The radixin FERM domain complexed with the CD44 cytoplasmic tail peptide has been crystallized. A diffraction data set from the complex was collected to 2.1 Å.
Keywords: ERM, FERM domain, CD44
Abstract
CD44 is an important adhesion molecule that specifically binds hyaluronic acid and regulates cell–cell and cell–matrix interactions. Increasing evidence has indicated that CD44 is assembled in a regulated manner into the membrane–cytoskeletal junction, a process that is mediated by ERM (ezrin/radixin/moesin) proteins. Crystals of a complex between the radixin FERM domain and the C-terminal cytoplasmic region of CD44 have been obtained. The crystal of the radixin FERM domain bound to the CD44 cytoplasmic tail peptide belongs to space group P212121, with unit-cell parameters a = 62.70, b = 66.18, c = 86.22 Å, and contain one complex in the crystallographic asymmetric unit. An intensity data set was collected to a resolution of 2.1 Å.
1. Introduction
CD44 is an important adhesion molecule that specifically binds hyaluronic acid, a major component of the extracellular matrix, and regulates cell–cell and cell–matrix interactions. CD44 has traditionally been referred to as the homing receptor of haematopoietic cells. Isoforms of CD44 have been identified that are derived from alternative splicing events of CD44 mRNA and are ubiquitously expressed in a variety of organs and cells, including cancer cells (Mackay et al., 1994 ▶; Martin et al., 2003 ▶). The juxtamembrane region of the CD44 cytoplasmic tail exists in two isoforms, possessing a short (five-residue) or long (72-residue) cytoplasmic tail. The isoform with the long cytoplasmic tail has been the subject of extensive research (Martin et al., 2003 ▶). The long cytoplasmic tail of CD44 has been shown to interact with actin filaments in various cells, a process thought to be mediated by ERM (ezrin/radixin/moesin) proteins (Hirao et al., 1996 ▶; Sainio et al., 1997 ▶; Yonemura et al., 1999 ▶; Akisawa et al., 1999 ▶). Increasing evidence has indicated that CD44 is assembled in a regulated manner into protein complexes at the membrane–cytoskeletal junction to serve to focus downstream signal transduction events that regulate cell growth and development (Thorne et al., 2004 ▶).
ERM proteins, which link membrane proteins to the actin cytoskeleton, play structural and regulatory roles at the polarized cell cortex (Yonemura et al., 1998 ▶; Bretscher, 1999 ▶; Mangeat et al., 1999 ▶). The N-terminal FERM (4.1, ezrin, radixin, moesin) domain of ERM proteins associates with the plasma membrane and directly binds the juxtamembrane region of the cytoplasmic tail of various type I transmembrane proteins such as cell-adhesion molecules, including ICAM-2 and CD44 (Yonemura et al., 1998 ▶; Alonso-Lebrero et al., 2000 ▶; Serrador et al., 2002 ▶; Snapp et al., 2002 ▶). The FERM domain also binds a type II membrane protein, the abundant microvillar neutral endopeptidase 24.11 (NEP; Iwase et al., 2004 ▶). In addition to these membrane proteins, the FERM domain binds intracellular proteins such as Na+/H+ exchanger regulatory factor (NHERF)/ERM-binding phosphoprotein 50 (EBP50) (Weinman et al., 1995 ▶; Reczek et al., 1997 ▶) and Rho GDP-dissociation inhibitor, which is responsible for the activation of Rho (Takahashi et al., 1997 ▶; Hamada et al., 2001 ▶).
The structural and biophysical properties of protein complexes comprising the radixin FERM domain and various target proteins have been extensively studied (Hamada, Matsui et al., 2000 ▶; Hamada et al., 2001 ▶; Terawaki et al., 2003 ▶; Takai et al., 2007 ▶) and the crystal structures of radixin FERM domain complexes with inositol-1,4,5-trisphosphate, ICAM-2, NHERF and NEP have been reported (Hamada, Shimizu et al., 2000 ▶; Hamada et al., 2003 ▶; Terawaki et al., 2006 ▶, 2007 ▶). The crystal structure of the radixin FERM domain bound to ICAM-2 revealed that ICAM-2 possesses the Motif-1 sequence motif RXXTYXVXXA (where X represents any amino acid) utilized for FERM binding, a sequence motif that is well conserved in related adhesion molecules comprising the immunoglobulin superfamily. However, the sequence motif is less conserved in CD44. Although protein–protein interactions play a critical role from a biological and medical perspective, precise details concerning the interaction between the FERM domain and CD44 remain unknown. Crystallographic studies of the protein complex were therefore performed in an effort to delineate the interactions between ERM proteins and CD44. Here, we report the crystallization and preliminary crystallographic investigation of the radixin FERM domain complexed with the cytoplasmic tail of CD44, referred to as the FERM–CD44 complex.
