The structure of S. aureus MenB, an enzyme in the biosynthetic pathway to vitamin K2, has been determined and compared with the enzyme derived from another important pathogen, M. tuberculosis.
Keywords: crotonase, synthase, vitamin biosynthesis, menaquinone, MenB
Abstract
Vitamin K2, or menaquinone, is an essential cofactor for many organisms and the enzymes involved in its biosynthesis are potential antimicrobial drug targets. One of these enzymes, 1,4-dihydroxy-2-naphthoyl-CoA synthase (MenB) from the pathogen Staphylococcus aureus, has been obtained in recombinant form and its quaternary structure has been analyzed in solution. Cubic crystals of the enzyme allowed a low-resolution structure (2.9 Å) to be determined. The asymmetric unit consists of two subunits and a crystallographic threefold axis of symmetry generates a hexamer consistent with size-exclusion chromatography. Analytical ultracentrifugation indicates the presence of six states in solution, monomeric through to hexameric, with the dimer noted as being particularly stable. MenB displays the crotonase-family fold with distinct N- and C-terminal domains and a flexible segment of structure around the active site. The smaller C-terminal domain plays an important role in oligomerization and also in substrate binding. The presence of acetoacetyl-CoA in one of the two active sites present in the asymmetric unit indicates how part of the substrate binds and facilitates comparisons with the structure of Mycobacterium tuberculosis MenB.
1. Introduction
Menaquinone (vitamin K2) is an important cofactor that is exploited in electron-transport pathways (Meganathan, 2001 ▶). The vitamin consists of a naphthoquinone moiety with a polyisoprenyl substituent, the length of which varies in different bacteria. Humans lack the enzymes that synthesize this vitamin and acquire it from diet or from intestinal bacteria and this absence contributes to the potential value of these enzymes as therapeutic targets, especially for important pathogenic bacteria such as Mycobacterium tuberculosis and Staphylococcus aureus.
The biosynthesis of menaquinone has been studied extensively in Escherichia coli (Bentley & Meganathan, 1982 ▶; Lin & Kuritzkes, 1987 ▶; Meganathan, 1996 ▶) and also in Bacillus subtilis and M. pheli (Rowland et al., 1995 ▶). The biosynthesis typically involves six to eight enzymes and in a number of cases the genes encoding these enzymes have been proven to be essential to the bacteria by genetic methods. The structures of three of the biosynthetic enzymes have been characterized: MenC and MenF from E. coli (Palmer et al., 1999 ▶; Thompson et al., 2000 ▶; Kolappan et al., 2007 ▶) and MenB from M. tuberculosis (Truglio et al., 2003 ▶; Johnston et al., 2005 ▶).
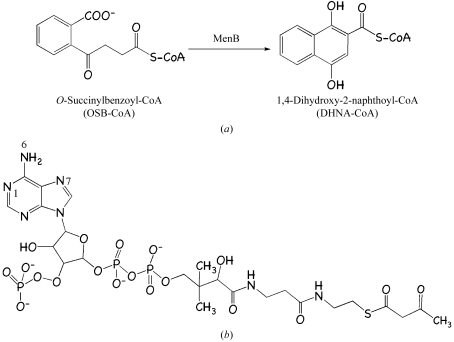
Our interest is in MenB, 1,4-dihydroxy-2-naphthoate synthase (EC 4.1.3.36). This enzyme converts O-succinylbenzoyl-CoA (the CoA ester of O-succinylbenzoic acid; OSB-CoA) to 1,4-dihydroxy-2-naphthoyl-CoA (the CoA ester of 1,4-dihydroxy-2-naphthoic acid; Fig. 1 ▶ a). The menB gene is essential in S. aureus (Forsyth et al., 2002 ▶), B. subtilis (Kobayashi et al., 2003 ▶) and Haemophilus influenzae (Akerley et al., 2002 ▶).
Figure 1.
(a) The reaction catalyzed by MenB. (b) Chemical structure of the ligand acetoacetyl-CoA.
