Abstract
Genetic disruption of the Saccharomyces cerevisiae C-4 sterol methyl oxidase ERG25 gene leads to sterol auxotrophy. We have characterized a suppression system that requires two mutations to restore viability to this disrupted strain. One suppressor mutation is erg11, which is blocked in 14α-demethylation of lanosterol and is itself an auxotroph. The second suppressor mutation required is either slu1 or slu2 (suppressor of lanosterol utilization). These mutations are leaky versions of HEM2 and HEM4, respectively; addition of exogenous hemin reverses the suppressing effects of slu1 and slu2. Suppression of erg25 by erg11 slu1 (or erg11 slu2) results in a slow-growing strain in which lanosterol, the first sterol in the pathway, accumulates. This result indicates that endogenously synthesized lanosterol can substitute for ergosterol and support growth. In the triple mutants, all but 1 (ERG6) of the 13 subsequent reactions of the ergosterol pathway are inactive. Azole antibiotics (clotrimazole, ketoconazole, and itraconazole) widely used to combat fungal infections are known to do so by inhibiting the ERG11 gene product, the 14α-demethylase. In this investigation, we demonstrate that treatment of the sterol auxotrophs erg25 slu1 or erg25 slu2 with azole antibiotics paradoxically restores viability to these strains in the absence of sterol supplementation via the suppression system we have described.
Keywords: fungi, sterol biosynthesis, azoles
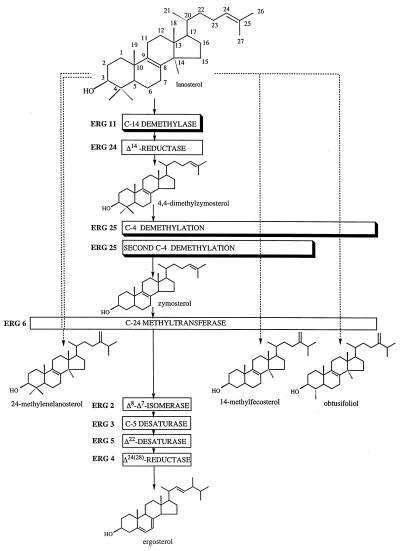
The yeast Saccharomyces cerevisiae has long served as a model system for studies in sterol biosynthesis. The sterol pathway is a branch of the isoprenoid pathway that also gives rise to several other essential compounds in the cell. Mutations in the sterol pathway prior to lanosterol, the first sterol molecule, lead to sterol auxotrophy. Most genes in yeast ergosterol synthesis have been identified and characterized by direct cloning or by analysis of mutations (1). Mutations from ERG6 to the end of the pathway (Fig. 1) have been determined to be nonessential for growth, because the sterol intermediates produced are able to substitute for ergosterol and permit growth. Mutations in the steps between lanosterol and zymosterol (Fig. 1), however, have been shown to result in sterol auxotrophy.
Figure 1.
Description of the late sterol biosynthetic pathway in S. cerevisiae wild type (—), erg11 (---), and erg11 erg25 slu (-- -- --). Shadowed boxes represent the enzymatic steps involved in this investigation. C-4 demethylation depends upon enzymes encoded by ERG25 and as yet two unidentified additional genes.
Lesions in the ergosterol biosynthetic pathway resulting in sterol auxotrophy are of particular interest in the development and application of antifungal drugs. Although there are a number of inhibitors of sterol biosynthesis (2), the azole antifungals are of critical importance in human health because of their widespread use (3). The ERG11 gene product, the cytochrome P450 lanosterol 14α-demethylase, represents the target site for the azoles. These drugs have been successfully used in both topical and systemic infections involving Candida albicans and other pathogenic fungi. Recent increases in fungal infection rates (4) due to advanced chemotherapies, invasive surgical techniques, transplantation technologies, and disease conditions affecting the immune system have resulted in increased azole use and the emergence of resistance (5, 6).
