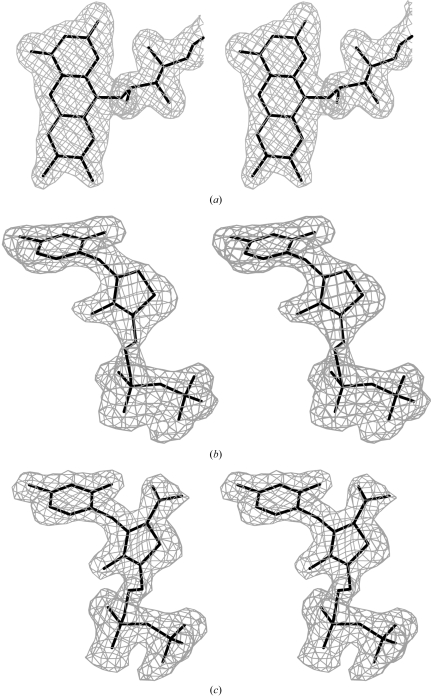
The crystal structures of pyruvate oxidase from A. viridans in complex with flavin adenine dinucleotide, thiamine diphosphate and the reaction intermediate 2-acetyl-thiamine diphosphate reveal details of substrate recognition and catalysis.
Keywords: pyruvate oxidase, Aerococcus viridans, flavin dinucleotide, thiamine diphosphate, 2-acetyl-thiamine diphosphate
Abstract
The crystal structures of pyruvate oxidase from Aerococcus viridans (AvPOX) complexed with flavin adenine dinucleotide (FAD), with FAD and thiamine diphosphate (ThDP) and with FAD and the 2-acetyl-ThDP intermediate (AcThDP) have been determined at 1.6, 1.8 and 1.9 Å resolution, respectively. Each subunit of the homotetrameric AvPOX enzyme consists of three domains, as observed in other ThDP-dependent enzymes. FAD is bound within one subunit in the elongated conformation and with the flavin moiety being planar in the oxidized form, while ThDP is bound in a conserved V-conformation at the subunit–subunit interface. The structures reveal flexible regions in the active-site tunnel which may undergo conformational changes to allow the entrance of the substrates and the exit of the reaction products. Of particular interest is the role of Lys478, the side chain of which may be bent or extended depending on the stage of catalysis. The structures also provide insight into the routes for electron transfer to FAD and the involvement of active-site residues in the catalysis of pyruvate to its products.
1. Introduction
Thiamine diphosphate (ThDP) dependent enzymes are present in various organisms and participate in diverse metabolic reactions. On the basis of the reactions they catalyze, ThDP-dependent enzymes have been classified into five families, which have been named after representative enzymes (Duggleby, 2006 ▶). The largest family is the pyruvate oxidases (POX) and is comprised of 2-ketoacid decarboxylating enzymes such as pyruvate decarboxylase (PDC), acetohydroxyacid synthase (AHAS), acetolactate synthase (ALS), N 2-(2-carboxyethyl)arginine synthase (CEAS), benzaldehyde aldolase (BAL), benzoylformate decarboxylase (BFDC), oxalyl-CoA-decarboxylase (OCDC) and indolepyruvate decarboxylase (IPDC). Recent studies on the POX family have provided valuable information on the mechanism of action of these enzymes and the catalytic role of ThDP. However, important details such as those pertaining to the changes in the active site and the involvement of the intermediates in the reaction pathway remain ambiguous.
POX is a unique enzyme in the sense that it utilizes not only ThDP but also the cofactor flavin adenine dinucleotide (FAD) for the oxidative decarboxylation of pyruvate. In the presence of oxygen and inorganic phosphate, the end products of the reaction are carbon dioxide, hydrogen peroxide and the high-energy metabolite acetyl phosphate. Owing to its attractive substrate and cofactors, POX is employed in a range of biosensors. For example, POX-based biosensors are being developed for the rapid detection of phosphate, which is detrimental to aquatic organisms (Mak et al., 2003 ▶), and are also being used in medical diagnosis to detect pyruvate (Zapata-Bacri & Burstein, 1988 ▶; Gavalas & Chaniotakis, 2000 ▶), the levels of which fluctuate in serum as a result of heart malfunction or poisoning.
At present, the POX-mediated reaction is believed to follow the pathway described by Tittmann and coworkers (Tittmann et al., 2005 ▶; Wille et al., 2006 ▶). It begins with the abstraction of a proton from the C2 atom of ThDP, which is facilitated by the 4′-amino group of ThDP. The resulting ylide attacks the carbonyl group of pyruvate to form the tetrahedral intermediate 2-lactyl-ThDP (LThDP). Decarboxylation results in the generation of the α-carbanion/enamine forms of 2-hydroxyethyl-ThDP (HEThDP). Through electron transfer to FAD, the enamine form is converted to a HEThDP radical and then to an anion–HEThDP adduct or to 2-acetyl-ThDP (AcThDP). The former is formed in the presence of phosphate and is cleaved to acetyl phosphate and ThDP.