The crystal structure of MenB from M. tuberculosis (MtbMenB) has been reported in the apo form and in complex with acetoacetyl-CoA or napthyl-CoA (Truglio et al., 2003 ▶; Johnston et al., 2005 ▶). These early studies confirmed MenB to be a member of the crotonase superfamily of enzymes, most of which are functional trimers or hexamers. A common feature of this group is that the substrates are CoA derivatives and the mechanism involves the stabilization of a thioester enolate by an oxyanion hole (Xiang et al., 1999 ▶; Gerlt & Babbitt, 2001 ▶).
Seeking to provide a template to support structure-based inhibitor development, we initiated a study of S. aureus 1,4-dihydroxy-2-naphthoate synthase (SaMenB). The substrate OSB-CoA is unstable; therefore, in order to obtain details of molecular interactions in the active site we studied the complex with acetoacetyl-CoA (Fig. 1 ▶ b).
2. Materials and methods
2.1. Cloning, expression and purification
The primers 5′-CAT-ATG-ACT-AAC-CGA-CAA-TGG-GAA-AC-3′ and 5′-GGA-TCC-TTA-TGG-GAA-TTT-AGG-GAA-TTG-3′ (Sigma–Aldrich) were used to amplify the menB gene from genomic DNA of S. aureus (ATCC35556; LGC Promochem). The primers included NdeI and BamHI restriction sites (shown in bold). The product of the PCR reaction was gel-purified (Qiagen) and ligated into the pCR-Blunt II-TOPO vector using the Zero Blunt Topo PCR Cloning Kit (Invitrogen). A restriction digest with NdeI and BamHI identified positive clones. Subsequently, the 822-base-pair fragment was gel-purified and ligated into a modified pET15b vector that generates a protein product carrying a tobacco etch virus (TEV) protease cleavage site. The integrity of the construct was confirmed by DNA sequencing. The plasmid pET15b-TEV-menB was heat-shock transformed into E. coli BL21(DE3) (Stratagene) and selected for on Luria–Bertani (LB) agar plates containing carbenicillin (50 mg l−1). A single colony was used to inoculate 15 ml LB broth with carbenicillin (50 mg l−1). This 15 ml culture was used to inoculate 1 l medium and grown at 310 K with shaking (200 rev min−1) until the A 550 reached 0.6 AU. At this point, the temperature was reduced to 298 K and expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside. Cells were harvested by centrifugation (Beckman JS-4.2, 3480g at 277 K for 35 min) after 14 h. The pellet was resuspended in 25 ml binding buffer (20 mM Tris–HCl pH 7.7, 250 mM NaCl) containing lysozyme (Sigma), one EDTA-free protease-inhibitor cocktail tablet (Roche) and DNase I (Roche Diagnostics) and lysed using a One-shot cell disruptor (Constant Cell Disruption Systems). The soluble and insoluble fractions were separated by centrifugation (Beckman JA-25.50, 48 400g at 277 K for 30 min) and the former filtered (0.2 µl) prior to loading onto a 5 ml HiTrap Chelating HP column (GE Healthcare) preloaded with nickel. A combination of step and linear gradients from 0 to 1000 mM imidazole were used to elute the protein. Fractions containing MenB were pooled then dialyzed into 20 mM Tris–HCl pH 7.7, 250 mM NaCl. The His tag was removed by proteolysis with His-tagged TEV protease for 3 h at 310 K. The uncleaved protein, the tag and TEV protease were removed by again passing over a HiTrap column (GE Healthcare). The high level of sample purity was confirmed by SDS–PAGE and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (data not shown). SaMenB was concentrated to 10 mg ml−1 (theoretical ∊0, 9190 M −1 cm−1) and used for crystallization.
2.2. Crystallization
Crystallization trials were carried out at 294 K using sitting drops and a range of commercial screens (Sigma, Jena Biosciences). Two crystal forms (cubes and rods) were obtained which grew in the same optimized conditions, (Fig. 2 ▶). The rods diffracted poorly, to only 6 Å, even at a synchrotron source and so were not used further. Large cubic crystals (maximum dimension 0.3 mm) were obtained by mixing 1 µl protein solution (10 mg ml−1) containing 3 mM acetoacetyl-CoA, 20 mM Tris–HCl pH 7.7, 250 mM NaCl with 1 µl reservoir solution (0.1 M Na HEPES pH 7.5, 1.6 M ammonium sulfate, 0.2 M NaCl). The crystal was soaked in a cryoprotectant (mother liquor plus 20% glycerol) and then flash-cooled by plunging into liquid nitrogen.