In a previous report (7) we described the cloning and characterization of the ERG25 gene (Fig. 1) encoding the C-4 sterol methyl oxidase, the enzyme responsible for the first step in the removal of the two methyl groups at the C-4 position. Null mutations of ERG25 resulted in sterol auxotrophy. A similar phenotype was also observed in erg11 (8, 9) and erg24 (10) mutants. erg11 and erg24 mutations have been shown to be subject to suppression which thereby restores viability to the cell. The suppressor for erg11 has been identified as a downstream erg3 mutation (ref. 11; Fig. 1). Suppression is hypothesized to occur because the erg11 mutation would produce C-14 methyl sterol intermediates that could not undergo complete C-5 desaturation (encoded by ERG3), resulting in the accumulation of a toxic sterol intermediate [14-methylergosta-8,24(28)-dien-3β-ol; ref. 12]. The erg3 gene product cannot initiate the desaturation step thus eliminating the formation of the diol. The fen1 mutation, which is the suppressor for erg24 (Δ14 reductase), allows cells normally accumulating ignosterol (ergosta-8,14-dien-3β-ol) to grow successfully with this intermediate (10, 13). The nature of the fen1 mutation has not been elucidated.
In this report we describe the characterization of suppressors of the erg25 mutation. The mechanism identified differs from those observed in erg11 and erg24 and involves two distinct mutations. Because one of these mutations can be mimicked by azoles, this mechanism of suppression has important implications in the development and application of antifungal compounds that target C-4 sterol demethylation in ergosterol biosynthesis. Other characteristics of ERG25 suppression have implications in the overall regulation of the enzymatic steps in sterol synthesis and in the utilization of alternative sterols in yeast.
MATERIALS AND METHODS
Strains and Plasmids.
The following yeast strains were used: WA1 (MATα ura3-52 leu2-3, 112 ade5 his7-2), WA6 (same as WA1 but MATa), TTY202-C (MATα ura3-52 leu2-3, 112 his− erg11::URA3, received from Jack Loper, Cincinnati). The erg25::URA3 disrupted strain CP512C was constructed as described (7). Briefly, a 0.4-kb XbaI fragment from plasmid pIU705 was deleted from within the ERG25 coding sequence and replaced by a 1.2-kb XbaI fragment containing the URA3 gene. A 2.2-kb DraI fragment containing ERG25 recombinogenic ends was used to disrupt a WA1/6 diploid and erg25::URA3 segregants were obtained. The erg25::LEU2 disrupted strain WA6–803 was generated in the same manner except the XbaI site of pIU705 was filled in with Klenow and ligated to a Klenow-filled 1.6-kb BamHI fragment containing the LEU2 gene derived from YDpL (14). A 2.6-kb DraI fragment was used to transform WA6, and transformants were grown anaerobically on ergosterol-containing medium. The disrupted version of ERG11 was obtained from plasmid p2500 (obtained from Jack Loper, ref. 9) as a 3.5-kb BamHI–HindIII fragment and was used to transform slu1 and slu2 strains (suppressor of lanosterol utilization). The slu1 mutant strain was derived as a segregant in a cross between a suppressed erg25::URA3 (CP512C-6S) crossed to WA1. Similarly, the slu2 strain was derived as a segregant from a cross between suppressed erg25::LEU2 erg11::URA3 (CP30A-5). pML77, containing a 4.7-kb BamHI–HindIII ERG11 derived from pVK11 (9), was used to complement erg11 disruptants.
Media.
Yeast sterol auxotrophs were grown anaerobically at 30°C on yeast complete medium (YPD, 1% yeast extract, 2% peptone, 2% glucose) supplemented with ergosterol (0.002%) and Tween 80 (0.5%). All other strains were grown on YPD aerobically. Ethanol at 2% replaced glucose (YPE) to distinguish slu mutants from wild type. Minimal medium consisted of 0.67% yeast nitrogen base, 2% glucose, and the addition of amino acid and nitrogenous base supplements as a 0.8% complete synthetic medium (CSM) addition (Bio 101). The azole antifungals, itraconazole (Janssen Pharmaceuticals), ketoconazole (Sigma), and clotrimazole (Sigma) were added at 1 μM to YPD plates. Ergosterol and hemin chloride (added to media at 26–130 μg/ml) were purchased from Sigma.
Transformations and DNA Sequencing.
A genomic library of S. cerevisiae in vector YCp50 was obtained from the laboratory of David Botstein (15) and used to clone slu1 and slu2. Plasmid DNA was extracted from yeast cells by standard methods (16) and transformed into Escherichia coli DH5α for plasmid amplification, restriction digests, and subcloning of DNA fragments. DH5α was grown in Luria–Bertani medium supplemented with ampicillin (50 mg/liter). DNA was isolated by the alkaline lysis method (17) and purified using Qiagen (Chatsworth, CA) columns. Sequencing was performed by the Biochemistry Biotechnology Facility at the Indiana University School of Medicine using the Applied Biosystems model 373 automated DNA sequencer. Sequencing reactions were performed using dye terminator chemistry with the Applied Biosystems Prism Kit.