Here, we describe the crystal structures of Aerococcus viridans pyruvate oxidase (AvPOX) with FAD, with both FAD and ThDP and with FAD and the AcThDP intermediate. Combined with the previously determined structures of Lactobacillus plantarum POX (LpPOX) in complex with LThDP, HEThDP and AcThDP (Muller & Schulz, 1993 ▶; Muller et al., 1994 ▶; Wille et al., 2005 ▶, 2006 ▶), our structures provide snapshots of different steps in the oxidative decarboxylation of pyruvate. Although most of the interactions of the cofactors and intermediates are similar in AvPOX and LpPOX, some notable changes were found in the present structures. The flavin moiety of FAD is planar in AvPOX, while it is bent in LpPOX. Regions close to the active site are flexible in the former but are rigid in the latter. Such regions may undergo considerable conformational changes to facilitate movement of the substrates and reaction products.
2. Materials and methods
2.1. Enzyme expression and purification
A plasmid containing the AvPOX gene was introduced into the expression host cell Escherichia coli DH1. Cells were grown at 310 K in BHI medium and were collected by centrifugation at 7000g for 10 min. Pellets were suspended in 10 mM phosphate buffer pH 6.5 containing 0.2% lysozyme. The suspension was incubated at 310 K for 2 h with continuous stirring. The supernatant obtained on centrifugation at 7000g for 20 min was loaded onto a DEAE-Sepharose FF column. The enzyme was eluted with a linear gradient of 0–0.5 M KCl in 10 mM phosphate buffer pH 6.5. Ammonium sulfate (13%) was added to the pooled enzyme fractions. The fractions were applied onto a Phenyl-Sepharose column and the enzyme was eluted with a linear gradient of 13–0% ammonium sulfate in 10 mM phosphate buffer pH 6.5. The fractions containing the enzyme were combined and concentrated by membrane filtration (Microcon YM3).
2.2. Crystallization
Lyophilized AvPOX sample was dissolved to a concentration of 40 mg ml−1 in 20 mM sodium phosphate buffer pH 7.0. Crystallizations were performed using the hanging-drop vapour-diffusion method at 293 K. For the AvPOX–FAD crystals, the droplets consisted of equal volumes (5 µl) of protein solution and reservoir solution containing 2.0 M ammonium sulfate in 20 mM sodium phosphate buffer pH 7.0. The AvPOX–FAD–ThDP crystals were obtained from similar droplets with the addition of 8 mM ThDP and 20 mM MgCl2 to the above conditions. The AvPOX–FAD–ThDP–Pyr crystals were prepared by soaking the above AvPOX–FAD–ThDP crystals in a solution containing 1.0 M pyruvate and 2.0 M ammonium sulfate for 3 h. Other crystals were obtained under conditions similar to those used for growing AvPOX–FAD–ThDP but with ammonium phosphate instead of ammonium sulfate. However, when these crystals were soaked in a solution containing 1.0 M pyruvate and 2.0 M ammonium phosphate they immediately dissolved. Other attempts to soak these crystals in solutions containing lower concentrations of pyruvate and ammonium phosphate resulted in the same dissolution of the crystals.
2.3. X-ray data collection and data processing
For data collection, all crystals were cryoprotected in solutions containing 2.0 M ammonium sulfate and 30% glycerol in 20 mM sodium phosphate buffer pH 7.0 and flash-frozen in liquid nitrogen. X-ray data were measured at 100 K using synchrotron radiation. For the AvPOX–FAD crystal, X-ray data were obtained at AR-NW12 (λ = 1.00 Å) of the Photon Factory (Ibaraki, Japan) and were recorded on an ADSC Quantum 210 CCD detector positioned 140 mm from the crystal. Images were obtained using a 0.3° oscillation range with 5 s exposure. The diffraction data for the AvPOX–FAD–ThDP crystal were measured at BL44XU (λ = 0.90 Å) of SPring-8 (Harima, Japan) in 0.5° oscillation frames with 10 s exposure times. The PX210 CCD detector was 300 mm from the crystal. The diffraction patterns of the AvPOX–FAD and AvPOX–FAD–ThDP crystals were indexed, merged and scaled with HKL-2000 (Otwinowski & Minor, 1997 ▶). Conversion of these data to structure-factor amplitudes was performed with TRUNCATE from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). Data collection from the AvPOX–FAD–ThDP–Pyr crystal was carried out at BL18B (λ = 1.00 Å) at Photon Factory. X-ray data were measured with an ADSC Quantum 4 CCD detector using 0.5° frames and 60 s exposure and with the detector 160 mm from the crystal. Diffraction patterns were processed with the program CrystalClear (Rigaku/MSC; Pflugrath, 1999 ▶). Intensity data were put on a relative scale and merged into independent reciprocal space using the programs SCALA and TRUNCATE in CCP4. The crystal data and statistics of data collection are summarized in Table 1 ▶.
