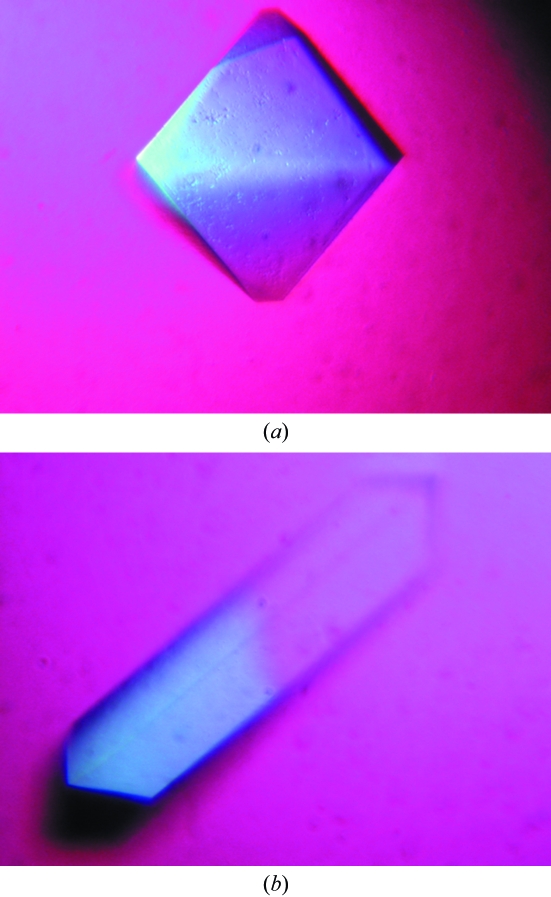
The β-xylosidase was crystallized using PEG 6000 as precipitant. 5% PEG 6000 yielded bipyramid-shaped tetragonal crystals diffracting to 1.55 Å resolution, and 13% PEG 6000 gave rectangular monoclinic crystals diffracting to 1.80 Å resolution.
Keywords: β-xylosidase, glycoside hydrolase family 43, Geobacillus thermoleovorans IT-08
Abstract
The main enzymes involved in xylan-backbone hydrolysis are endo-1,4-β-xylanase and β-xylosidase. β-Xylosidase converts the xylo-oligosaccharides produced by endo-1,4-β-xylanase into xylose monomers. The β-xylosidase from the thermophilic Geobacillus thermoleovorans IT-08, a member of glycoside hydrolase family 43, was crystallized at room temperature using the hanging-drop vapour-diffusion method. Two crystal forms were observed. Bipyramid-shaped crystals belonging to space group P43212, with unit-cell parameters a = b = 62.53, c = 277.4 Å diffracted to 1.55 Å resolution. The rectangular crystals belonged to space group P21, with unit-cell parameters a = 57.94, b = 142.1, c = 153.9 Å, β = 90.5°, and diffracted to 1.80 Å resolution.
1. Introduction
The major components of the plant cell wall are cellulose, hemicellulose and lignin (Timell, 1982 ▶). In contrast to cellulose, which only contains glucose, hemicellulose contains many different sugar monomers such as xylose, mannose, galactose, rhamnose and arabinose. Xylose is the sugar monomer that is present in the largest amount in hemicellulose, mostly in the form of xylan, which is a complex heteropolysaccharide consisting of a backbone of β-(1,4)-linked xylose residues and side chains consisting of arabinose, glucuronic acid or its 4-O-methyl ether, which can be decorated with ferulic, acetic and p-coumaric acids. The frequency and composition of the branches are dependent on the source of the xylan (Beg et al., 2001 ▶; Saha, 2003 ▶).
The complete degradation of xylan requires a large variety of cooperatively acting enzymes. The main enzymes involved in xylan-backbone hydrolysis are endo-1,4-β-xylanase and β-xylosidase. Endo-1,4-β-xylanase (EC 3.2.1.8) cleaves xylan into β-d-xylopyranosyl oligosaccharides. β-Xylosidase (EC 3.2.1.37) cleaves these xylo-oligomers into xylose monomers. Removal of the side groups is catalyzed by α-l-arabinofuranosidases, α-d-glucuronidases, acetylxylan esterases, ferulic acid esterases and p-coumaric acid esterases (Subramaniyan & Prema, 2002 ▶; Polizeli et al., 2005 ▶).
