Chloride intracellular channel 2 (CLIC2) belongs to a family of intracellular chloride-channel proteins that can exist in a soluble form. The expression, purification and crystallization in two different crystal forms of human CLIC2 is reported.
Keywords: chloride intracellular channel 2, chloride-channel proteins
Abstract
The chloride intracellular channel (CLIC) family of proteins are unusual in that they can exist in either an integral membrane-channel form or a soluble form. Here, the expression, purification, crystallization and preliminary diffraction analysis of CLIC2, one of the least-studied members of this family, are reported. Human CLIC2 was crystallized in two different forms, both in the presence of reduced glutathione and both of which diffracted to better than 1.9 Å resolution. Crystal form A displayed P212121 symmetry, with unit-cell parameters a = 44.0, b = 74.7, c = 79.8 Å. Crystal form B displayed P21 symmetry, with unit-cell parameters a = 36.0, b = 66.9, c = 44.1 Å. Structure determination will shed more light on the structure and function of this enigmatic family of proteins.
1. Introduction
The first member of the chloride intracellular channel (CLIC) family was identified based on intracellular chloride-channel activity and was purified by affinity for a chloride-channel inhibitor (Landry et al., 1993 ▶). This led to the identification of a number of homologues that all contain a conserved region of approximately 240 residues. Consistent with their original identification as chloride channels, a number of family members are able to insert into artificial membranes in vitro and form ion channels with varying degrees of anion selectivity. Surprisingly, CLIC-family proteins can also exist in a soluble form and do not possess any obvious hydrophobic transmembrane segments, features that are reminiscent of many bacterial pore-forming toxins (Cromer et al., 2002 ▶). Based on a very weak sequence similarity between omega glutathione S-transferase (GST) and the conserved region of CLIC proteins, it was hypothesized that in their soluble form CLIC proteins adopt the canonical GST fold (Dulhunty et al., 2001 ▶). Structure determination of both CLIC1 (Harrop et al., 2001 ▶) and CLIC4 (Littler et al., 2005 ▶; Li et al., 2006 ▶) has confirmed this hypothesis and also revealed that CLIC1 can covalently bind glutathione via a conserved cysteine (Cys24; Harrop et al., 2001 ▶) in a similar manner to omega-class GSTO1-1 (Board et al., 2000 ▶). The structure adopted by cytosolic GSTs and soluble CLICs comprises two domains: an N-terminal mixed α/β thioredoxin-like domain and an all-α-helical C-terminal domain. There is good evidence that the N-terminus can translocate across membranes (Tonini et al., 2000 ▶), a step in channel formation that would require some unfolding of the N-terminal domain. Consistent with this concept, oxidation of CLIC1 favours channel formation and leads to an alternative form of soluble CLIC1 that is dimeric, with considerable unfolding of the N-terminal domain and the formation of an intramolecular disulfide between Cys59 and the conserved Cys24 (Littler et al., 2004 ▶).
CLIC2 is a relatively poorly studied member of the CLIC family. The N-terminal domain of CLIC2 lacks the cysteine equivalent to Cys59 of CLIC1, but contains another cysteine Cys33 that together with the conserved Cys30 (equivalent to Cys24 in CLIC1) forms a CxxC motif similar to glutaredoxin. Based on these cysteine-residue differences, we hypothesized that CLIC2 may undergo differential redox regulation and conformational change relative to CLIC1. To investigate this hypothesis, we have undertaken structural studies of human CLIC2 and report here the production of well diffracting crystals that have enabled the determination of the structure of CLIC2, which will be reported elsewhere.
2. Experimental procedures and results
2.1. Cloning, expression and purification
CLIC2 was expressed with a His-tagged ubiquitin fused at the N-terminus. Human CLIC2 was amplified from the EST clone AI129485 and ligated between the BamHI and PstI sites of the pQE-30 vector (Qiagen, Clifton Hills, Australia) to produce pQECLIC2 as described previously (Board et al., 2004 ▶). The CLIC2 insert was subcloned as a SacII/HindIII fragment into pHUE (Catanzariti et al., 2004 ▶) to create a His6-ubiquitin-CLIC2 fusion protein. This protein was expressed in BL21(DE3) cells grown overnight in the presence of 0.1 mM isopropyl β-thiogalactoside and processed using the methods described by Whittington et al. (1999 ▶). The recombinant protein was purified by immobilized metal-affinity chromatography with Ni–agarose as described previously for His-tagged GSTs (Whittington et al., 1999 ▶). Following dialysis to remove imidazole, the His6-ubiquitin tag was cleaved by digestion with a ubiquitin-specific protease (Baker et al., 1994 ▶; Catanzariti et al., 2004 ▶) and both the protease and His6-ubiquitin tag were removed by immobilized metal-affinity chromatography with Ni–agarose (Catanzariti et al., 2004 ▶). The protein was further purified by gel filtration on a Pharmacia fast protein liquid-chromatography Superose 12 column equilibrated with 50 mM HEPES, 10% glycerol pH 7.0.