Table 1. Crystal data and statistics of data collection and refinement.
Values in parentheses are for the outer resolution shell.
| AvPOX–FAD | AvPOX–FAD–ThDP | AvPOX–FAD–ThDP–Pyr | |
|---|---|---|---|
| Crystal data | |||
| Space group | I222 | I222 | I222 |
| Unit-cell parameters (Å) | |||
| a | 77.6 | 78.1 | 78.0 |
| b | 105.3 | 106.1 | 105.5 |
| c | 155.3 | 155.4 | 155.9 |
| Z† | 1 | 1 | 1 |
| Data collection | |||
| Maximum resolution (Å) | 1.58 | 1.80 | 1.95 |
| Outer shell resolution range (Å) | 1.64–1.58 | 1.86–1.80 | 2.02–1.95 |
| No. of observed reflections | 476332 | 284643 | 335885 |
| No. of unique reflections | 84186 | 56020 | 45577 |
| Completeness (%) | 95.5 (77.4) | 92.5 (91.8) | 98.4 (98.4) |
| Rmerge‡ (%) | 8.2 (37.9) | 5.4 (33.4) | 8.1 (28.4) |
| I/σ(I) | 21.8 (2.0) | 11.0 (1.8) | 6.8 (1.9) |
| Refinement | |||
| Resolution range (Å) | 6.0–1.6 | 10–1.8 | 10–2.0 |
| Protein atoms | 4598 | 4590 | 4590 |
| Ligand molecules | |||
| FAD | 1 | 1 | 1 |
| ThDP | — | 1 | — |
| AcThDP | — | — | 1 |
| Mg2+ | — | 1 | 1 |
| SO42− | 4 | 4 | — |
| Water | 692 | 563 | 689 |
| R factor§ (%) | 18.3 | 16.3 | 19.5 |
| Rfree¶ (%) | 20.6 | 19.6 | 24.3 |
Number of subunits in the asymmetric unit.
R
merge = 100 × 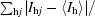
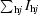 , where I
hj is the jth measurement of the intensity of reflection h and 〈I
h〉 is its mean value.
, where I
hj is the jth measurement of the intensity of reflection h and 〈I
h〉 is its mean value.
R factor = 100 × 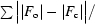
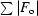 , where |F
o| and |F
c| are the observed and calculated structure-factor amplitudes, respectively.
, where |F
o| and |F
c| are the observed and calculated structure-factor amplitudes, respectively.
Calculated using a random set containing 10% of observations that were not included throughout refinement (Brünger, 1992 ▶).
2.4. Structure determination and refinement
Initial phases were derived by molecular replacement with the program AMoRe (Navaza, 1994 ▶) using the structure of the wild-type LpPOX structure (PDB code 1pox; Muller et al., 1994 ▶) as a search model. The structure was constructed on electron-density maps displayed in QUANTA (Accelrys Inc.) After rigid-body refinement, the atomic parameters were refined by the restrained maximum-likelihood least-squares technique in REFMAC5 from CCP4 (Murshudov et al., 1997 ▶). The structure was revised by interpreting omit maps at every residue. Sulfate and water molecules found in |F o| − |F c| maps (>2.5σ) were included in the subsequent structure refinements. The final refinements were performed with the program CNS (Brünger et al., 1998 ▶). Electron density for FAD, ThDP and a ThDP intermediate were observed in the active sites. These ligands were introduced into the models and were included in the structure refinements. In the final refinements, the geometrical restraints imposed on the ligands were completely released to examine their deformability.
The statistics of refinement for all three crystals are given in Table 1 ▶. Fig. 1 ▶ shows omit maps of the ligands, which were drawn with the program O (Jones et al., 1991 ▶). The figures showing the structural details were drawn with PyMOL (DeLano Scientific; http://www.pymol.org). Structural comparisons of the present structures with those of Zymomonas mobilis PDC (ZmPDC; Dobritzsch et al., 1998 ▶), yeast AHAS (Pang et al., 2002 ▶), Klebsiella pneumoniae ALS (Pang et al., 2004 ▶), Streptomyces clavuligerus CEAS (Caines et al., 2004 ▶), Pseudomonas fluorescens BAL (Mosbacher et al., 2005 ▶), P. putida BFDC (Hasson et al., 1998 ▶), Oxalobacter formigenes OCDC (Berthold et al., 2005 ▶) and Enterobacter cloacae IPDC (Schütz et al., 2003 ▶) were carried out using Swiss-PdbViewer v.3.7 (http://www.expasy.org/spdbv/).
Figure 1.
