Abstract
A previous study of the retinitis pigmentosa mutation L125R and two designed mutations at this site, L125A and L125F, showed that these mutations cause partial or total misfolding of the opsins expressed in COS cells from the corresponding mutant opsin genes. We now report on expression and characterization of the opsins from the following retinitis pigmentosa mutants in the transmembrane domain of rhodopsin that correspond to six of the seven helices: G51A and G51V (helix A), G89D (helix B), A164V (helix D), H211P (helix E), P267L and P267R (helix F), and T297R (helix G). All the mutations caused partial misfolding of the opsins as observed by the UV/visible absorption characteristics and by separation of the expressed opsins into fractions that bound 11-cis-retinal to form the corresponding mutant rhodopsins and those that did not bind 11-cis-retinal. Further, all the mutant rhodopsins prepared from the above mutants, except for G51A, showed strikingly abnormal bleaching behavior with abnormal metarhodopsin II photointermediates. The results show that retinitis pigmentosa mutations in every one of the transmembrane helices can cause misfolding of the opsin. Therefore, on the basis of these and previous results, we conclude that defects in the packing of the transmembrane helices resulting from these mutations are relayed to the intradiscal domain, where they cause misfolding of the opsin by inducing the formation of a disulfide bond other than the native Cys-110—Cys-187 disulfide bond. Thus, there is coupling between packing of the helices in the transmembrane domain and folding to a tertiary structure in the intradiscal domain.
Keywords: G-protein-coupled receptor, signal transduction, 11-cis-retinal, disulfide bond, metarhodopsin II
A large number of point mutations in rhodopsin that occur in all three [intradiscal, transmembrane (TM), and cytoplasmic] domains in rhodopsin are associated with retinitis pigmentosa (RP) (2–6). Previous work with designed (7, 8) and naturally occurring RP mutants (9) in the intradiscal domain has demonstrated partial or total misfolding of the corresponding opsins upon expression of the mutant genes in COS-1 cells. Operationally, misfolding in opsin has been defined as loss of the ability to bind 11-cis-retinal (7). This in turn has been concluded to be due to the formation in the intradiscal domain of a disulfide bond other than the native Cys-110—Cys-187 disulfide bond (7–10). A recent study of the RP mutation, L125R, in helix C of the TM domain and two additional mutations at this site, L125A and L125F, also demonstrated misfolding (11). These results led to the conclusion that defects in the packing of the helices are relayed to the intradiscal domain, where they also cause misfolding by inducing the formation of an abnormal disulfide bond. In undertaking the present work we argued that helix C, where the Leu-125 mutations are located, may be a special case in causing misfolding. This helix contains the conserved charge pair (Glu-134⋅Arg-135), the counterion to the protonated Schiff base (Glu-113), and Cys-110, a participant in the native disulfide bond. Consequently, we wished to determine if coupling between folding in the two domains is a basic characteristic such that certain mutations in every one of the seven TM helices could cause misfolding.
We have now studied the following RP mutations in the TM domain that, together with the previously studied mutations in helix C, represent all seven of the TM helices. The mutations studied (Fig. 1) are G51V (12), G51A (5), G89D (4, 12), A164V (13), H211P (14), P267L (14), P267R (15), and S297R (15). The mutant opsin genes corresponding to all the above mutations were expressed in COS cells and the opsins produced were purified. They all were found to be mixtures of retinal-binding and non-retinal-binding fractions as seen from the UV/visible (UV/Vis) absorption spectral characteristics. The mixtures were separated into fractions that reconstituted with 11-cis-retinal to form the mutant rhodopsins and fractions that did not bind 11-cis-retinal. The degrees of misfolding varied with the mutants, H211P and P267R forming mostly misfolded opsins. Furthermore, when the purified 500-nm chromophore-forming mutant rhodopsins were tested for their bleaching properties (metarhodopsin II formation and its decay) these were strikingly abnormal except for G51A. Thus, the results show that RP mutations in every one of the TM helices can cause misfolding of the opsin, and they substantiate the earlier conclusion that there is coupling between the packing of the helices to form the TM domain and folding in the intradiscal domain to a tertiary structure. While the correctly folded rhodopsin structure contains the disulfide bond between Cys-110 and Cys-187, the misfolded structures contain a different intradiscal disulfide bond.