Puspaningsih (2004 ▶) isolated a xylanolytic gene cluster (called pTP510) from the thermophilic bacterium Geobacillus thermoleovorans IT-08 (GenBank accession Nos. DQ345777, DQ387046 and DQ387047). The β-xylosidase gene (xyl; GenBank accession No. DQ345777) was subcloned into the pET101/D-TOPO expression vector. The native β-xylosidase (Xyl; GenPept accession No. ABC75004) contains 511 amino-acid residues with a calculated molecular weight of 57 993 Da. The optimum pH and temperature of this enzyme are pH 6.0 and 328 K, respectively. The enzyme cleaves various xylans, as well as the artificial substrate p-nitrophenyl-β-d-xylopyranoside (Puspaningsih, unpublished work). Of potential biotechnological interest is that the xylose produced by the β-xylosidase enzyme can be used as a raw material for the bioethanol and xylitol industry (Kuhad & Singh, 1993 ▶; Olsson & Hahn-Hagerdal, 1996 ▶).
Based on amino-acid sequence similarities and conserved catalytic residues, β-xylosidases have been grouped into seven different glycoside hydrolase (GH) families in the CAZy database (http://www.cazy.org/; Henrissat & Davies, 1997 ▶). These families are GH3, GH30, GH39, GH43, GH51, GH52 and GH54. The β-xylosidase from G. thermoleovorans IT-08 (Xyl) belongs to the GH43 family, which is part of the GH-F clan. The enzymes in this family share a three-dimensional structure consisting of a five-bladed β-propeller fold (Nurizzo et al., 2002 ▶). They hydrolyze their substrate using a mechanism that leads to inversion of the anomeric configuration via a single nucleophilic displacement (Sinnott, 1990 ▶; Brux et al., 2006 ▶). The catalytic mechanism of family GH43 β-d-xylosidases involves a Glu as a general acid, an Asp as a general base and another Asp that modulates the pK a of the general acid and keeps it in the correct orientation relative to the substrate (Brux et al., 2006 ▶). These residues are conserved in all β-d-xylosidases in the family.
The three-dimensional structures of several members of the GH43 β-xylosidase family have been determined: the 1,4-β-xylosidases from Bacillus halodurans C-125 (PDB code 1yrz; A. A. Fedorov, E. V. Fedorov & S. C. Almo, unpublished), B. subtilis (PDB code 1yif; Y. Patskovsky & S. C. Almo, unpublished work), Clostridium acetobutylicum (PDB code 1yi7; A. Teplyakov, E. Fedorov, G. L. Gilliland, S. C. Almo & S. K. Burley, unpublished work) and G. stearothermophilus (PDB code 2exh; Brux et al., 2006 ▶). Their primary structures are considerably similar to each other, with identities of around 52–67%. In contrast, the sequence of Xyl is more divergent, with an identity of 32% to the nearest structurally characterized homologue (1yi7). Therefore, we consider it of interest to relate the functional properties of Xyl to its structure, with comparison to other GH43 β-xylosidases. Understanding the interaction between Xyl and its substrate at the molecular level may enable the improvement of the enzyme for industrial applications, such as increasing its pH optimum and thermostability. In the present study, we describe the purification, crystallization and preliminary X-ray analysis of the protein.
2. Experimental
2.1. Overexpression and purification of Xyl
The xyl gene (GenBank accession No. DQ345777) from G. thermoleovorans IT-08 was subcloned into the linear pET101/D-TOPO vector (Invitrogen) using the forward primer 5′-CACCATGGAATATTCTAACCCAGTAATTAAAGG-3′ and the reverse primer 5′-TATTTCAGGAATATATTTAAACCAATCAAAA-3′. The construct, called pET-xyl, was overexpressed as a His-tagged protein in Escherichia coli BL21 (DE3). The overexpressed protein (543 amino-acid residues with a calculated molecular weight of 61 533 Da) comprises the complete Xyl protein (511 residues) followed by 32 additional residues at the C-terminus (KGELNSKLEGKPIPNPL LGLDSTRTGHHHHHH), which contain a V5 epitope and a 6×His extension (both shown in bold). A pre-culture was grown overnight in LB medium containing ampicillin (100 µg ml−1) at 310 K and used for a 1% inoculation of 6 l fresh LB medium containing 100 µg ml−1 ampicillin. Expression of Xyl was induced with 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) at an OD600 of 0.7–0.8. The cells were harvested after an additional 3 h of growth. The cell pellet was resuspended in buffer A (50 mM phosphate buffer pH 8.0, 250 mM NaCl, 5 mM imidazole and 5 mM β-mercaptoethanol) using 4 ml buffer per gram of cells. After addition of protease-inhibitor cocktail (Roche Diagnostics, one tablet per 20 ml suspension) and DNAse I (Roche Diagnostics, catalytic amount), the cell suspension was lysed by two passages through a French Press (Fisher Scientific) at 55 MPa. Cell debris was removed by centrifugation at 35 000g for 20 min at 277 K. The supernatant was heated to 323 K for 1 h and centrifuged again to remove the E. coli proteins. The Xyl-containing supernatant was then loaded onto an ∼1 ml Ni–NTA agarose column (Qiagen) pre-equilibrated with buffer A and incubated overnight at 280 K. After discarding the flowthrough, the column was washed with five column volumes of buffer A and eluted with five column volumes of buffer B (50 mM phosphate pH 8.0, 250 mM NaCl, 100 mM imidazole and 1 mM β-mercaptoethanol). The Xyl fraction was diluted ten times with buffer C (25 mM Tris–HCl buffer pH 8.0) and applied onto a 1 ml Resource Q anion-exchange column (Pharmacia Biotech) pre-equilibrated with the same buffer. The column was eluted with a linear NaCl gradient (0–0.5 M) in buffer C using ÄKTA FPLC (Amersham Biosciences). Xyl eluted at ∼100 mM NaCl. The purified Xyl solution was concentrated to 14 mg ml−1 in a Microsep 10K Omega concentrator (Pall Corporation) and stored at 253 K.