Sterol Analyses.
Sterols were isolated as nonsaponifiables as described (7). Gas chromatography (GC) analyses of nonsaponifiables were analyzed on a HP5890 series II equipped with the Hewlett–Packard chemstation software package. The capillary column (HP-5) was 15 m × 0.25 mm × 0.25 mm film thickness and was programmed from 195°C to 300°C (3 min at 195°C, then an increase at 5.5°C/min until the final temperature of 300°C was reached and held for 4 min). The linear velocity was 30 cm/sec using nitrogen as the carrier gas and all injections were run in the splitless mode. GC/mass spectrometry (MS) analyses were done using a Varian 3400 gas chromatograph interfaced to a Finnigan–MAT (San Jose, CA) SSQ 7000 mass spectrometer. The GC separations were done on a fused silica column, DB-5 15 m × 0.32 mm × 0.25 mm film thickness programmed from 50°C to 250°C at 20°C/min after a 1-min hold at 50°C. The oven temperature was then held at 250°C for 10 min before programming the temperature to 300°C at 20°C/min. Helium was the carrier gas with a linear velocity of 50 cm/sec in the splitless mode. The mass spectrometer was in the electron impact ionization mode at an electron energy of 70 eV, an ion source temperature of 150°C, and scanning from 40 to 650 atomic mass units at 0.5-sec intervals.
RESULTS AND DISCUSSION
Isolation of erg25 Suppressor Mutations.
The erg25 mutant of S. cerevisiae is an ergosterol auxotroph and requires ergosterol supplementation for growth. Typically, yeast cells are able to take up ergosterol anaerobically but aerobic uptake requires a heme-deficient (hem; refs. 18 and 19) or upc (uptake of cholesterol; ref. 20) background. We have observed that erg25 mutants plated aerobically on YPD medium from YPD plus ergosterol show low levels of growth after several days of incubation. This growth has been shown to be the result of the accumulation of suppressor mutations. The sterol profile of a suppressor strain, CP512C-6S, shows 93% lanosterol and 7% 24-methylenelanosterol (Table 1) rather than 4,4-dimethylzymosterol typical of erg25 (7). The accumulation of such high levels of C-14 methyl sterols implicated a block at the upstream C-14 demethylase encoded by the ERG11 gene (Fig. 1). However, erg11 mutants have been reported to accumulate not only lanosterol but also significant amounts of 14-methylfecosterol and obtusifoliol (refs. 8 and 21; Fig. 1).
Genetic Analysis of Suppression.
The suppressor strain was crossed to a wild-type strain and analyzed for the presence of erg11 and erg11 erg25 segregants. Of 30 tetrads, which included only 11 complete 4-spored asci, 11 segregants were identified as erg11 erg25 double mutants on the basis of lanosterol accumulation and the segregation of uracil prototrophy (erg25::URA3). Of these 11 segregants, only 7 could grow on YPD without sterol supplementation, suggesting that a second, independently segregating gene was required for suppression. The sterol profiles of all 11 isolates were identical regardless of the ability to grow on YPD without sterol supplementation.
The erg11 segregants resulting from the suppressor–wild-type cross were found to grow only under anaerobic conditions with ergosterol supplementation and to accumulate lanosterol, 14-methylfecosterol, and obtusifoliol (Table 1). They also failed to complement a known erg11-disrupted strain, TTY202C, but could be complemented using a ERG11-containing plasmid, pML77. Lastly, among the presumably wild-type segregants were isolates which grew slower on YPD and failed to grow on YPE after 48 h of incubation. These isolates were found to contain a mutation, slu1, which is required along with erg11 for suppression of the erg25 defect.
To assess the nature of the slu mutation, an erg11 erg25 double mutant strain was created by crossing erg25::LEU2 × erg11::URA3. This cross was designed to obtain the double mutant that required ergosterol for growth and was prototrophic for uracil and leucine. This strain, CP30A, was plated on YPD and allowed to accumulate suppressors. One colony, CP30A-5, was then crossed to a wild type. Twenty-eight tetrads were isolated and 23 of 47 erg11 erg25 double mutants grew without sterol supplementation. All erg11 erg25 double mutants accumulated principally lanosterol. Again, a slow growing, presumably wild-type segregant was obtained that was able to grow well on YPD medium but very slowly on YPE. This segregant was designated as slu2. The slu1 × slu2 cross complemented suggesting that these mutants were not alleles.