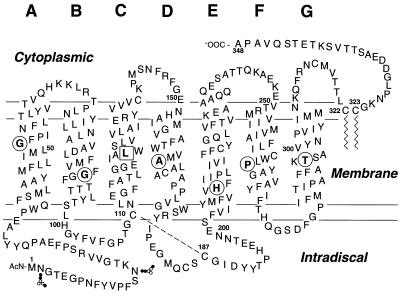
Figure 1.
Secondary structure model of bovine rhodopsin, showing the sites, circled, of the RP mutations in TM helices A, B, and D–G studied in the present work. The mutation, L125R, boxed, in helix C has been studied previously (11). The amino acids that replace the natural amino acids in the RP mutants are as follows: Gly-51 → Ala or Val; Gly-89 → Asp; Ala-164 → Val; His-211 → Pro; Pro-267 → Leu or Arg; and Thr-297 → Arg. The jagged lines at Cys-322 and Cys-323 indicate the palmitoyl groups in native rhodopsin at these residues, whereas the disulfide bond between Cys-110 and Cys-187 is indicated by the broken line. The interrupted horizontal lines indicate the approximate boundaries of the seven TM helices. The small circles in the N-terminal region indicate Asn-linked carbohydrates.
MATERIALS AND METHODS
Materials.
11-cis-Retinal was a gift from Rosalie Crouch (University of South Carolina and the National Eye Institute, National Institutes of Health). Dodecyl maltoside (DM) was from Anatrace (Maumee, OH). Anti-rhodopsin monoclonal antibody rho-1D4 was purified from myeloma cell lines provided by R. S. Molday (University of British Columbia, Vancouver) and was coupled to cyanogen bromide-activated Sepharose 4B (Sigma) as described (16). The buffers used are as follows; buffer A, 137 mM NaCl/2.7 mM KCl/1.8 mM KH2PO4/10 mM NaHPO4, pH 7.2; buffer B, buffer A plus 5 mM ATP, 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and 1% (wt/vol) DM; buffer C, 10 mM Tris⋅HCl, pH 8.0, containing 2 mM ATP, 2 mM MgCl2, and 0.05% DM; buffer D, buffer A plus 0.05% DM; buffer E, 2 mM NaH2PO4 at pH 6.0, and 0.05% DM; and buffer F, 2 mM NaH2PO4 at pH 6.0, 0.05% DM, and 150 mM NaCl.
Construction of the Opsin Gene Mutants G51A, G51V, G89D, A164V, H211P, P267R, P267L, and T297R.
G51A, G51V, G89D, A164V, and H211P were produced by restriction fragment replacement, the synthetic DNA duplexes containing the changed codons replacing the counterparts in the synthetic rhodopsin gene PMT4 vector (17, 18). The codons used for the amino acids replacing the native amino acids were: A, GCT; V, GTT; D, GAT; and P, CCA. The mutants G51A and G51V involved replacement of the BclI–HindIII restriction fragment in the gene (nucleotides 147–209), the mutant G89D involved replacement of the restriction fragment BglII–NcoI (nucleotides 253–307), the mutant A164V contained replacement of the fragment AhaII–SfiI (nucleotides 474–508), and the H211P mutant involved replacement of the fragment AvaII–MscI (nucleotides 634–680).
A two-step technique was used for preparation of the mutants P267R, P267L, and T297R. PCR mutagenesis was used to create a unique larger fragment flanked by MluI and NotI (nucleotides 758-1055). This fragment was then subcloned into the wild-type PMT4 vector at these sites. The PCR protocol used was as follows. For the mutants P267R and P267L, the complementary sets of oligonucleotides (for 267R, 5′-CTGCTGGCTGCGATATGCTGGTG-3′, for 267L, 5′-CTGCTGGCTGCTATATGCTGGTG-3′) flanking the mutation site were used as primers. For the mutant T297R, the two complementary oligonucleotides, corresponding to the sequence 5′-CTTTGCCAAGAGGTCTGCCGTC-3′ were used as the primers. The reaction mixture contained in 5 μl of reaction buffer [100 mM KCl/60 mM (NH4)2SO4/200 mM Tris⋅HCl, pH 8.0/20 mM MgCl2/1% Triton X-100 and 100 μg/ml nuclease-free BSA], 1 μl of wild-type construct (50 ng), 1 μl of each primer (125 ng), 1 μl of dNTPs (10 mM), 40 μl of water, and 1 μl of Pfu DNA polymerase (2.5 units; Stratagene). The cycling protocol used involved 95°C for 30 sec, followed by 55°C for 1 min and 68°C for 10 min. Sixteen cycles were performed. The products were then digested with the restriction enzyme DpnI (New England Biolabs) for 3 hr to remove native wild-type strands, leaving only the synthesized strands. Ten microliters of the PCR product was then used to transform 125 μl of DH5α competent cells (≈2 × 109 cells), followed by DNA extraction from selected clones. All DNA sequences were confirmed by the dideoxynucleotide chain-termination method (19).