Stereoviews of omit maps contoured at the 2.5σ level for (a) the flavin moiety of FAD, (b) ThDP and (c) AcThDP bound in the AvPOX–FAD, AvPOX–FAD–ThDP and AvPOX–FAD–ThDP–Pyr structures, respectively.
3. Results
3.1. Overall structures
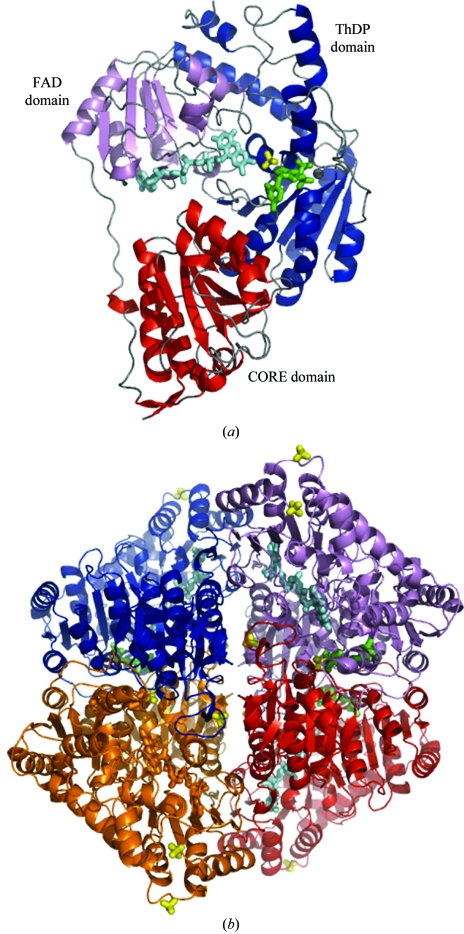
The AvPOX monomer consists of three regions, denoted CORE (Asn4–Lys186), FAD (Tyr187–Glu337) and ThDP (Ser338–Lys592) domains (Fig. 2 ▶ a). The core scaffold of each subunit is comprised of six parallel β-sheets, which are surrounded by α-helices. Two subunits of the dimer are related by crystallographic twofold symmetry. The active site is formed by residues from the ThDP domain of one subunit and from the CORE domain of the neighbouring subunit. Another dimer is related by further crystallographic twofold symmetry; thus, the complete tetramer has 222 symmetry (Fig. 2 ▶ b). These features are maintained in all three crystal structures. Superimpositions of corresponding Cα atoms among the three structures of AvPOX gave r.m.s.d. values ranging from 0.26 to 0.39 Å, suggesting that binding of FAD, ThDP and the ThDP intermediate has little effect on the overall protein conformation. Except for the three N-terminal residues, some side chains at the surface of the protein and several residues in a loop region (Asp469–Tyr496) near the active site, the electron densities were well defined over the entire structures of the three AvPOX structures.
Figure 2.
The overall structure of AvPOX–FAD–ThDP. (a) The monomeric unit of AvPOX–FAD–ThDP showing the CORE (red), FAD (violet) and ThDP (blue) domains. (b) The tetrahedral AvPOX–FAD–ThDP homotetramer with 222 symmetry. Subunits A, B, C and D are coloured red, blue, violet and orange, respectively. FAD, ThDP and sulfate ions are shown as cyan, green and yellow stick representations, respectively, while the magnesium ions are shown as grey spheres.
3.2. ThDP binding
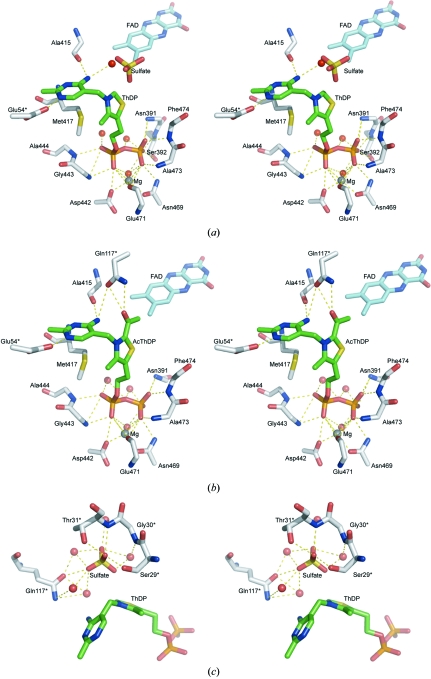
In the AvPOX–FAD–ThDP structure, the ThDP cofactor is bound in a cleft between the ThDP domain of one subunit and the CORE domain of the adjacent subunit, although most of the contacts are made with the ThDP domain (Fig. 3 ▶ a). The diphosphate moiety of ThDP forms hydrogen bonds with the main-chain N atoms of Asn391, Ser392, Gly443, Ala444 and Phe474 and with the side-chain atoms of Ser392, Asp442 and Asn469. The diphosphate terminus is anchored by Mg2+, which is octahedrally coordinated by the main-chain O atom of Glu471, the side-chain atoms of Asp442 and Asn469 and a water molecule (Fig. 3 ▶ a). Several of the residues interacting with the diphosphate group are from the 441GDGX 24NN469 motif, which is found in all ThDP-dependent enzymes (Hawkins et al., 1989 ▶). In AvPOX, however, the second-to-last residue of the motif is replaced by serine. The pyrimidine ring forms two hydrogen bonds with the enzyme: N1′ to Glu54* OE2 (where the asterisk denotes a residue in the adjacent subunit) and N4′ to Ala415 O. The side chain of Met417 protrudes from the surface, forcing the thiazolium and pyrimidine rings to adopt a V-shaped conformation (the torsional angles ϕT and ϕP are 92° and −62°, respectively). This V-conformation results in the distance between the 4′-amino nitrogen and C2 atoms being 3.2 Å.