GC/MS Analyses of slu1 and slu2.
slu1 strains accumulated principally ergosta-5,7-dien-3β-ol, ergosterol, and lanosterol (Table 1). Lesser amounts of 14-methylfecosterol (≈0.2%) and trace amounts of a 14-methyldiol sterol were also noted. The large accumulation of the ergosta-5,7-dien-3β-ol and the multiple C-14 methyl sterol species indicated blocks in the two cytochrome P450 gene products of ERG5 and ERG11, respectively (22). A similar situation was seen in the sterol analysis of slu2 mutants. This strain accumulated lanosterol, 14-methylfecosterol, and fecosterol (or under more extended growth conditions ergosta-5,7-dien-3β-ol) as its principal sterols again indicating defects in the two cytochrome P450 requiring steps in sterol biosynthesis. Based on the inability to grow efficiently on ethanol and the sterol profiles of slu1 and slu2, defects in heme biosynthesis were implicated. This result was confirmed when the addition of hemin (130 μg/ml) to the growth medium of slu2 resulted in wild-type levels (50–70%) of ergosterol synthesis and normal growth on ethanol.
Cloning and Characterization of slu1 and slu2.
slu1 and slu2 were cloned using a YCp50 based Saccharomyces library (15). Transformants were scored for growth on YPE medium within 48 h. A slu1-complementing plasmid containing a 10-kb genomic DNA insert was subcloned as separate HindIII fragments (3 kb and 1.3 kb) into pRS316 (23) for DNA sequencing. DNA sequence analysis indicated that the 1.3-kb DNA fragment contained only the HEM2 (encodes δ-aminolevulinate dehydratase) gene, and this plasmid was able to complement slu1 mutants. A 5-kb DNA BamHI fragment from the slu2 complementing plasmid (originally containing 12 kb of genomic insert) was also subcloned into pRS316 for DNA sequencing, and sequence analysis indicated that this genomic fragment was adjacent to the HEM4 gene (encodes uroporphyrin III synthase). A 1.5-kb BamHI DNA fragment from the same plasmid and containing only the entire HEM4 gene complemented slu2 mutants. slu1 and slu2 are complemented exclusively by HEM2 and HEM4, respectively. Coupled with the fact that hemin restores wild-type ergosterol levels and growth on ethanol, these complementation results provide strong evidence that slu1 and slu2 are mutations in HEM2 and HEM4, respectively.
slu1 and slu2 allow erg11 erg25 double mutants to grow on YPD plates without sterol supplementation. The slu mutants differ from true heme mutants in that they are leaky and provide sufficient heme for some ergosterol synthesis as well as the synthesis of unsaturated fatty acid and methionine. They also differ in that heme mutants are respiratory deficient whereas slu mutants grow very slowly on nonfermentable energy sources. The erg11 erg25 slu mutants have a different sterol profile than erg11 in that the former strains accumulate no sterols (Table 1) beyond lanosterol, with the exception of a small amount of the 24-methylenelanosterol (Fig. 1). However, in erg11 mutants, downstream interconversions do occur as evidenced by the accumulation of 14-methylfecosterol and obtusifoliol. Growth utilizing exclusively lanosterol is rare in biological systems and, particularly in yeast, has been the subject of some controversy. Using a yeast mutant, GL7, unable to grow without sterol supplementation, Nes et al. (24) demonstrated some growth on lanosterol whereas Buttke and Bloch (25) reported that lanosterol did not support the growth of GL7. However, the present investigation clearly demonstrates that endogenously synthesized lanosterol can indeed support yeast growth in a suppressed erg25 strain. Lastly, Buttke and Van Cleave (26) recently demonstrated growth on lanosterol in a human T cell line (A3.01) deficient in cholesterol biosynthesis.
Creating erg11 erg25 slu Triple Mutants.