Expression of the Mutant Opsin Genes and Purification of the Mutant Rhodopsin and Misfolded Opsins.
The procedure for the transient transfection of PMT4 vectors carrying the opsin genes in COS-1 cells and their growth has been described (16). The cells were harvested 52–56 hr after transfection, washed with buffer A, and incubated with 5 μM 11-cis-retinal (in the dark) for 3 hr at 4°C. They were then solubilized in buffer B by nutating at 4°C for 1 hr and centrifuged at 35,000 rpm for 30 min at 4°C. The solubilized protein was then purified by immunoaffinity adsorption on rho-1D4-Sepharose (200 μl with binding capacity of 160 μg of rhodopsin) (16).
(i) Elution of the total retinal-binding (mutant rhodopsins) and the non-retinal-binding opsins.
The Sepharose was initially washed with 50 bed vol of buffer C, followed by a further 50 bed vol of buffer D. Elution was performed using 2-bed-vol portions of buffer D in the presence of 70 μM C′1–C′9 peptide (corresponding to the carboxyl terminus of rhodopsin). This eluted both the reconstituted mutant rhodopsins and the non-retinal-binding opsins from the Sepharose.
(ii) Separation of the mutant rhodopsins and the corresponding non-retinal-binding opsins (10).
The initial step for selective elution of the reconstituted mutant rhodopsins was identical to that described in i; however, in addition, 50 bed vol of buffer E was used to wash the column prior to elution in the presence of the peptide. The chromophore-forming fraction was eluted with 2-bed-vol portions of buffer E in the presence of 70 μM peptide. A total of at least 15 bed vol were used. This was followed by elution with buffer F. This eluted the non-retinal-binding opsins.
UV/Vis Absorption Spectroscopy.
UV/Vis absorption spectra were recorded using a Perkin–Elmer λ-6 UV/Vis spectrophotometer, equipped with water-jacketed cuvette holders connected to a circulating water bath. All spectra were recorded with a bandwidth of 2 nm, a response time of 1 sec, and a scan speed of 200 nm/min at 20°C. The molar extinction values (ɛ500) for the mutant rhodopsins were calculated from the absorption at 440 nm after acidification as described (20). The value used for wild type is 40,600 M−1⋅cm−1. For the photobleaching experiments the samples were illuminated with a 150-W fiber optic light (Fiber Lite A-200; Dolan-Jenner, Woburn, MA) equipped with a >495-nm long-pass filter for 10 sec and various time intervals up to 18 min. The corresponding bleached spectrum was recorded immediately after illumination.
Rate of Metarhodopsin II Decay as Measured by Retinal Release.
The rate of metarhodopsin II decay was measured, after illumination of the samples, by following the rate of fluorescence increase, which corresponds to the rate of retinal release (21), using a solution of 2 μg of the protein in 200 μl of buffer E. The samples were bleached at 20°C for 30 sec and the fluorescence increase was measured. The excitation and emission wavelengths were 295 nm (slit = 0.25 nm) and 330 nm (slit = 12 nm), respectively.
RESULTS
Isolation and Purification of the RP Mutant Proteins G51A, G51V, G89D, A164V, P267L, and T297R.
The procedures used for purification and separation of the proteins, namely, mutant rhodopsins resulting from binding of 11-cis-retinal to the correctly folded opsins and the misfolded non-retinal-binding fractions, have been described in Materials and Methods. The results obtained for the above mutants are described below.
Mutants G51A and G51V (Helix A) (Fig. 2 A and B, respectively).