The purified protein was dialysed into 50 mM HEPES pH 7.5 and 100 mM NaCl and concentrated to 7.25 mg ml−1 for crystal form A and to 15 mg ml−1 in 20 mM Tris–HCl pH 7.5, 50 mM NaCl for crystal form B. The purified protein was essentially completely monomeric in solution, as indicated by gel-filtration chromatography on a Superdex 75 10/300 chromatography column (GE Biosciences) in 50 mM sodium phosphate pH 7.4 and 100 mM sodium chloride (data not shown), and was greater than 95% pure as determined by SDS–PAGE.
2.2. Crystallization
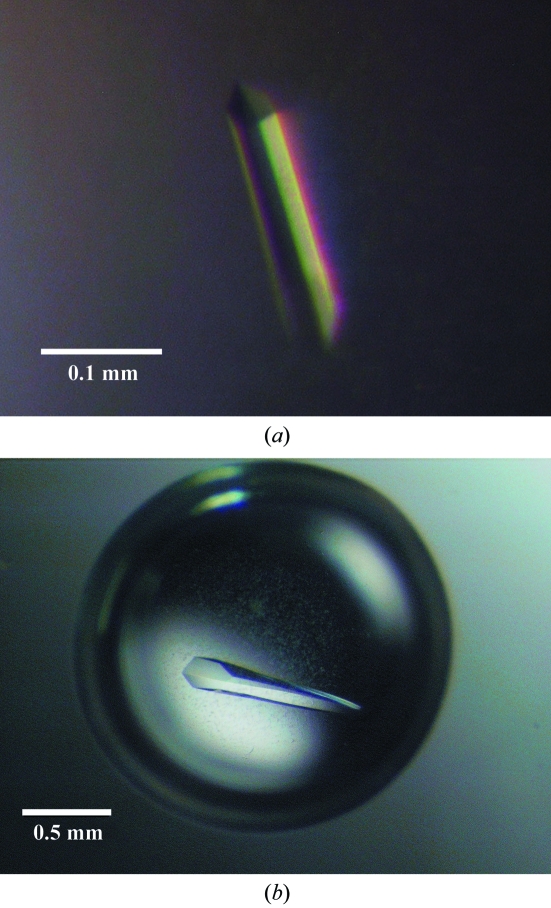
All crystallization experiments were carried out using the hanging-drop vapour-diffusion technique using 24-well Linbro tissue-culture plates (ICN Inc.) at 292 K. Drops were formed by mixing equal volumes (1 µl) of protein solution and reservoir solution. Two different crystal forms were found (Fig. 1 ▶) using different batches of purified protein at different concentrations and with slightly different reservoir solutions. For crystal form A, the protein concentration was 7.25 mg ml−1 and the reservoir solution contained 35–50%(v/v) PEG 400, 100 mM Tris–HCl pH 8.0–9.2 and 5 mM reduced glutathione (GSH). Crystals appeared after 2–3 d. For crystal form B, the protein concentration was 15 mg ml−1 and the reservoir solution contained 30–32%(v/v) PEG 400, 100 mM Tris–HCl pH 7.5 and 5 mM GSH. Crystals appeared after 3 d and were used immediately for X-ray data collection. GSH was found to be a necessary ingredient for both crystal forms.
Figure 1.
Crystals of CLIC2 in (a) crystal form A and (b) crystal form B.
2.3. Data collection
Both crystal forms were frozen in the buffer from the crystallization drop, as the concentration of PEG 400 was sufficient to prevent ice-crystal formation. Crystals were mounted in cryo-loops (Hampton Research, CA, USA) and transferred directly into a stream of nitrogen gas maintained at 100 K. For crystal form A, X-ray diffraction data were collected on BioCARS beamline 14-ID-B at the Advanced Photon Source, Chicago, USA (Table 1 ▶). For crystal form B, X-ray diffraction data were collected in-house using a Rigaku RU200H generator equipped with mirror optics (Xenocs) and a MAR Research 345 mm imaging-plate detector. Diffraction data were integrated and scaled using HKL (Otwinowski & Minor, 1997 ▶) for crystal form A and MOSFLM (Collaborative Computational Project, Number 4, 1994 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶) for crystal form B. Data statistics are shown in Table 1 ▶. Both crystal forms diffracted to better than 1.9 Å resolution. Crystal form A displayed P212121 symmetry, with unit-cell parameters a = 44.0, b = 74.7, c = 79.8 Å. Crystal form B displayed P21 symmetry, with unit-cell parameters a = 36.0, b = 66.9, c = 44.1 Å, β = 99.9°.
Table 1. Crystal data and X-ray diffraction data-collection statistics.