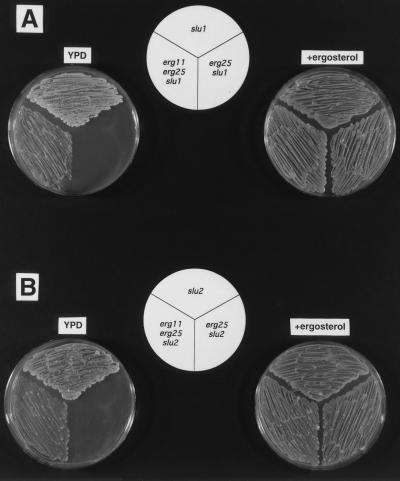
To verify that slu1 and slu2 are indeed required for suppression, we sequentially transformed slu1 and slu2 with disrupted versions of erg25 and erg11. The growth characteristics of the constructed double and triple mutants are presented in Fig. 2. Fig. 2A indicates that erg25 slu1 mutants grow on YPD with ergosterol supplementation but fail to grow on YPD alone. The same is demonstrated in Fig. 2B for erg25 slu2. However, when erg25 slu1 and erg25 slu2 are transformed with erg11 to generate the triple mutants, both erg11 erg25 slu1 (Fig. 2A) and erg11 erg25 slu2 (Fig. 2B) are able to grow on YPD without supplementation. All experiments to test viability and growth responses of the erg11erg25 slu mutants on YPD plates resulted from pregrowth of each strain on YPD plus ergosterol grown anaerobically. Using this protocol, slu2 suppresses erg11erg25 more efficiently than slu1. The slu2 strain is more similar in sterol profile to a true heme mutant than slu1 (Table 1). Although the triple mutants can be sequentially patched from YPD to YPD plates after 48 h growth, the triple mutants in liquid YPD cultures often failed to grow. This problem could be remedied by the addition of 0.1% Tween 80. Lastly, only erg11erg25 slu2 was able to grow with confluence on solid CSM (containing Difco Noble agar), but this growth took 96 h. However, both triple mutants responded to as little as 1 ng/ml ergosterol, which resulted in growth on CSM solid medium after 48 h, suggesting that sterol uptake is occuring in these strains.
Figure 2.
(A) Growth responses of slu1, erg25 slu1, and erg11 erg25 slu1 grown aerobically on YPD and anaerobically on YPD with ergosterol. The triple mutant (erg11 erg25 slu1) is suppressed on YPD unlike the erg25 slu1 double mutant. (B) The same result with slu2, erg25 slu2, and erg11 erg25 slu2. All plates were grown for 48 h.
Azole Antibiotics Rescue erg25 slu Double Mutants.
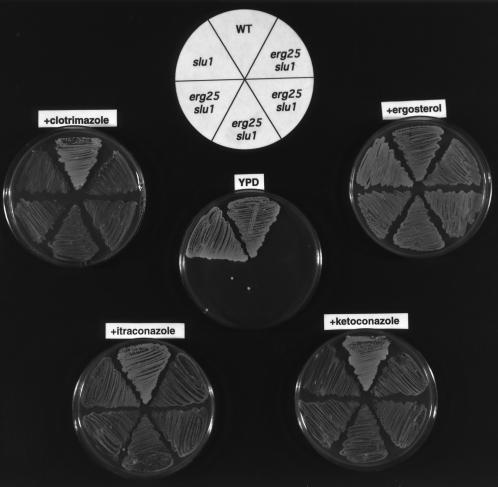
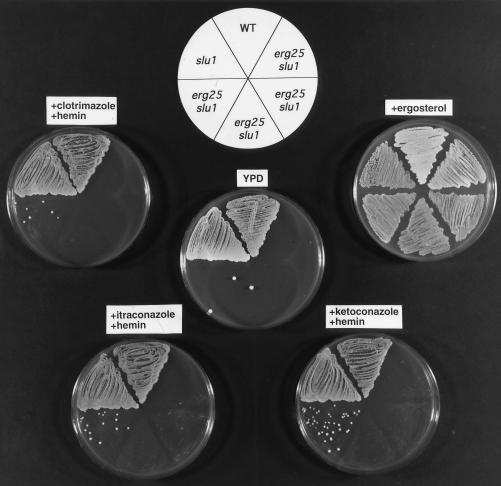
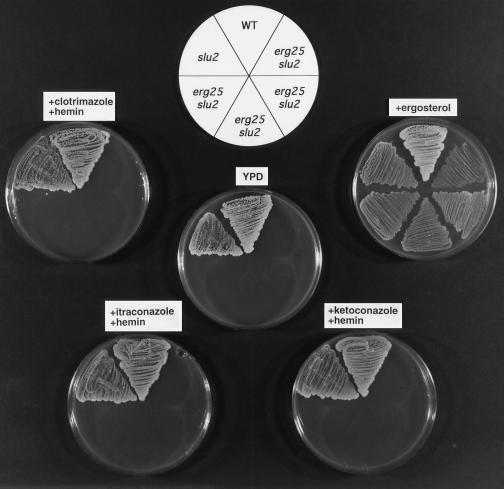
The fact that both erg11 and slu1 (or slu2) are cosuppressors of erg25 suggested that azole antibiotics, which block C-14 demethylation, might be able to function as a replacement cosuppressor for erg11 and elicit suppression with slu1 or slu2. The data presented in Fig. 3 shows that wild-type and slu1 strains grow on YPD with and without ergosterol supplementation. The four erg25 slu1 transformants fail to grow on YPD alone, but in the presence of 1 μM azole (clotrimazole, itraconazole, or ketoconazole) growth of erg25 slu1 can be demonstrated. The growth patterns presented in Fig. 4 show a similar response for erg25 slu2 to the presence of azoles, although slu2 is not as effective a cosuppressor as slu1.