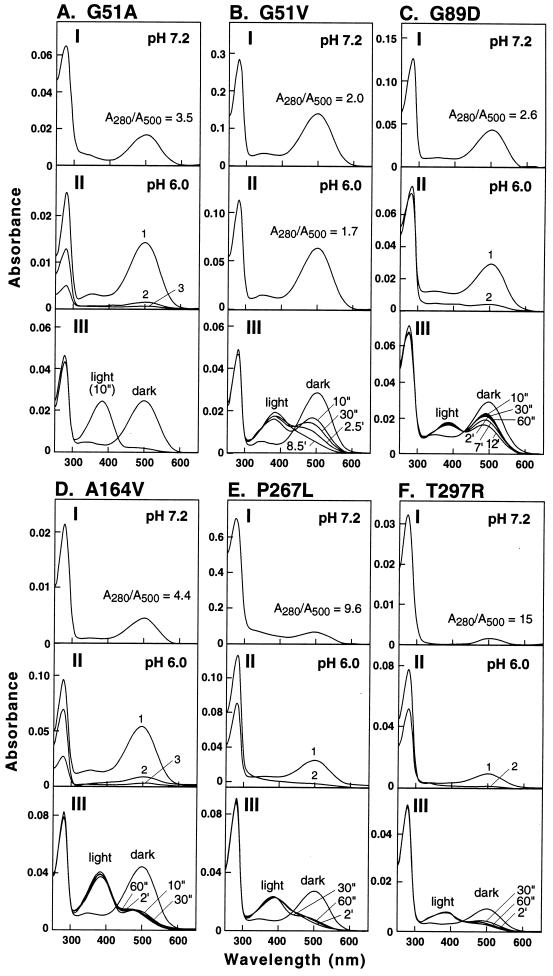
Figure 2.
Spectral properties of the mutant proteins (A–F). In every case, the three panels I–III show the following: (I) UV/Vis spectrum of the total protein (mixture of the 11-cis-retinal-reconstituted mutant rhodopsin and the non-retinal-binding opsin) eluted from the immunoaffinity column at pH 7.2 by buffer A; ratios of absorbance at 280 nm and 500 nm are indicated. (II) Fractions of proteins eluted from the immunoaffinity column first at pH 6.0 in 2 mM phosphate buffer followed by fractions 2 or 2 and 3 eluted at the same pH but in the presence of 150 mM NaCl. (III) Changes in absorption spectrum observed at different periods of illumination (", sec; ′, min).
When eluted at pH 7.2, the proteins from G51A (Fig. 2I) showed an A280/A500 ratio of 3.5, higher than that for the wild-type rhodopsin (close to 1.6; ref. 22). This indicated the presence of misfolded opsin, and fractional elution at pH 6 confirmed this conclusion. The first fraction eluted at pH 6 in the absence of sodium chloride showed an A280/A500 ratio of 1.7, whereas subsequent fractions 2 and 3 showed non-retinal-binding opsin. All fractions were positive to immunoblotting with the anti-rhodopsin antibody 1D4. The protein from the mutant G51V (Fig. 2B) also showed a small proportion of the corresponding misfolded opsin (I and II). The bleaching properties of the purified mutant rhodopsins G51A and G51V (III, Fig. 2 A and B) were very different. While the mutant rhodopsin, G51A, showed bleaching typical of wild-type rhodopsin, that from G51V showed very abnormal behavior. Thus, after illumination for 10 sec, two absorption peaks were observed (Fig. 2. B III) and only slowly on further illumination did the longer-wavelength-absorbing peak shift to the 380-nm absorbance, characteristic of metarhodopsin II. The decay rate of this photointermediate monitored after a total illumination period of 30 sec showed a t1/2 time of 15.7 min for G51V and 23.2 min for G51A (Table 1).
Table 1.
Metarhodopsin II decay rates for the purified reconstituted mutant rhodopsins
| Protein | Decay rate t1/2, min |
|---|---|
| Wild type (COS) | 13.4 |
| G51A | 23.2 |
| G51V | 15.7 |
| G89D | 29.1 |
| A164V | 2.3 |
| P267L | 6.9 |
| T297R | 11.6 |
Mutant G89D (Helix B) (Fig. 2C).
As seen from the UV/Vis spectrum, the mutant protein eluted at pH 7.2 (I) did not correspond to pure mutant rhodopsin, and further fractionation at pH 6 gave the A280/A500 ratio of 2.2 as well as a non-retinal-binding fraction (II) that was positive to immunoblotting with the anti-rhodopsin antibody. The purified mutant rhodopsin also showed very abnormal bleaching behavior (III). In addition the metarhodopsin II decay was much slower (t1/2, 29.1 min) (Table 1).