Figure 3.
Stereoviews of the active sites, showing the interactions involving (a) ThDP and magnesium in AvPOX–FAD–ThDP, (b) AcThDP and magnesium in AvPOX–FAD–ThDP–Pyr and (c) sulfate in AvPOX–FAD–ThDP. The FAD, ThDP/AcThDP and sulfate molecules are shown as cyan, green and yellow stick representations, respectively. The magnesium ions and the water molecules are shown as grey and red spheres, respectively. All residues marked with an asterisk belong to the symmetry-related subunit. The dashed lines indicate hydrogen-bonding and ionic interactions.
3.3. AcThDP binding
In the initial |F o| − |F c| electron-density map of AvPOX–FAD–ThDP–Pyr calculated with the phase angles derived from the model of AvPOX–FAD–ThDP, electron densities were found extending from the C2 atom of ThDP into the active site, indicating the formation of a ThDP intermediate. A well defined density for the cofactor intermediate is also observed in the final omit electron-density map (Fig. 1 ▶ c), which shows that the five atoms of the thiazolium ring, as well as the atoms of the extension, are all in the same plane. The geometrical restraints on this part were released in the final stages of refinement, but the atomic arrangement was not changed, reconfirming the planarity. The intermediate could thus either be the HEThDP enamine or AcThDP. We assigned the trapped intermediate as AcThDP. It has been reported that in the absence of phosphate, AcThDP is the predominant intermediate. The crystallization solution for AvPOX–FAD–ThDP–Pyr contained only a very small amount of phosphate, which may have been removed after the long soak in a solution containing no phosphate.
AcThDP is bound to the enzyme in an identical fashion to ThDP and the interactions of the diphosphate moiety with the enzyme and the Mg2+ ion are similar to those observed in the AvPOX–FAD–ThDP structure (Fig. 3 ▶ b). The interactions with one of the phosphate groups are changed because the side chain of the Ser392 residue is shifted away from the phosphate by about 3 Å. The thiazolium and pyrimidine rings of AcThDP also adopt the V-conformation (ϕT = 94° and ϕP = −65°). The carbonyl oxygen of the acetyl moiety forms hydrogen bonds to the side-chain atoms of the Gln117* residue of the neighbouring subunit. The position of the Gln117* residue is the same in the other two AvPOX structures. Four residues, Ser29*, Gly30*, Ser77* and Gly78*, interact indirectly with the acetyl group of the intermediate by hydrogen bonding through water molecules. The methyl group of the acetyl moiety makes a van der Waals contact with the C7 methyl group of FAD.
3.4. FAD conformation
The cofactor FAD is bound in a cleft surrounded by all three domains in the subunit and adopts the elongated conformation. There are numerous hydrogen-bond formations and van der Waals interactions between the enzyme and FAD. Adenine is bound tightly to the enzyme by hydrogen bonds to the main-chain N atom of Ala321 and the side-chain atom of Asp320. The ribose ring is held in place by hydrogen bonds to the the side-chain atoms of Asp301. The phosphate groups of FAD form hydrogen bonds to the main-chain atoms of Ile216 and Asn281 and the side-chain atoms of Thr239, Lys241 and Ser280. The flavin moiety also forms several hydrogen bonds to the enzyme; these contacts are to the main-chain atoms of Thr257, Val260 and Pro412. In the oxidized form of LpPOX, the flavin moiety is bent about the N5–N10 axis (Muller & Schulz, 1993 ▶; Muller et al., 1994 ▶; Wille et al., 2005 ▶, 2006 ▶). However, in the present oxidized AvPOX (as evidenced by the bright yellow colour of the crystals), the flavin is planar. This supports the assumption that the conformation of the flavin moiety does not affect its redox potential (Wille et al., 2005 ▶).