Figure 2.
Two crystal forms of SaMenB. The maximum dimension of the samples is 0.3 mm.
2.3. Data collection and analysis
Diffraction data were recorded at the European Synchrotron Radiation Facility (ESRF) on station ID 23-2. A MAR CCD was used to collect the data, using X-rays of wavelength 0.87300 Å. The crystals belonged to space group P213, with a unit-cell edge of 120.0 Å. Data were processed using MOSFLM (Leslie, 1992 ▶) and SCALA (Evans, 2006 ▶) from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▶). A Matthews coefficient (Matthews, 1968 ▶) of 2.40 Å3 Da−1 suggested an asymmetric unit containing two subunits with approximately 50% bulk solvent.
2.4. Structure determination
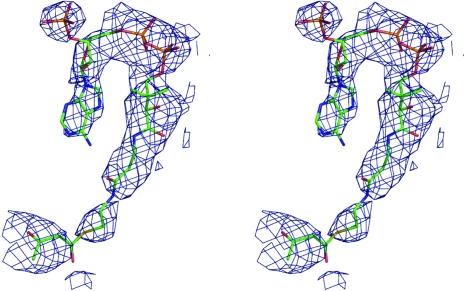
The structure was solved by molecular replacement (Phaser; Storoni et al., 2004 ▶) using a polyalanine model of one subunit from the MtbMenB structure (PDB code 1q52), which shares 50% sequence identity, as the search model. Two subunits were positioned and then refined with REFMAC5 (Murshudov et al., 1997 ▶). The calculation of R free used 5% of the data. Electron-density and difference density maps, all σA-weighted (Read, 1986 ▶), were inspected and the model was improved using Coot (Emsley & Cowtan, 2004 ▶). A simulated-annealing run was performed using CNS (Brünger et al., 1998 ▶) followed by REFMAC5, incorporating translation, libration and screw-rotation displacements (TLS) refinement (Winn et al., 2001 ▶). Since only low-resolution data were available, a conservative approach to the identification of waters was adopted. One of the active sites contained density for acetoacetyl-CoA (Fig. 3 ▶) and incorporation of this ligand constituted the final part of model building.
Figure 3.
The OMIT difference density map associated with acetoacetyl-CoA. The map (blue mesh) was calculated with coefficients |F o − F c| and αc and contoured at 3σ. F o represents the observed structure factors, F c represents the calculated structure factors and αc represents the calculated phases for which ligand contributions were omitted. The ligand atoms are shown as sticks and coloured as follows: C, pink; N, blue; O, red; P, orange; S, yellow.
Strict noncrystallographic symmetry restraints were initially applied to the two subunits in the asymmetric unit. These were gradually relaxed during refinement and then removed when the model was completed. Model geometry was analyzed using PROCHECK (Laskowski et al., 1993 ▶). All residues are within the allowed regions of a Ramachandran plot. Secondary structure was assigned using a combination of PROMOTIF (Hutchinson & Thornton, 1996 ▶), PROCHECK and visual inspection. Figures were prepared with PyMOL (The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA). Crystallographic statistics are given in Table 1 ▶.
Table 1. Crystallographic statistics.