2. Materials and methods
2.1. Protein preparation
The region of cDNA coding for the mouse radixin FERM domain (FERM; residues 1–310; Funayama et al., 1991 ▶; Hamada, Shimizu et al., 2000 ▶) was subcloned into pET49b(+) plasmid (Novagen) using the SmaI and EcoRI restriction-enzyme sites. FERM was expressed in Rosetta2(DE3)pLysS cells (Novagen) as a fusion protein with glutathione-S-transferase (GST). Cells were grown at 310 K in Luria–Bertani (LB) medium containing 50 µg ml−1 kanamycin and 50 µg ml−1 chloramphenicol. When the absorbance at 660 nm (OD660) of the cell culture reached 0.6, isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a concentration of 1 mM to induce expression of the FERM gene. Cells were grown for an additional 2 h following IPTG induction and were then collected by centrifugation at 4000 rev min−1 (Beckman J2-M1 JA10 rotor) for 15 min at 277 K. Wet cells expressing FERM were suspended in 50 mM MES buffer pH 6.8 containing 500 mM NaCl, 1 mM dithiothreitol (DTT) and 1 mM EDTA and then disrupted by sonication at 277 K. The soluble portion of the cell extract was purified according to the methods described by Hamada, Shimizu et al. (2000 ▶). Glutathione Sepharose 4B resin-bound GST-fusion protein was cleaved from the resin using 4 units ml−1 HRV3C protease (Novagen) for 18 h at 277 K. The expected peptide of the construct begins after two extra residues (Gly-Pro) that are a part of the HRV3C protease recognition sequence. As the final step of purification, gel filtration was performed using a Superdex 200 gel-filtration column (GE Healthcare) with 20 mM MES buffer pH 6.8 containing 300 mM NaCl and 1 mM DTT (stock buffer). Purified FERM was concentrated to 36 mg ml−1 using Amicon Ultra centrifugal filter devices (10 000 MWCO, Millipore). Concentrated FERM was divided into 20–50 µl aliquots in 0.5 ml tubes (Eppendorf) and immediately frozen in liquid nitrogen. Frozen samples were stored at 193 K until use. The identity of the purified protein was confirmed using matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI–TOF MS; PerSeptive Inc.) and N-terminal sequence analysis (M492; Applied Biosystems).
2.2. Peptide preparation and binding assay
The CD44 peptide region selected for the binding assay and crystallization was determined according to previous studies (Yonemura et al., 1998 ▶; Legg & Isacke, 1998 ▶). The synthesized peptide corresponding to the juxtamembrane region of the mouse CD44 cytoplasmic tail, CD44cyt(2–21), 293-SRRRCGQKKKLVINGGNGTV-312 based on the sequence data (Nottenburg et al., 1989 ▶), was purchased from Toray Research Center (Tokyo Japan). CD44cyt(2–21) consists of 20 N-terminal residues encompassing the reported binding region (Yonemura et al., 1998 ▶; Legg & Isacke, 1998 ▶). CD44cyt(2–21) was dissolved at a concentration of 12 mM in stock buffer, divided into 25 µl aliquots in 0.5 ml tubes (Eppendorf) and then stored at 193 K until use.
N-terminally biotinylated CD44cyt(2–21) [bio-CD44cyt(2–21)] was purchased from Toray Research Center (Tokyo, Japan) and dissolved at a concentration of 5 mM in HBS-EP buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20) (BIAcore) containing 1 mM DTT. Surface plasmon resonance (SPR) measurements were carried out using a BIAcore 3000 Biosensor (BIAcore) to investigate the interaction between bio-CD44cyt(2–21) and FERM, with a running buffer of HBS-EP containing 1 mM DTT. Bio-CD44cyt(2–21) was immobilized onto the surface of an SA sensor chip (sensor chip SA, BIAcore), which was regenerated with 0.01 M HEPES buffer pH 7.4 containing 1 M NaCl and 1 mM DTT. Purified FERM was injected onto the peptide surfaces at 298 K. Kinetic parameters were evaluated using the BIA evaluation software (BIAcore).