3.5. Sulfate-binding sites
Four sulfate anions are also observed in the AvPOX–FAD and AvPOX–FAD–ThDP crystals. In AvPOX–FAD–ThDP, a sulfate anion is located near the thiazolium ring of ThDP (Fig. 3 ▶ c). The sulfate anion is bound to the Ser29*, Thr31* and Gln117* residues of a neighbouring subunit. The anion also interacts indirectly with Gly30* through water mediation. One of the sulfate O atoms interacts with the C2 atom of ThDP. The three other sulfate anions in AvPOX–FAD–ThDP are located in equivalent positions in AvPOX–FAD. Two of these common sulfate-binding sites are on the surface of the tetramer, while the remaining common binding site is in a crevice surrounded by three subunits. The sulfate anion present only in AvPOX–FAD is in a location similar to that of one of the phosphate groups of ThDP in AvPOX–FAD–ThDP.
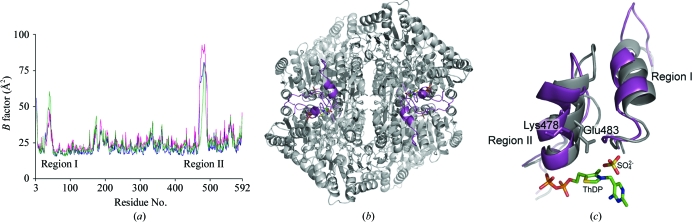
3.6. Flexible regions
In general, temperature factors (B factors) characterize the conformational flexibility of amino-acid residues. Fig. 4 ▶(a) shows plots of the observed B factors versus the Cα atoms of the residues in the three AvPOX structures. The graph demonstrates a high degree of flexibility of two regions near the active site. The first region (region I; residues 27–46) is located near the N-terminus, while the second (region II; residues 469–496) includes residues interacting with the diphosphate moiety of ThDP. Examination of the electron densities in these regions revealed that while the residues in region I fit reasonably well into the densities, those in region II are disordered.
Figure 4.
Flexible regions in AvPOX. (a) Average B factors of the main-chain atoms are shown for AvPOX–FAD (blue), AvPOX–FAD–ThDP (magenta) and AvPOX–FAD–ThDP–Pyr (green). (b) A top view of the tunnels for substrate entry to the active site in AvPOX–FAD–ThDP. The locations of the flexible regions are coloured in magenta. (c) Side view of the tunnel with the flexible regions in AvPOX–FAD–ThDP (magenta) superimposed onto the corresponding regions in LpPOX (grey; Wille et al., 2006 ▶). ThDP, FAD, sulfate, Lys478 and Glu483 are shown as stick representations. Lys478 in AvPOX corresponds to Glu483 in LpPOX.
4. Discussion
4.1. Comparison with other POX-family enzymes
Superimpositions of the 570 corresponding Cα atoms between the LpPOX structure (Muller et al., 1994 ▶) and any of the three AvPOX structures show similarities in the overall main-chain folding, with r.m.s.d. values ranging from 0.82 to 0.90 Å. In the AvPOX–FAD–ThDP structure, the enzyme is in a state where it is waiting for the substrate pyruvate. The enzyme in this state was compared with related enzymes belonging to the POX family (Table 2 ▶). Although AHAS from yeast has a low sequence identity (24%) to AvPOX, the 326 corresponding Cα atoms superimposed with an r.m.s.d. of 1.44 Å (Pang et al., 2002 ▶). Both POX and AHAS have an essential requirement for the cofactor FAD, although FAD lacks the redox function in the latter enzyme. All other POX-family enzymes showed a good equivalence, with deviations of less than 2 Å for the corresponding Cα atoms. The positions of the residues in the FAD domain of AvPOX correlate particularly well with those of the residues in the middle domain (also referred to as the β or R domains) of the other enzymes.
Table 2. Comparisons of the sequences, structures and flexible regions of the POX family of enzymes.
The A subunits from the PDB files were used in all cases.
| Protein | Identity (%) | Similarity (%) | R.m.s.d.† (Å) | Region II residues |
|---|---|---|---|---|
| AvPOX (1v5f) | — | — | — | 469–496‡ |
| LpPOX (1pox) | 47 | 66 | 0.96 (568) | 474–501 |
| ZmPDC (1zpd) | 26 | 46 | 1.55 (338) | 467–488 |
| AHAS (1jsc) | 24 | 45 | 1.44 (316) | 577–604‡ |
| ALS (1ozf) | 23 | 44 | 1.77 (322) | 474–500 |
| CEAS (1upb) | 22 | 42 | 1.51 (309) | 490–518 |
| BAL (2ag0) | 22 | 40 | 1.55 (337) | 475–503 |
| BFDC (1bfd) | 21 | 40 | 1.52 (348) | 455–482 |
| OCDC (2c31) | 21 | 38 | 1.63 (351) | 479–504 |
| IPDC (1ovm) | 21 | 38 | 1.62 (293) | 462–498 |
Calculated by superimposing the corresponding Cα atoms. Values in parentheses indicate the number of aligned Cα atoms.