Values in parentheses are for the highest resolution shell.
| Unit-cell parameter (Å) | a = 120.0 |
| Oscillation range/Δ (°) | 45/1 |
| Resolution range (Å) | 84.8–2.9 (3.06–2.90) |
| No. of reflections | 50883 |
| Multiplicity | 3.9 |
| Completeness (%) | 100 (100) |
| 〈I/σ(I)〉 | 7.8 (1.7) |
| Rsym (%) | 8.9 (45.0) |
| Wilson B (Å2) | 71.8 |
| DPI† (Å) | 0.605 |
| Protein residues | 517 |
| Water molecules | 64 |
| Acetoacetyl-CoA molecules | 1 |
| Rwork/Rfree (%) | 24.7 (34.1)/32.8 (37.7) |
| Average B values (Å2) | |
| Overall | 31.4 |
| Main chain | 30.6 |
| Side chains | 32.2 |
| Water molecules | 37.2 |
| Acetoacetyl-CoA | 69.4 |
| R.m.s.d. bond lengths (Å) | 0.006 |
| R.m.s.d. bond angles (°) | 1.138 |
Diffraction-component precision index (Cruickshank, 1999 ▶).
2.5. Quaternary structure analyses
Analytical gel-filtration experiments were conducted on a Superose 10/300 analytical gel-filtration column (GE Healthcare) pre-equilibrated with the respective buffers and calibrated with molecular-weight standards (blue dextran, >2000 kDa; BSA, 67 kDa; carbonic anhydrase, 29.5 kDa; cytochrome c, 12.5 kDa; GE Healthcare; data not shown). Apo SaMenB was run in 20 mM Tris–HCl pH 7.7, 250 mM NaCl.
Samples for analytical ultracentrifugation were prepared in the same buffers (see above) to concentrations of 0.25, 0.5 and 1.0 mg ml−1. Sedimentation-velocity experiments were performed (wavelength of 280 nm at 45 000 rev min−1 and 293 K with an AN50-TI rotor) using a Beckman Coulter XL-1 analytical ultracentrifuge. Samples were centrifuged simultaneously and A 280 measurements were taken at 5 min intervals for 16 h. The resultant data were analyzed using the programs SEDFIT and SEDNTERP (Schuck, 2000 ▶; Lebowitz et al., 2002 ▶).
3. Results and discussion
3.1. Quaternary structure
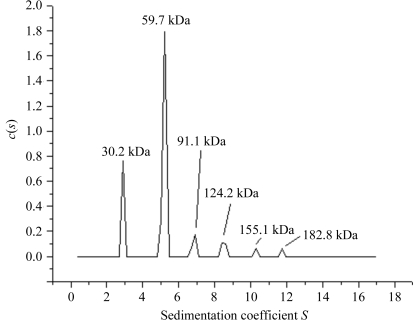
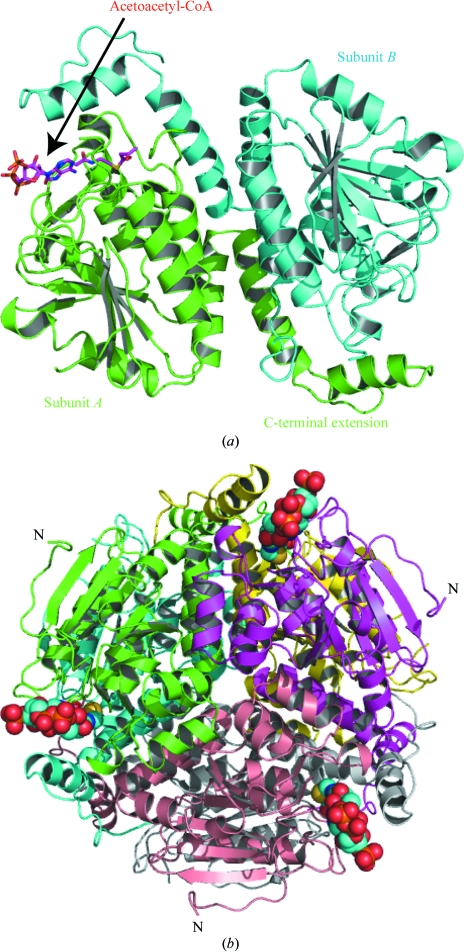
Size-exclusion chromatography indicates that SaMenB, like MtbMenB, forms a hexamer. The estimated molecular weight of 185 kDa agrees with the theoretical value of 192 kDa for such an assembly. However, analytical ultracentrifugation (sedimentation-velocity experiment) revealed six different states ranging from monomers to hexamers (Fig. 4 ▶). The dimeric form, as the most abundant, would be considered to be more stable compared with the other forms under the conditions of the experiment. SaMenB crystallized in space group P213 and the asymmetric unit contains a dimer (Fig. 5 ▶ a). The crystallographic threefold axis generates the hexameric assembly, which is depicted in Fig. 5 ▶(b). This information suggests that the SaMenB hexamer might be considered as a trimer of dimers rather than a dimer of trimers as reported for MtbMenB (Truglio et al., 2003 ▶; Johnston et al., 2005 ▶). However, it is interesting to note that monomeric and pentameric forms of the enzyme are observed in solution, suggesting that the assemblies are not particularly stable. Presumably, the presence of monomer allows complexation with one or two dimers to produce the trimeric and pentameric forms, respectively.