2.3. Crystallization
Purified FERM and CD44cyt(2–21) were mixed in a 1:5 molar ratio [0.3 mM FERM and 1.5 mM CD44cyt(2–21)] with stock buffer (protein solution 1). Initial screening of the crystallization conditions was carried out with the sitting-drop vapour-diffusion method at 277 K using a HYDRA II (Scrum) and commercial crystallization screening kits (Qiagen; PEGs I, PEGs II, JCSG, pH Clear, MB Class, MB Class II, Classics, Classics Lite, ProComplex and the PACT suite). FERM–CD44cyt(2–21) complex crystals were obtained within 3 d by mixing 0.2 µl protein solution 1 with 0.2 µl PACT suite reservoir solution No. 88 [0.1 M bis-Tris buffer pH 8.5 containing 20% polyethylene glycol 3350 (PEG 3350) and 0.2 M potassium thiocyanate]. Crystallization conditions were then refined by changing the pH (between 8.0 and 8.7 in increments of 0.1) and the PEG 3350 concentration (15–25% in 5% increments).
2.4. X-ray data collection
Crystals for diffraction studies were transferred stepwise into a cryoprotective solution consisting of 0.05 M Tris buffer pH 8.6 containing 0.15 M NaCl, 0.1 M potassium thiocyanate, 20% PEG 3350 and 20% PEG 200 for flash-cooling. X-ray diffraction data were collected from the FERM–CD44cyt(2–21) complex crystal using a Rigaku MSC Jupiter 210 detector installed on beamline BL38B1 at SPring-8, Harima, Japan. All data were processed using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
A quantitative analysis of peptide binding to the purified FERM protein was performed using an SPR sensor (BIAcore). CD44cyt(2–21) binds the radixin FERM domain with a dissociation constant (K d) of 0.95 µM (data not shown). The crystals obtained from the initial screening appeared to be clustered and stacked. A microseeding method was then employed in an effort to obtain single crystals suitable for X-ray diffraction.
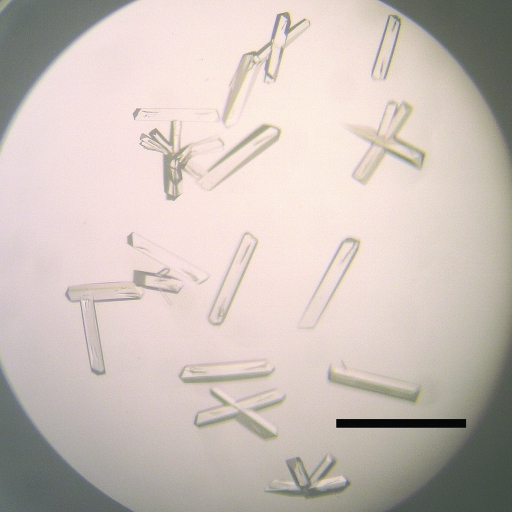
Crystals for microseeding were obtained using the hanging-drop vapour-diffusion method at 277 K. FERM and CD44cyt(2–21) were mixed in a 1:20 molar ratio [0.3 mM FERM and 6 mM CD44cyt(2–21)] with stock buffer (protein solution 2). The drop was comprised of 1 µl protein solution 2 and 1 µl reservoir solution containing 0.1 M Tris pH 8.6, 25% PEG 3350 and 0.2 M potassium thiocyanate. Clustered crystals were obtained after 3 d. These crystals were crushed in 10 µl 0.1 M Tris pH 8.6 containing 15% PEG 3350 and 0.2 M potassium thiocyanate (seed solution) using a glass homogenizer (Radnoti). The solution was then diluted 5000-fold with the same buffer. Microseeding was carried out by the sitting-drop vapour-diffusion method at 277 K. A 0.5 µl aliquot of the diluted seed solution was mixed with 2.5 µl protein solution 2 and 2 µl reservoir solution. Rod-like crystals (Fig. 1 ▶) of the FERM–CD44cyt(2–21) complex were obtained within 10 d. These rod-like crystals belong to space group P212121, with unit-cell parameters a = 62.70, b = 66.18, c = 86.22 Å.
Figure 1.
Rod-shaped crystals of the complex formed by FERM with CD44cyt(2–21) obtained by microseeding. The scale bar indicates 0.5 mm.