2.2. Crystallization of Xyl
The protein was diluted to a concentration of 7 mg ml−1 with buffer C containing 100 mM NaCl. Initial crystallization trials were performed with the JCSG-plus Screen (Molecular Dimensions Ltd), Structure Screens I and II (Molecular Dimensions Ltd) and Wizard Screens I and II (Emerald Biosystems) using the sitting-drop vapour-diffusion method. The crystallization screens were set up using an Oryx 6 crystallization robot (Douglas Instruments) by mixing 0.15 µl protein solution with 0.15 µl reservoir solution on CrystalClear Strips 96-well sitting-drop plates from Hampton Research. All crystallization trials were performed at 293 K.
The most promising initial crystallization conditions were obtained with the JCSG-plus condition containing 0.1 M HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer pH 7.0 and 10%(w/v) PEG 6000 as a precipitant in the reservoir. This condition was optimized using the hanging-drop vapour-diffusion method. 1 µl protein solution was mixed with 1 µl well solution and subsequently positioned over wells containing 0.5 ml well solution. The pH and PEG concentration were varied. The best crystals were obtained from setups with well solutions containing 0.1 M HEPES buffer pH 7.0 and a PEG 6000 concentration of either 5 or 13%(w/v). These conditions yielded crystals with different morphologies: bipyramid-shaped and rectangular, respectively.
2.3. Data collection and processing
X-ray diffraction data were collected at 100 K on beamlines BM16 (wavelength 0.99992 Å) and ID23-2 (wavelength 0.87260 Å) at the European Synchrotron Radiation Facility (Grenoble, France) using an ADSC Quantum Q210r CCD detector and a 225 mm MAR Mosaic CCD detector, respectively. The crystals were cryoprotected with 20%(v/v) glycerol and flash-cooled in a cold nitrogen stream. The monoclinic crystal was rotated through 180° with an oscillation range of 0.5° per frame; the tetragonal crystal was rotated through 200° with an oscillation range of 1.0° per frame. All data were processed using the programs MOSFLM (Leslie, 1991 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results
Two Xyl crystal forms were obtained from PEG 6000 solutions in 0.1 M HEPES buffer pH 7.0. Both diffracted X-rays to high resolution (see Table 1 ▶). Bipyramid-shaped crystals (Fig. 1 ▶ a) grew from 5%(w/v) PEG 6000 to a maximum dimension of 0.25 mm and belong to space group P41212 or P43212. These crystals contain one 61.5 kDa molecule (including the V5 epitope and 6×His tag) per asymmetric unit (V M is 2.2 Å3 Da−1). Rectangular-shaped crystals (Fig. 1 ▶ b) grew within one week from 13%(w/v) PEG 6000 to dimensions of 0.2 × 0.03 × 0.03 mm. They belong to space group P21 and contain four molecules per asymmetric unit (V M is 2.6 Å3 Da−1).
Table 1. Data-collection statistics.
Values in parentheses are for the last resolution shell.
| Bipyramidal crystal | Rectangular crystal | |
|---|---|---|
| ESRF beamline | ID23-2 | BM16 |
| Wavelength | 0.87260 | 0.99992 |
| Space group | P43212 | P21 |
| Unit-cell parameters | ||
| a (Å) | 62.53 | 57.94 |
| b (Å) | 62.53 | 142.1 |
| c (Å) | 277.4 | 153.9 |
| α (°) | 90 | 90 |
| β (°) | 90 | 90.52 |
| γ (°) | 90 | 90 |
| Resolution (Å) | 1.55 (1.63–1.55) | 1.80 (1.90–1.80) |
| Rmerge† | 0.146 (0.761) | 0.085 (0.489) |
| Rp.i.m.‡ | 0.040 (0.280) | 0.046 (0.258) |
| Total No. of observations | 1150089 (94995) | 844701 (111617) |
| Total No. of unique reflections | 81489 (11687) | 210751 (27382) |
| Mean I/σ(I) | 14.2 (2.3) | 12.1 (2.8) |
| Completeness (%) | 100.0 (100.0) | 91.9 (81.8) |
| Multiplicity | 14.1 (8.1) | 4.0 (4.1) |
R
merge = 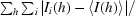
 .