Figure 4.
Analytical ultracentrifugation of SaMenB. The concentration used was 1 mg ml−1. The corresponding molecular weights for each peak are shown. The calculated molecular weights for a monomer, dimer, trimer, tetramer, pentamer and hexamer are 32, 64, 96, 128, 160 and 192 kDa, respectively.
Figure 5.
(a) The asymmetric unit of SaMenB. Subunits are coloured green and cyan. Acetoacetyl-CoA is bound in one active site. The ligand is shown as in Fig. 3 ▶. (b) Ribbon diagram of the MenB hexamer viewed down the crystallographic threefold axis. The subunits are coloured green and cyan (the asymmetric unit), magenta, yellow, grey and wheat. The ligand atoms are coloured as in Fig. 3 ▶ but shown as van der Waals spheres. The N-terminus is marked for three subunits.
3.2. Subunit structure
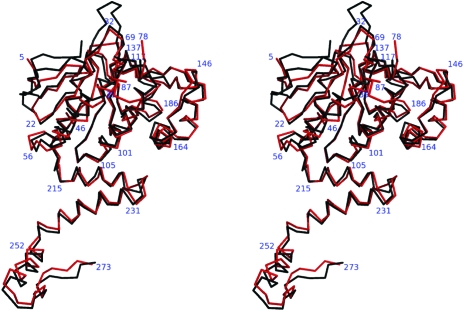
MenB is a member of the crotonase superfamily (enoyl-CoA hydratases/isomerases) of enzymes. The subunit fold consists of an N-terminal domain of about 215 residues in an α/β structure (ten β-strands, seven α-helices) and a smaller C-terminal domain of about 60 residues arranged into three α-helices, which extend out from the main domain (Figs. 5 ▶ and 6 ▶). Some parts of both subunits could not be modelled since there was no interpretable electron density. These sections are residues 1–5 and 79– 86, the latter of which is close to the active site, and the implication is that these are particularly flexible regions. The two subunits within the asymmetric unit of SaMenB are similar, irrespective of whether a ligand occupies the active site or not, and a superposition of 261 Cα positions results in a root-mean-square deviation of 0.23 Å. It is therefore only necessary to give certain details relating to one subunit and we select that which has acetoacetyl-CoA bound. The structure of SaMenB agrees closely with that of MtbMenB. The superposition of these structures gives a root-mean-square deviation of 1.2 Å for the overlay of 249 Cα positions (Fig. 6 ▶). SaMenB is about 40 residues shorter than MtbMenB. The M. tuberculosis enzyme has an N-terminal extension and insertions between β1 and β2 and after β3 and β4 (Fig. 6 ▶). These differences are distant from the active site.
Figure 6.
Least-squares superposition of Cα atoms of MtbMenB (black) and SaMenB (red). The numbers correspond to selected residues of SaMenB.