For MALDI–TOF MS analysis, the crystals obtained from one drop (5 µl) were washed with 10 µl reservoir solution thoroughly (ten times) and were redissolved in 10 µl 0.01 M HEPES pH 7.4 containing 0.15 M NaCl and 1 mM DTT. The solution was desalted using ZipTipC18 tips (Millipore) and added to a matrix of 10 mg ml−1 3,5-dimethoxy-4-hydroxycinnamic acid dissolved in 50% or 30% acetonitrile containing 0.1% trifluoroacetic acid. MALDI–TOF MS of resolved crystals gave peaks at 36 885.2 Da corresponding to the radixin FERM domain (calculated molecular weight 36 880.6 Da) and 2171.5 Da corresponding to the CD44cyt(2–21) peptide (calculated molecular weight 2171.5 Da), indicating that the crystals contained both FERM and CD44cyt(2–21). It seems probable that the asymmetric unit of the crystal contains one 1:1 FERM–CD44cyt(2–21) complex with a Matthews coefficient of 2.3 Å3 Da−1. This corresponds to 46.3% solvent content by volume, which is reasonable for protein crystals. The crystallographic data and intensity data-processing statistics are summarized in Table 1 ▶. Efforts are currently being directed towards solution of the crystal structure using conventional molecular-replacement methods.
Table 1. X-ray diffraction data of the FERM–CD44cyt(2–21) complex.
Values in parentheses are for the outer resolution shell.
| Complex | FERM–CD44cyt(2–21) |
| Beamline (SPring-8) | BL38B1 |
| Detector | MAR CCD |
| Wavelength (Å) | 1.00 |
| Temperature (K) | 100 |
| Oscillation range (°) | 180 [180 × 1° images] |
| Exposure time (s) | 25 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 62.70, b = 66.18, c = 86.22 |
| Resolution (Å) | 2.1 (2.17–2.10) |
| Reflections (total/unique) | 156148/21492 |
| Completeness (%) | 100.0 (100.0) |
| Mosaicity | 0.5–0.8 |
| 〈I/σ(I)〉 | 12.2 (4.9) |
| Rmerge† (%) | 5.0 (36.1) |
R
merge = 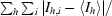
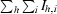 , where I
h,i is the ith observed intensity of reflection h and 〈I
h〉 is the average intensity over symmetry-equivalent measurements.
, where I
h,i is the ith observed intensity of reflection h and 〈I
h〉 is the average intensity over symmetry-equivalent measurements.
Acknowledgments
We would like to thank J. Tsukamoto for technical support in performing MALDI–TOF MS analysis and N-terminal sequence analysis of the proteins and peptides. We gratefully acknowledge Sachiko Tsukita and Shoichiro Tsukita for providing the mouse radixin cDNA and CD44 cDNA. We also acknowledge K. Hasegawa, H. Sakai and M. Yamamoto at SPring-8 for help with data collection at synchrotron beamline BL38B1. This work was supported in part by a CREST grant to TH from the Japan Science and Technology Agency and by a Grant-in-Aid for Scientific Research (A) and for Scientific Research on Priority Areas to TH from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. ST was supported by a Center of Excellence (COE) postdoctoral research fellowship with a Grant-in-Aid for the 21st Century COE Research from MEXT. RM was supported by a postdoctoral research fellowship for Young Scientists from the Japanese Society for the Promotion of Science.