.
R
p.i.m. = 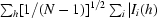 −
− 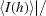
 .
.
Figure 1.
Photographs of crystals of Xyl from G. thermoleovorans IT-08. (a) Bipyramid-shaped crystal (space group P43212); (b) rectangular-shaped crystal (space group P21).
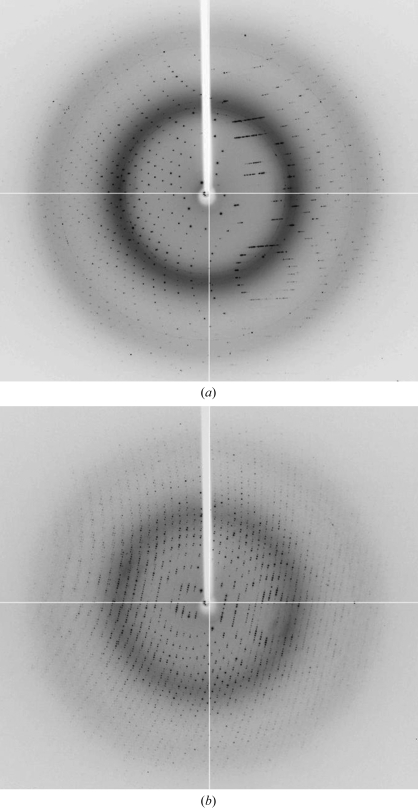
The bipyramid-shaped tetragonal crystals diffract to at least 1.55 Å, while data to 1.80 Å resolution could be collected from the rectangular monoclinic crystals (Fig. 2 ▶). Table 1 ▶ lists some pertinent details of the data collection. Preliminary molecular-replacement studies using 1yif (Y. Patskovsky & S. C. Almo, unpublished work) as a starting structure confirmed the presence of four molecules per asymmetric unit in the monoclinic crystal form. The tetragonal crystals contain one molecule per asymmetric unit and molecular-replacement analysis demonstrated the space group to be P43212.
Figure 2.
X-ray diffraction images. Full-view examples of a frame collected from crystals of space group P43212 (a) and P21 (b).
At present, we are refining and analyzing the two crystal forms. We hope that the binding of substrates and substrate analogues will help us in understanding the molecular basis of the catalytic activity of the enzyme and may guide us in improving the enzyme for industrial applications.
Acknowledgments
This research was supported by a mobility grant from The Scientific Programme Netherlands–Indonesia of the Royal Netherlands Academy of Sciences. We thank members of the Protein Crystallography Group for assistance with crystallization, data collection and processing.
References
- Beg, Q. K., Kapoor, M., Mahajan, L. & Hoondal, G. S. (2001). Appl. Microbiol. Biotechnol.56, 326–338. [DOI] [PubMed]
- Brux, C., Ben-David, A., Shallom-Shezifi, D., Leon, M., Niefind, K., Shoham, G., Shoham, Y. & Schomburg, D. (2006). J. Mol. Biol.359, 97–109. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol.7, 637–644. [DOI] [PubMed]
- Kuhad, R. C. & Singh, A. (1993). Crit. Rev. Biotechnol.13, 151–172.
- Leslie, A. G. W. (1991). Crystallographic Computing 5: From Chemistry to Biology, edited by D. Moras, A. D. Podjarny & J. C. Thierry, pp. 50–61. Oxford University Press.
- Nurizzo, D., Turkenburg, J. P., Charnock, S. J., Roberts, S. M., Dodson, E. J., McKie, V. A., Taylor, E. J., Gilbert, H. J. & Davies, G. J. (2002). Nature Struct. Biol.9, 665–668. [DOI] [PubMed]
- Olsson, L. & Hahn-Hagerdal, B. (1996). Enzyme Microb. Technol.18, 312–331.
- Polizeli, M. L. T. M., Rizzatti, A. C. S., Monti, R., Terenzi, H. F., Jorge, J. A. & Amorim, D. S. (2005). Appl. Microbiol. Biotechnol.67, 577–591. [DOI] [PubMed]
- Puspaningsih, N. N. T. (2004). PhD thesis. Bogor Agricultural University, Indonesia.
- Saha, B. C. (2003). J. Ind. Microbiol. Biotechnol.30, 279–291. [DOI] [PubMed]
- Sinnott, M. L. (1990). Chem. Rev.90, 1171–1202.
- Subramaniyan, S. & Prema, P. (2002). Crit. Rev. Biotechnol.22, 33–46. [DOI] [PubMed]
- Timell, T. E. (1982). Wood Sci. Technol.16, 83–122.