Accessible surface areas were calculated for both MtbMenB and SaMenB using the protein–protein interactions server (Luscombe et al., 1998 ▶; http://www.biochem.ucl.ac.uk/bsm/PP/server/index.html). The hexameric assembly is similar for each enzyme and is constructed from two types of subunit–subunit associations. One results in the formation of the dimer and the other then generates the hexamer. In MtbMenB the surface-accessible area calculated for the dimer interface is 1470 Å2 (10.4% of the total surface area of a subunit in isolation) and residues here participate in 14 hydrogen bonds and two salt bridges. The surface-accessible area used by a single subunit for the trimer interface is 1340 Å2 (9.5%) and involves 15 hydrogen bonds and a single salt bridge. In SaMenB, the dimer interface involves 14 hydrogen bonds and two salt bridges, while the trimer interface uses ten hydrogen bonds. The accessible surface area at the dimer interface is significantly increased to 1990 Å2 (13.8% of the total surface area) compared with MtbMenB and this observation is consistent with the abundance of dimeric SaMenB seen in the analytical centrifugation experiments. The trimer interface accessible area is 1270 Å2 (8.8% of that of a subunit), providing a rationale for the stability of the MenB hexamer.
3.3. The active site
Residues from the N-terminal domain of one subunit and the C-terminal domain of the adjacent subunit in the asymmetric unit create the MenB active site (Truglio et al., 2003 ▶). Here, a loop of about ten amino acids (residues 79–86) at one side of the active site is disordered. A similar observation has been made for the MtbMenB structures (Truglio et al., 2003 ▶; Johnston et al., 2005 ▶) and we presume that this loop is flexible and speculate that this property may assist binding and/or release of substrate/product. It is conceivable that the CoA moiety of the substrate binds first and the OSB segment is then oriented for catalysis.
The structures with acetoacetyl-CoA indicate the binding mode of a fragment of substrate. The acetoacetyl-CoA is bound in a shallow surface cleft, where it displays a similar conformation and comparable interactions with the active-site residues as observed in MtbMenB complexes (Truglio et al., 2003 ▶). The adenine moiety forms hydrophobic interactions with Ala36, Val33 and Phe258. The carbonyl O of Gly75 forms hydrogen-bonding interactions with the adenine N6. The side chain of Ser35 donates a hydrogen bond to the adenine N7 and Arg34 interacts with the ligand diphosphate (data not shown). These amino-acid residues are strictly conserved between SaMenB and MtbMenB. Also conserved between SaMenB and MtbMenB is a glutamine at positions 77 and 107, respectively. In MtbMenB the adenine N1 accepts a hydrogen bond donated from the amide of Gln107, but in the lower resolution structure of SaMenB this interaction is missing since the polypeptide adopts a slightly different conformation. This part of the SaMenB structure is poorly ordered and immediately precedes the loop for which no electron density was observed.
We do not have structural information on the binding mode of that part of the substrate where catalysis occurs, namely the O-succinylbenzoyl moiety of the substrate. On the basis of the active-site conformation of MtbMenB and molecular modelling, a binding mode and the residues important for recognition and processing of the OSB segment and the enzyme mechanism have been proposed (Truglio et al., 2003 ▶). It has been suggested that the OSB tail binds in a deep hydrophobic pocket near Gly161 of MtbMenB (Truglio et al., 2003 ▶). In SaMenB the residues lining this pocket are Gly75, Leu94, Leu97, Gly121, Ser149, Asp151, Asp151, Thr242, Gly239 and Tyr246 and they are strictly conserved in MtbMenB, whilst Val96 is changed to an isoleucine (Ile136), a conservative alteration, in MtbMenB.
A plausible mechanism of action for MenB has been described (Meganathan, 2001 ▶) consistent with modelling based on an MtbMenB structure (see Fig. 10 in Truglio et al., 2003 ▶). Five residues positioned at the site of catalysis are of particular interest and in MtbMenB they are Gly105, Gly161, Ser190, Asp192 and Tyr287. Asp192 initiates catalysis by inducing the abstraction of a succinyl proton by the benzoate, which results in the formation of an oxyanion. The oxyanion is likely to be resonance-stabilized as a carbanion assisted by proximity to the amides of Gly161 and Gly105. Ring closure followed by elimination of a water molecule to produce the keto form of the product may follow. At this stage Tyr287 and Ser149 help in reducing the ring, which results in the formation of the product. These five residues are strictly conserved in SaMenB as Gly75, Gly121, Ser149, Asp151 and Tyr246.