Contain disordered regions.
4.2. Role of the flexible regions in catalysis
The two flexible regions occupy the walls of the tunnel leading to the active site (Fig. 4 ▶ b). Both AHAS and OXC were reported to have disordered residues in the same region (Pang et al., 2002 ▶; Berthold et al., 2005 ▶). In the structures of AHAS in complex with various herbicides, these regions become ordered and close the active site (McCourt et al., 2005 ▶), whereas in the OXC structure they are already folded up. In the remaining POX-family enzymes, however, the residues corresponding to these flexible regions have very low B factors, indicating stability. Closer examination of the present AvPOX structures and the LpPOX structures revealed several differences in the conformations of the flexible regions (Muller & Schulz, 1993 ▶; Muller et al., 1994 ▶; Wille et al., 2005 ▶, 2006 ▶). In the LpPOX structures these regions are more compact compared with those in AvPOX (Fig. 4 ▶ c). This is a consequence of more extensive contacts in the residues occupying the base of the flexible regions in LpPOX; such contacts may lead to their stability. Another stabilizing factor in LpPOX is the additional contact between the Glu483 side chain and the thiazolium moiety of ThDP (or the extension from the thiazolium moiety of the ThDP intermediates). In the LpPOX structures, Glu483 adopts an extended conformation and appears to close the active site. In AvPOX, the main chain of the Lys478 residue occupies a position similar to that of LpPOX Glu483, but its side chain is bent and as such is incapable of forming interactions with ThDP or AcThDP. Lys478 may adopt the ‘open’ or ‘closed’ conformations, acting as a lid to allow the reaction to proceed in the case of the former or to stop in the case of the latter. In a study performed on ZmPDC variants, it was shown that mutation of Glu473, the residue that corresponds to AvPOX Lys478, slows covalent addition of pyruvate and decarboxylation of LThDP (Tittmann et al., 2003 ▶). Further mutation studies on these flexible regions in AvPOX, especially on Lys478, are necessary to determine their roles in pyruvate catalysis.
4.3. Catalytic mechanism
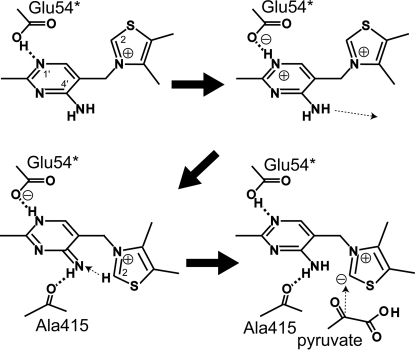
The role of the pyrimidine ring in the reactions of ThDP-dependent enzymes has been widely studied for several decades (Jordan & Mariam, 1978 ▶; Nemeria et al., 2007 ▶). As shown in Fig. 5 ▶, the N1′ atom is hydrogen bonded to the carboxyl group of the conserved Glu54* residue and such an interaction could induce proton release from the N4′ amino group. The resulting N4′ imino group then forms a hydrogen bond with the carbonyl group of the Ala415 residue. This triggers the transfer of protons from the thiazolium C2 atom to the N4′ atom and from the N1′ atom to Glu54*. The C2 atom becomes highly nucleophilic and is favourable for attack by the methoxyl group of pyruvate. This substrate is allowed to enter the active-site tunnel by movements of the flexible regions, particularly of the Lys478 side chain. As a result, the LThDP intermediate is formed, the structure of which was recently reported (Wille et al., 2006 ▶). Using the present AvPOX structures, a model of LThDP and several active-site residues in AvPOX was successfully built. The model shows that Gly30* and Gln117* may form interactions with LThDP. Lys478, adopting the extended conformation, has its side-chain amide group in a position that is also favourable for interaction with LThDP.
Figure 5.
Proton movements upon ThDP binding to facilitate pyruvate attack.
Decarboxylation of LThDP occurs and the resulting CO2 is expelled from the active site, possibly with Lys478 adopting the open conformation. The α-carbanion/enamine forms of HEThDP are formed, coupled with the transfer of an electron to FAD. Assuming that the hydroxyethyl moiety of HEThDP occupies the same position as the acetyl moiety of the trapped AcThDP in the AvPOX–FAD–ThDP–Pyr structure, electron transfer could proceed via three routes, consistent with previous suggestions (Muller & Schulz, 1993 ▶; Muller et al., 1994 ▶; Pang et al., 2002 ▶). Firstly, the methyl group of the hydroxyethyl moiety of HEThDP may directly transfer an electron to the methyl group on the C7 atom of the flavin. Secondly, an electron may be passed via Phe116* to C7. Thirdly, an electron may be relayed to the methyl group on the C7 of flavin via Phe474.
