriented with its A and B rings directed into the active-site cavity at the opposite side of the C and D rings of 5β-pregnane-3α,20α-diol and HPD (Fig. 5 ▶ c). This is likely to be a consequence of the asymmetric nature of the four rings forming the frame of the steroid substrate molecules. Owing to the difference in the orientation of the A and B rings of androstenedione, its C3 carbonyl group was within hydrogen-bonding distance of Lys31 (2.6 Å), which is known to play an important role in stabilizing the substrate in the active site for efficient catalysis, as demonstrated recently with epi-T (Faucher et al., 2007 ▶). Similar to the 3-keto/3α-hydroxy substrates, the methyl groups of androstenedione are sandwiched between the aromatic rings of Tyr224 and Trp227. The C17 carbonyl group on the D ring is within hydrogen-bonding distance of the catalytic residues His117 (3.3 Å) and Tyr55 (2.9 Å) and the C17 C atom was separated by 3.3 Å from the C4 atom on the nicotinamide ring of the coenzyme. These results are consistent with the findings of a recent study on the crystal structure of the AKR1C21–NADP+–epi-T ternary complex, which identified Lys31 as an important residue for substrate stability during catalysis (Faucher et al., 2007 ▶).
4. Conclusion
The biological importance of the 17α-hydroxysteroid activity of AKR1C21 was the impetus for this study since AKR1C21 is the only member of the AKR1C family that is known to efficiently catalyse stereospecific reduction of the ketone group at the C17 position of the steroid molecule. In order to determine the factors that influence the bifunctionality of AKR1C21, three substrates were docked into its active site. The results showed that C3 and C17 steroid substrates were oriented differently in the active site and hence were involved in hydrogen bonding to different residues lining the substrate-binding cavity. Our models suggest that Lys31 (as shown for the 17-keto steroid) and Gly225 and Gly226 (as shown for the 3α-hydroxysteroid and 3-keto steroid) are the key residues involved in substrate recognition as well as product release. This is consistent with the recent findings of a study on the crystal structure of the AKR1C21–NADP+–epi-T ternary complex, which identified Lys31 as an important residue for substrate stability during catalysis (Faucher et al., 2007 ▶). Knowledge of the additional residues involved in substrate binding may be valuable for the design of AKR1C21 inhibitors, which is important owing to its pivotal role in the intracrine regulation of hormonal steroids.
Supplementary Material
PDB reference: 3(17)α-hydroxysteroid dehydrogenase, 2p5n, r2p5nsf
Acknowledgments
The authors wish to thank Armin Wagner and Andre Mitschler for their assistance during data collection at SLS. This work was supported by an Australian Research Council Linkage International Award to OEK and AH. UD is the recipient of a Monash Graduate School PhD Scholarship.