In summary, we have generated a soluble recombinant source of SaMenB and determined the structure at 2.9 Å resolution. The active site and interactions with acetoacetyl-CoA are highly conserved compared with those observed in higher resolution structures of MtbMenB typically reported at near 2.0 Å resolution. The high degree of conservation between the two enzymes and the better order of the MtbMenB samples suggests that the latter provides a better template for structure-based methods to develop enzyme inhibitors. In the future, it will be important to crystallize MenB in complex with ligands that mimic the OSB component of the substrate in order to obtain data that are pertinent to the enzyme mechanism.
Supplementary Material
PDB reference: MenB–acetoacetyl-CoA complex, 2uzf, r2uzfsf
Acknowledgments
This work was supported by The Wellcome Trust and BBSRC (UK) Structural Proteomics of Rational Targets. We thank the ESRF for beam time and D. Flott for support.
References
- Akerley, B. J., Rubin, E. J., Novick, V. L., Amaya, K., Judson, N. & Mekalanos, J. J. (2002). Proc. Natl Acad. Sci. USA, 99, 966–971. [DOI] [PMC free article] [PubMed]
- Bentley, R. & Meganathan, R. (1982). Microbiol. Rev.46, 241–280. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cruickshank, D. W. J. (1999). Acta Cryst. D55, 583–601. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Forsyth, R. A. et al. (2002). Mol. Microbiol.43, 1387–1400. [DOI] [PubMed]
- Gerlt, J. A. & Babbitt, P. C. (2001). Annu. Rev. Biochem.70, 209–246. [DOI] [PubMed]
- Hutchinson, E. G. & Thornton, J. M. (1996). Protein Sci.5, 212–220. [DOI] [PMC free article] [PubMed]
- Johnston, J. M., Arcus, V. L. & Baker, E. N. (2005). Acta Cryst. D61, 1199–1206. [DOI] [PubMed]
- Kobayashi, K. et al. (2003). Proc. Natl Acad. Sci. USA, 100, 4678–4683.
- Kolappan, S., Zwahlen, J., Zhou, R., Truglio, J. J., Tonge, P. J. & Kisker, C. (2007). Biochemistry, 46, 946–953. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lebowitz, J., Lewis, M. S. & Schuck, P. (2002). Protein Sci.11, 2067–2079. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Lin, E. C. C. & Kuritzkes, D. (1987). Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, edited by F. C. Neidhardt, Y. Kasahara & G. A. Rechnitz, pp. 202–221. Washington: American Society for Microbiology.
- Luscombe, N. M., Laskowski, R. A., Westhead, D. R., Milburn, D., Jones, S., Karmirantzou, M. & Thornton, J. M. (1998). Acta Cryst. D54, 1132–1138. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Meganathan, R. (1996). Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed., edited by F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter & H. E. Umbarger, pp. 642–656. Washington: American Society for Microbiology.
- Meganathan, R. (2001). Vitam. Horm.61, 173–218. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Palmer, D. R., Garrett, J. B., Sharma, V., Meganathan, R., Babbitt, P. C. & Gerlt, J. A. (1999). Biochemistry, 38, 4252–4258. [DOI] [PubMed]
- Read, R. J. (1986). Acta Cryst. A42, 140–149.
- Rowland, S. L., Errington, J. & Wake, R. G. (1995). Gene, 164, 113–116. [DOI] [PubMed]
- Schuck, P. (2000). Biophys. J.78, 1606–1619. [DOI] [PMC free article] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Thompson, T. B., Garrett, J. B., Taylor, E. A., Meganathan, R., Gerlt, J. A. & Rayment, I. (2000). Biochemistry, 39, 10662–10676. [DOI] [PubMed]
- Truglio, J. J., Theis, K., Feng, Y., Gajda, R., Machutta, C., Tonge, P. J. & Kisker, C. (2003). J. Biol. Chem.278, 42352–42360. [DOI] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Xiang, H., Luo, L., Taylor, K. L. & Dunaway-Mariano, D. (1999). Biochemistry, 38, 7638–7652. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: MenB–acetoacetyl-CoA complex, 2uzf, r2uzfsf