Mutant A164V (Helix D) (Fig. 2D).
The UV/Vis spectrum (I) indicated a larger proportion of the misfolded mutant opsin as well as a slight blue shift in the absorption maximum (λmax, 498 nm). The blue shift increased with an increase in size of the side chain in amino acid. This was more apparent when a larger substitution was made at this position (A164I, λmax, 494 nm; A164L, λmax, 488 nm, unpublished data). As seen in II, fractionation of the proteins by elution at pH 6 raised the A280/A500 absorption ratio to the normal wild-type level, and the subsequent peaks eluted in the presence of salt showed the non-retinal-binding fractions. The bleaching behavior was abnormal in that bimodal absorption peaks were observed that seemed to be refractory to further illumination. The metarhodopsin II state was very unstable, with a t1/2 of 2.3 min (Table 1).
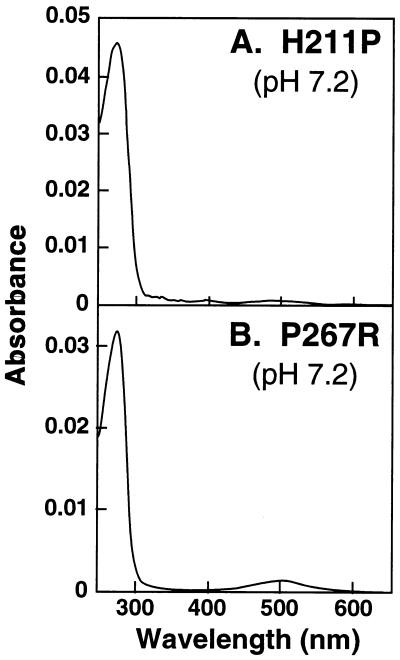
Mutant H211P (Helix E) (Fig. 3A).
Figure 3.
UV/Vis absorption spectra of the total mutant proteins eluted at pH 7.2 from immunoaffinity columns. (A) H211P. (B) P267R.
As seen from the absorption spectrum, this amino acid replacement caused misfolding of the opsin essentially completely.
Mutants P267R (Fig. 3B) and P267L (Fig. 2E) (Helix F).
The mutant P267R caused misfolding essentially completely and the replacement of the amino acid to leucine also caused extensive misfolding (Fig. 2E I). The mutant rhodopsin purified at pH 6 (II) while having the wild-type λmax at 500 nm showed abnormal bleaching behavior (III). Metarhodopsin II decay was faster, with a t1/2 of 6.9 min (Table 1).
Mutant T297R (Helix G) (Fig. 2F).
As for the other known RP mutations inserting arginine, for example L125R, this mutation also caused mostly misfolding (I). However, the small amount of the retinal-binding fraction could be purified (II) with some improvement in the A280/A500 absorption ratio. This fraction again showed abnormal bleaching behavior (III) and a mildly decreased metarhodopsin II decay rate (t1/2,11.6 min) (Table 1).
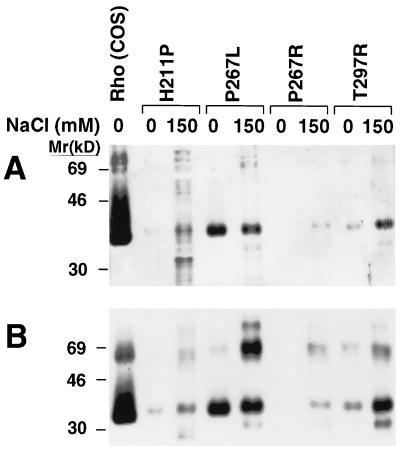
SDS/PAGE Analysis of Purified Proteins Expressed from the Mutant Genes H211P, P267L, P267R, and T297R.
Purified retinal-binding (eluates from the antibody column with no salt) and non-retinal-binding fractions (eluates in the presence of 150 mM salt) from the four mutants were studied by SDS/PAGE (Fig. 4). The proteins were visualized by silver staining (Fig. 4A) and by immunoblotting with anti-rhodopsin antibody 1D4 (Fig. 4B). All the proteins seen on silver staining were also immunopositive.
Figure 4.
SDS/PAGE and immunoblot analysis of the proteins expressed from the RP mutants H211P, P267R, P267L, and T297R. (A) Silver-stained gel with wild-type rhodopsin (COS) in lane 1. The no-salt elution fraction (0 NaCl) and 150 mM NaCl elution fractions are shown. (B) Corresponding 1D4 monoclonal antibody immunoblot for a gel run in parallel to that in A.