References
- Bauman, D. R., Steckelbroeck, S. & Penning, T. M. (2004). Drug News Perspect.17, 563–574. [DOI] [PubMed]
- Bellemare, V., Faucher, F., Breton, R. & Luu-The, B. (2005). BMC Biochem.6, 12. [DOI] [PMC free article] [PubMed]
- Bennett, M. J., Albet, R. H., Jez, J. M., Ma, H., Penning, T. M. & Lewis, M. (1997). Structure, 5, 799–812. [DOI] [PubMed]
- Catlin, D. H., Leder, B. Z., Ahrens, B. D., Hatton, C. K. & Finkelstein, J. S. (2002). Steroids, 67, 559–564. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Couture, J. F., Legrand, P., Cantin, L., Luu-The, V., Labrie, F. & Breton, R. (2003). J. Mol. Biol.331, 593–604. [DOI] [PubMed]
- Darmanin, C. & El-Kabbani, O. (2000). Bioorg. Med. Chem. Lett.10, 1101–1104. [DOI] [PubMed]
- Darmanin, C. & El-Kabbani, O. (2001). Bioorg. Med. Chem. Lett.11, 3133–3136. [DOI] [PubMed]
- El-Kabbani, O., Ishikura, S., Wagner, A., Schulze-Briese, C. & Hara, A. (2005). Acta Cryst. F61, 688–690. [DOI] [PMC free article] [PubMed]
- El-Kabbani, O., Judge, K., Ginell, S. L., Myles, D. A. A., DeLucas, L. J. & Flynn, T. G. (1995). Nature Struct. Biol.2, 687–692. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Faucher, F., Cantin, L., Pereira de Jesus-Tran, K., Lemieux, M., Luu-The, V., Labrie, F. & Breton, R. (2007). J. Mol. Biol.369, 525–540. [DOI] [PubMed]
- Faucher, F., Pereira de Jesus-Tran, K., Cantin, L., Luu-The, V., Labrie, F. & Breton, R. (2006). J. Mol. Biol.364, 747–763. [DOI] [PubMed]
- Hoog, S. S., Pawlowski, J. E., Alzari, P. M., Penning, T. M. & Lewis, M. (1994). Proc. Natl Acad. Sci. USA, 91, 2517–2521. [DOI] [PMC free article] [PubMed]
- Hyndman, D., Bauman, D. R., Heredia, V. V. & Penning, T. M. (2003). Chem. Biol. Interact.143–144, 499–525. [DOI] [PubMed]
- Ishikura, S., Usami, N., Nakajima, S., Kameyama, A., Shiraishi, H., Carbone, V., El-Kabbani, O. & Hara, A. (2004). Biol. Pharm. Bull.27, 1939–1945. [DOI] [PubMed]
- Jez, J. M., Bennett, M. J., Schlegel, B. P., Lewis, M. & Penning, T. M. (1997). Biochem. J.326, 625–636. [DOI] [PMC free article] [PubMed]
- Jez, J. M. & Penning, T. M. (2001). Chem. Biol. Interact.130–132, 499–525. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- McPherson, A. (1985). Methods Enzymol. 114, 112–120. [DOI] [PubMed]
- Morris, K. D. W., Moorefield, C. N. & Amin, J. (1999). Mol. Pharmacol.56, 752–759. [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nahoum, V., Gangloff, A., Legrand, P., Zhu, D. W., Cantin, L., Zhorov, B. S., Luu-The, V., Labrie, F., Breton, R. & Lin, S. (2001). J. Biol. Chem.276, 42091–42098. [DOI] [PubMed]
- Nakagawa, M., Tsukada, F., Nakayama, T., Matsuura, K., Hara, A. & Sawada, H. (1989). J. Biochem. (Tokyo), 106, 633–638. [DOI] [PubMed]
- Oppermann, U., Filling, C., Hult, M., Shafqat, N., Wu, X., Lindh, M., Shafqat, J., Nordling, E., Kallberg, Y., Persson, B. & Jornvall, H. (2003). Chem. Biol. Interact.143–144, 247–253. [DOI] [PubMed]
- Penning, T. M. (2003). Human Reprod. Update, 9, 193–205. [DOI] [PubMed]
- Penning, T. M., Burczynski, M. E., Jez, J. M., Hung, C., Lin, H., Haiching, M. A., Moore, M., Palackal, N. & Ratnam, K. (2000). Biochem. J.351, 67–77. [DOI] [PMC free article] [PubMed]
- Rondeau, J. M., Tete-Favier, F., Podjarny, A., Reymann, J. M., Barth, P., Biellman, J. F. & Moras, D. (1992). Nature (London), 355, 469–472. [DOI] [PubMed]
- Rupprecht, R. & Holsboer, F. (1999). Trends Neurosci.22, 410–416. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Wilson, D. K., Nakano, T., Petrash, J. M. & Quiocho, F. A. (1995). Biochemistry, 34, 14323–14330. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: 3(17)α-hydroxysteroid dehydrogenase, 2p5n, r2p5nsf