Once again, the active-site lid opens for the phosphate to enter, which may occupy the sulfate-binding site near the thiazolium ring found in the AvPOX–FAD–ThDP structure. The phosphate may also form contacts with Ser29*, Gly30*, Thr31* and Gln117*, as well as with Lys478 in the extended form. Similar contacts between phosphate and LpPOX have been reported (Wille et al., 2006 ▶). The phosphate attacks HEThDP and the anionic phosphate–radical adduct is formed, which subsequently facilitates the transfer of the second electron to FAD. The final products, acetyl phosphate and hydrogen peroxide, are then released to regenerate the ThDP cofactor.
Supplementary Material
PDB reference: AvPOX–FAD, 2dji, r2djisf
PDB reference: AvPOX–FAD–ThDP, 1v5f, r1v5fsf
PDB reference: AvPOX–FAD–ThDP–Pyr, 1v5g, r1v5gsf
Acknowledgments
We thank M. Suzuki, N. Igarashi and A. Nakagawa for help with data collection. This work was supported in part by Grants-in-Aid for the Protein 3000 Project for Metabolic Proteins (S. Kuramitsu) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Berthold, C. L., Moussatche, P., Richards, N. G. J. & Lindqvist, Y. (2005). J. Biol. Chem.280, 41645–41654. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Caines, M. E. C., Elkins, J. M., Hewitson, K. S. & Schofield, C. J. (2004). J. Biol. Chem.279, 5685–5692. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dobritzsch, D., König, S., Schneider, G. & Lu, G. (1998). J. Biol. Chem.273, 20196–20204. [DOI] [PubMed]
- Duggleby, R. G. (2006). Acc. Chem. Res.39, 550–557. [DOI] [PubMed]
- Gavalas, V. G. & Chaniotakis, N. A. (2000). Anal. Chim. Acta, 427, 271–277.
- Hasson, M. S., Muscate, A., McLeish, M. J., Polovnikova, L. S., Gerlt, J. A., Kenyon, G. L., Petsko, G. A. & Ringe, D. (1998). Biochemistry, 37, 9918–9930. [DOI] [PubMed]
- Hawkins, C. F., Borges, A. & Perham, R. N. (1989). FEBS Lett.255, 77–82. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Jordan, F. & Mariam, Y. H. (1978). J. Am. Chem. Soc.100, 2534–2540.
- McCourt, J. A., Pang, S. S., Guddat, L. W. & Duggleby, R. G. (2005). Biochemistry, 44, 2330–2338. [DOI] [PubMed]
- Mak, W. C., Chan, C., Barford, J. & Renneberg, R. (2003). Biosens. Bioelectron.19, 233–237. [DOI] [PubMed]
- Mosbacher, T. G., Mueller, M. & Schulz, G. E. (2005). FEBS J.272, 6067–6076. [DOI] [PubMed]
- Muller, Y. A. & Schulz, G. E. (1993). Science, 259, 965–967. [DOI] [PubMed]
- Muller, Y. A., Schumacher, G., Rudolph, R. & Schulz, G. E. (1994). J. Mol. Biol.237, 315–335. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Nemeria, N., Chakraborty, S., Bakyal, A., Korotchkina, L. G., Patel, M. S. & Jordan, F. (2007). Proc. Natl Acad. Sci. USA, 104, 78–82. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pang, S. S., Duggleby, R. G. & Guddat, L. W. (2002). J. Mol. Biol.317, 249–262. [DOI] [PubMed]
- Pang, S. S., Duggleby, R. G., Schowen, R. L. & Guddat, L. W. (2004). J. Biol. Chem.279, 2242–2253. [DOI] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Schütz, A., Sandalova, T., Ricagno, S., Hübner, G., König, S. & Schneider, G. (2003). Eur. J. Biochem.270, 2312–2321. [DOI] [PubMed]
- Tittmann, K., Golbik, R., Uhlemann, K., Khailova, L., Schneider, G., Patel, M., Jordan, F., Chipman, D. M., Duggleby, R. G. & Hübner, G. (2003). Biochemistry, 42, 7885–7891. [DOI] [PubMed]
- Tittmann, K., Wille, G., Golbik, R., Weidner, A., Ghisla, S. & Hübner, G. (2005). Biochemistry, 44, 13291–13303. [DOI] [PubMed]
- Wille, G., Meyer, D., Steinmetz, A., Hinze, E., Golbik, R. & Tittmann, K. (2006). Nature Chem. Biol.2, 324–328. [DOI] [PubMed]
- Wille, G., Ritter, M., Weiss, M. S., König, S., Mäntele, W. & Hübner, G. (2005). Biochemistry, 44, 5086–5094. [DOI] [PubMed]
- Zapata-Bacri, A. M. & Burstein, C. (1988). Biosensors, 3, 227–237. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AvPOX–FAD, 2dji, r2djisf
PDB reference: AvPOX–FAD–ThDP, 1v5f, r1v5fsf
PDB reference: AvPOX–FAD–ThDP–Pyr, 1v5g, r1v5gsf