Finally, the proteins from H211P and P267R, which were mostly misfolded (Fig. 3) exhibited little eluted protein in the absence of NaCl, with the majority of proteins being eluted with 150 mM NaCl.
DISCUSSION
Earlier studies on some RP mutations in the TM domain of rhodopsin suggested misfolding of the opsin (12, 14). A recent detailed study of mutations at the Leu-125 locus (11) and the results now described demonstrate that amino acid replacements in every one of the TM helices can cause misfolding of the opsin. Misfolding has been shown by separation of the mixtures of opsins that were produced into fractions that fail to bind 11-cis-retinal (misfolded opsins) and the mutant rhodopsins that result from the binding of the retinal to the correctly folded opsins. All the proteins were positive to immunoblotting with the anti-rhodopsin antibody 1D4.
The purified fractions of mutant rhodopsins that formed the normal 500-nm chromophore in the dark state all showed strikingly abnormal bleaching behavior (III in Fig. 2 B–F), except for the mutant rhodopsin G51A (III in Fig. 2A) and the metarhodopsin II photointermediates showed abnormal decay rates. These data all highlight the conclusion that there is indeed a conformational change in the TM domain upon light activation, with requirements for interhelical interactions between amino acids that are different from those for 11-cis-retinal binding in the dark state. Clearly, in the above mutant rhodopsins, while the requirements for 11-cis-retinal binding in the dark are met, the requirements for the optimal metarhodopsin II conformation are not met.
Not all amino acid substitutions in a TM α-helix may be expected to cause misfolding. The consequences of such substitutions will be determined by the nature of the amino acid and by its orientation within the helix. An amino acid (i) may face an adjacent helical bundle and participate in specific interhelical interactions, (ii) may be in the retinal-binding pocket, or (iii) may face the phospholipid bilayer. Indeed, moving a mutation to neighboring positions in the same helix can provide some insights into the orientation of an amino acid within a helix. In previous work the RP mutant P171L was found to bind 11-cis-retinal poorly, whereas the mutant P170L, with the same mutation in the neighboring position, formed rhodopsin chromophore almost normally (23). Similarly, transferring the mutation L125R to a number of proximal positions showed striking effects on the phenotypes in these mutants (11).
The important conclusion from the present work is the coupling between the packing of the helices to form the TM domain and the folding in the intradiscal domain to a tertiary structure. It is appropriate to summarize the findings during the last 10 years that have led to this conclusion. (i) Karnik et al. (24) showed by systematic replacement by serine, one at a time, of the 10 cysteine residues in bovine opsin, that Cys-110 and Cys-187 are essential for the formation of the correct functional rhodopsin structure. (ii) Karnik and Khorana (25) showed that these two cysteines formed a disulfide bond and pointed out that this disulfide bond is conserved in most of the G-protein-coupled receptors. (iii) Ridge et al. (10), choosing a bovine opsin mutant that contained only the three intradiscal cysteines, Cys-110, Cys-185, and Cys-187, demonstrated partial misfolding of the corresponding opsin expressed in COS cells. Importantly, they separated the misfolded opsin from the correctly folded opsin, the 11-cis-retinal-binding fraction. They showed further that the misfolded opsin also contained a disulfide bond like the correctly folded opsin. (iv) A wide variety of mutants in the intradiscal domain prepared by designed mutagenesis (7) as well as mutants causing partial or total misfolding and in certain cases misfolding were confirmed by separation of the correctly folded and misfolded opsins. (v) As described in this paper, mutations in every one of the TM domains can cause misfolding. (vi) Misfolding caused by mutations in both the intradiscal and the TM domains has been, and still is, attributed to the formation of an intradiscal disulfide bond other than the native disulfide bond. This conclusion follows from the fact that the covalent disulfide bond formation is the only step that represents an irreversible commitment on the part of the opsin molecule undergoing folding. Experiments to accurately identify the disulfide bond(s) in the misfolded opsins remain to be done.
Finally, it is noted that the coupling between the two domains in in vivo folding concluded above has been demonstrated previously to be essential for signal transduction by rhodopsin after light activation (26, 27). We believe that such couplings will prove to be a general feature of the G-protein-coupled receptors.