References
- Alonso-Lebrero, J. L., Serrador, J. M., Domínguez-Jiménez, C., Barreiro, O., Luque, A., del Pozo, M. A., Snapp, K., Kansas, G., Schwartz-Albiez, R., Furthmayr, H., Lozano, F. & Sánchez-Madrid, F. (2000). Blood, 95, 2413–2419. [PubMed]
- Akisawa, N., Nishimori, I., Iwamura, T., Onishi, S. & Hollingsworth, M. A. (1999). Biochem. Biophys. Res. Commun.258, 395–400. [DOI] [PubMed]
- Bretscher, A. (1999). Curr. Opin. Cell Biol.11, 109–116. [DOI] [PubMed]
- Funayama, N., Nagafuchi, A., Sato, N., Tsukita, S. & Tsukita, S. (1991). J. Cell Biol.115, 1039–1048. [DOI] [PMC free article] [PubMed]
- Hamada, K., Matsui, T., Tsukita, S., Tsukita, S. & Hakoshima, T. (2000). Acta Cryst. D56, 922–923. [DOI] [PubMed]
- Hamada, K., Seto, A., Shimizu, T., Matsui, T., Takai, Y., Tsukita, S., Tsukita, S. & Hakoshima, T. (2001). Acta Cryst. D57, 889–890. [DOI] [PubMed]
- Hamada, K., Shimizu, T., Matsui, T., Tsukita, S., Tsukita, S. & Hakoshima, T. (2000). EMBO J.19, 4449–4462. [DOI] [PMC free article] [PubMed]
- Hamada, K., Shimizu, T., Yonemura, S., Tsukita, S., Tsukita, S. & Hakoshima, T. (2003). EMBO J.22, 502–514. [DOI] [PMC free article] [PubMed]
- Hirao, M., Sato, N., Kondo, T., Yonemura, S., Monden, M., Sasaki, T., Takai, Y., Tsukita, S. & Tsukita, S. (1996). J. Cell Biol.135, 37–51. [DOI] [PMC free article] [PubMed]
- Iwase, A., Shen, R., Navarro, D. & Nanus, D. M. (2004). J. Biol. Chem.279, 11898–11905. [DOI] [PubMed]
- Legg, J. W. & Isacke, C. M. (1998). Curr. Biol.8, 705–708. [DOI] [PubMed]
- Mackay, C. R., Terpe, H. J., Stauder, R., Marston, W. L., Stark, H. & Gunthert, U. (1994). J. Cell Biol.124, 71–82. [DOI] [PMC free article] [PubMed]
- Mangeat, P., Roy, C. & Martin, M. (1999). Trends Cell Biol.9, 187–192. [DOI] [PubMed]
- Martin, T. A., Harrison, G., Mansel, R. E. & Jiang, W. G. (2003). Crit. Rev. Oncol. Hematol.46, 165–186. [DOI] [PubMed]
- Nottenburg, C., Rees, G. & St John, T. (1989). Proc. Natl Acad. Sci. USA, 86, 8521–8525. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Reczek, D., Berryman, M. & Bretscher, A. (1997). J. Cell Biol.139, 169–179. [DOI] [PMC free article] [PubMed]
- Sainio, M., Zhao, F., Heiska, L., Turunen, O., den Bakker, M., Zwarthoff, E., Lutchman, M., Rouleau, G. A., Jaaskelainen, J., Vaheri, A. & Carpen, O. (1997). J. Cell Sci.110, 2249–2260. [DOI] [PubMed]
- Serrador, J. M., Urzainqui, A., Alonso-Lebrero, J. L., Cabrero, J. R., Montoya, M. C., Vicente-Manzanares, M., Yáñez-Mó, M. & Sánchez-Madrid, F. (2002). Eur. J. Immunol.32, 1560–1566. [DOI] [PubMed]
- Snapp, K. R., Heitzig, C. E. & Kansas, G. S. (2002). Blood, 99, 4494–4502. [DOI] [PubMed]
- Takahashi, K., Sasaki, T., Mammoto, A., Takaishi, K., Kameyama, T., Tsukita, S., Tsukita, S. & Takai, Y. (1997). J. Biol. Chem.272, 23371–23375. [DOI] [PubMed]
- Takai, Y., Kitano, K., Terawaki, S., Maesaki, R. & Hakoshima, T. (2007). Acta Cryst. F63, 49–51. [DOI] [PMC free article] [PubMed]
- Terawaki, S., Kitano, K. & Hakoshima, T. (2007). J. Biol. Chem.282, 19854–19862. [DOI] [PubMed]
- Terawaki, S., Maesaki, R. & Hakoshima, T. (2006). Structure, 14, 777–789. [DOI] [PubMed]
- Terawaki, S., Maesaki, R., Okada, K. & Hakoshima, T. (2003). Acta Cryst. D59, 177–179. [DOI] [PubMed]
- Thorne, R. F., Legg, J. W. & Isacke, C. M. (2004). J. Cell Sci.117, 373–380. [DOI] [PubMed]
- Weinman, E. J., Steplock, D., Wang, Y. & Shenolikar, S. (1995). J. Clin. Invest.95, 2143–2149. [DOI] [PMC free article] [PubMed]
- Yonemura, S., Hirao, M., Doi, Y., Takahashi, N., Kondo, T., Tsukita, S. & Tsukita, S. (1998). J. Cell Biol.140, 885–895. [DOI] [PMC free article] [PubMed]
- Yonemura, S., Tsukita, S. & Tsukita, S. (1999). J. Cell Biol.145, 1497–1509. [DOI] [PMC free article] [PubMed]