Values in parentheses are for the highest resolution bin.
| Crystal | Form A | Form B |
|---|---|---|
| Space group | P212121 | P21 |
| Unit-cell parameters (Å, °) | a = 44.0, b = 74.7, c = 79.8 | a = 36.0, b = 66.9, c = 44.1, β = 99.9 |
| Resolution (Å) | 1.85 (1.92–1.85) | 1.86 (1.95–1.86) |
| No. of crystals | 1 | 1 |
| Oscillation range (°) | 360 (360 × 1° images) | 300 (300 × 1° images) |
| Temperature (K) | 100 | 100 |
| Wavelength (Å) | 1.0875 | 1.54182 |
| No. of observations | 732050 | 102405 |
| No. of unique reflections | 23144 | 16303 |
| Multiplicity | 10.5 (4.9) | 6.0 (5.3) |
| Data completeness (%) | 93.9 (62.0) | 99.5 (84.3) |
| I/σ(I) | 31.3 (2.8) | 24.2 (5.1) |
| Rmerge† (%) | 6.8 (30.6) | 4.7 (31.5) |
R
merge = 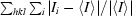 , where I
i is the intensity of the ith measurement of an equivalent reflection with indices hkl.
, where I
i is the intensity of the ith measurement of an equivalent reflection with indices hkl.
The structures of CLIC2 in both crystal forms have now been determined by molecular replacement using the published CLIC1 structure (Harrop et al., 2001 ▶) as a probe and have been reported elsewhere (Cromer et al., 2007 ▶). The atomic coordinates and structure factors (PDB codes 2r4v and 2r5g for crystal forms A and B, respectively) have been deposited in the Protein Data Bank at the Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, USA (http://www.rcsb.org). These structures will provide further insight into the structure and function of this intriguing family of proteins.
Acknowledgments
We thank Harry Tong and other staff at BioCARS for their help with data collection during our visit to the Advanced Photon Source. This work, including the use of the BioCARS sector, was supported by the Australian Synchrotron Research Program, which is funded by the Commonwealth of Australia under the Major National Research Facilities Program. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Energy Research. This work was also supported by grants from the National Health and Medical Research Council of Australia (NHMRC) and the Australian Research Council (ARC) to PGB, BAC and MWP. MWP is an ARC Federation Fellow and NHMRC Honorary Fellow.
References
- Baker, R. T., Smith, S. A., Marano, R., McKee, J. & Board, P. G. (1994). J. Biol. Chem.269, 25381–25386. [PubMed]
- Board, P. G. et al. (2000). J. Biol. Chem.275, 24798–24806. [DOI] [PubMed]
- Board, P. G., Coggan, M., Watson, S., Gage, P. W. & Dulhunty, A. F. (2004). Int. J. Biochem. Cell Biol.36, 1599–1612. [DOI] [PubMed]
- Catanzariti, A. M., Soboleva, T. A., Jans, D. A., Board, P. G. & Baker, R. T. (2004). Protein Sci.13, 1331–1339. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cromer, B. A., Gorman, M. A., Hansen, G., Adams, J. J., Coggan, M., Littler, D. R., Brown, L. J., Mazzanti, M., Breit, S. N., Curmi, P. M. G., Dulhunty, A. F., Board, P. G. & Parker, M. W. (2007). In the press.
- Cromer, B. A., Morton, C. J., Board, P. G. & Parker, M. W. (2002). Eur. Biophys. J.31, 356–364. [DOI] [PubMed]
- Dulhunty, A., Gage, P., Curtis, S., Chelvanayagam, G. & Board, P. G. (2001). J. Biol. Chem.276, 3319–3323. [DOI] [PubMed]
- Harrop, S. J. et al. (2001). J. Biol. Chem. 276, 44993–45000. [DOI] [PubMed]
- Landry, D., Sullivan, S., Nicolaides, M., Redhead, C., Edelman, A., Field, M., al-Awqati, Q. & Edwards, J. (1993). J. Biol. Chem.268, 14948–14955. [PubMed]
- Li, Y.-F., Li, D.-F., Zeng, Z.-H & Wang, D.-C. (2006). Biochem. Biophys. Res. Commun.343, 1272–1278. [DOI] [PubMed]
- Littler, D. R., Assaad, N. N., Harrop, S. J., Brown, L. J., Pankhurst, G. J., Luciani, P., Aguilar, M.-I., Mazzanti, M., Berryman, M. A., Breit, S. N. & Curmi, P. M. G. (2005). FEBS J.272, 4996–5007. [DOI] [PubMed]
- Littler, D. R., Harrop, S. J., Fairlie, W. D., Brown, L. J., Pankhurst, G. J., Pankhurst, S., DeMaere, M. Z., Campbell, T. J., Bauskin, A. R., Tonini, R., Mazzanti, M., Breit, S. N. & Curmi, P. M. G. (2004). J. Biol. Chem.279, 9298–9305. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tonini, R., Ferroni, A., Valenzuela, S. M., Warton, K., Campbell, T. J., Breit, S. N. & Mazzanti, M. (2000). FASEB J.14, 1171–1178. [DOI] [PubMed]
- Whittington, A., Vichai, V., Webb, G., Baker, R., Pearson, W. & Board, P. G. (1999). Biochem. J.337, 141–151. [PMC free article] [PubMed]