Acknowledgments
We have benefitted greatly from discussions with Prof. U. L. RajBhandary (Biology Department, Massachusetts Institute of Technology) and our laboratory colleagues. We thank Ms. Judy Carlin for her sustained assistance in the preparation of the manuscript. This work was supported by National Institutes of Health Grant GM28289, fellowships from the Ministerio de Educacion y Ciencia and Direccio General de Recerca de Catalunya from Spain (P.G.), and National Cancer Institute Training Grant 2 T32 CA09112 (X.L.). J.H. is the recipient of a Howard Hughes Medical Institute Physician Postdoctoral Fellowship.
ABBREVIATIONS
- TM
transmembrane
- RP
retinitis pigmentosa
- DM
dodecyl maltoside. Mutant rhodopsins with amino acid substitutions are designated by the one letter abbreviations for amino acids. The amino acid to the left of the residue number is the original, whereas that to the right is the substituted amino acid
Footnotes
This is paper 24 in the series “Structure and function in rhodopsin.” Paper 23 is ref. 1.
References
- 1.Thurmond R L, Creuzenet C, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berson E L. Invest Ophthalmol Vis Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]
- 3.Dryja T P, Berson E L. Invest Ophthalmol Vis Sci. 1995;36:1197–1200. [PubMed] [Google Scholar]
- 4.Sung C-H, Davenport C M, Hennessey J C, Maumenee I H, Jacobson S G, Heckenlively J R, Nowakowski R, Fishman G, Gouras P, Nathans J. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macke J P, Davenport C M, Jacobson S G, Hennessey J C, Gonzalez-Fernandez F, Conway B P, Heckenlively J, Palmer R, Maumenee I H, Sieving P, Gouras P, Good W, Nathans J. Am J Hum Genet. 1993;53:80–89. [PMC free article] [PubMed] [Google Scholar]
- 6.Inglehearn C F, Keen T J, Bashir R, Jay M, Fitzke F, Bird A C, Crombie A, Bhattacharya S S. Hum Mol Genet. 1992;1:41–45. doi: 10.1093/hmg/1.1.41. [DOI] [PubMed] [Google Scholar]
- 7.Doi T, Molday R S, Khorana H G. Proc Natl Acad Sci USA. 1990;87:4991–4995. doi: 10.1073/pnas.87.13.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anukanth A, Khorana H G. J Biol Chem. 1994;269:19738–19744. [PubMed] [Google Scholar]
- 9.Liu X, Garriga P, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridge K D, Lu Z, Liu X, Khorana H G. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 11.Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1996;93:4560–4564. doi: 10.1073/pnas.93.10.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dryja T P, Hahn L B, Cowley G S, McGee T L, Berson E L. Proc Natl Acad Sci USA. 1991;88:9370–9374. doi: 10.1073/pnas.88.20.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs S, Kranich H, Denton M J, Zrenner E, Bhattacharya S S, Humphries P, Gal A. Hum Mol Genet. 1994;3:1203. doi: 10.1093/hmg/3.7.1203. [DOI] [PubMed] [Google Scholar]
- 14.Sung C, Davenport C M, Nathans J. J Biol Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- 15.Souied E, Gerber S, Rozet J, Bonneau D, Dufier J, Ghazi I, Philip N, Soubrane G, Coscas G, Munnich A, Kaplan J. J Hum Mol Genet. 1994;3:1433–1434. doi: 10.1093/hmg/3.8.1433. [DOI] [PubMed] [Google Scholar]
- 16.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferretti L, Karnik S S, Khorana H G, Nassal M, Oprian D D. Proc Natl Acad Sci USA. 1986;83:599–603. doi: 10.1073/pnas.83.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke R R, Sakmar T P, Oprian D D, Khorana H G. J Biol Chem. 1988;267:2119–2122. [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrens D L, Khorana H G. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 22.Hong K, Knudsen P J, Hubbell W L. Methods Enzymol. 1982;81:144–150. doi: 10.1016/s0076-6879(82)81024-0. [DOI] [PubMed] [Google Scholar]
- 23.Kaushal S, Khorana H G. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 24.Karnik S S, Sakmar T P, Chen H-B, Khorana H. Proc Natl Acad Sci USA. 1998;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karnik S S, Khorana H G. J Biol Chem. 1990;265:17520–17524. [PubMed] [Google Scholar]
- 26.Davidson F F, Loewen P C, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaushal S, Ridge K D, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]