Figure 3.
Growth responses of wild-type (WA1), slu1, and four erg25 slu1 transformants grown on YPD, YPD plus ergosterol, and YPD plus 1 μM azoles (clotrimazole, ketoconazole, or itraconazole) to indicate that azoles mimic erg11 mutations and rescue the erg25 slu1 double mutant. All growth was aerobic except on ergosterol where growth was anaerobic. All plates were grown for 48 h.
Figure 4.
Growth responses of wild-type (WA1), slu2, and four erg25 slu2 transformants grown under the same conditions as in Fig. 3.
Reversal of slu1 and slu2 Cosuppression by Hemin.
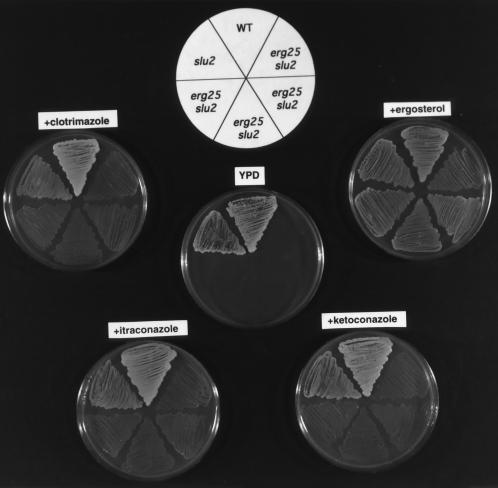
The data presented thus far indicates that a leaky hem2 or hem4 (slu1 or slu2) is a required cosuppressor for erg25. If low heme levels permit suppression, one would predict that exogenous addition of heme would diminish the effects of slu1 or slu2 on suppression. The growth patterns in Figs. 5 and 6 demonstrate that the addition of hemin at 13–26 μg/ml to azole-containing plates reverses the growth-suppressing effects of azoles on erg25 slu1 and erg25 slu2 strains, demonstrating that hemin reverses the slu phenotype. When hemin is provided in YPD medium to erg11 erg25 slu1 (or slu2) triple mutants, growth is significantly retarded (data not shown) but not completely inhibited. The lack of absolute consistency in the ability of hemin to reverse the cosuppressor activity of slu1 and slu2 can be attributed to the sterol composition of each of the starting strains and the ability of hemin to completely reverse the low heme phenotype.
Figure 5.
Growth responses of wild-type (WA1), slu1, and four erg25 slu1 transformants grown on YPD, YPD plus ergosterol, and YPD plus azoles (1 μM clotrimazole, ketoconazole, or itraconazole) and hemin (26 μg/ml) which reverses the growth stimulatory effects of the azoles. All growth was aerobic except on ergosterol where growth was anaerobic.
Figure 6.
Growth responses of wild-type (WA1), slu2, and four erg25 slu2 transformants grown under the same conditions as in Fig. 5.
The sterol accumulation profiles shown in Table 1 indicate that cells which produce only 4,4-dimethylzymosterol do not grow. The data presented in Figs. 5 and 6 utilize erg25 slu1 and erg25 slu2, both of which accumulate 4,4-dimethylzymosterol. The addition of hemin and azoles (an erg11 simulation) would prevent further 4,4-dimethylzymosterol synthesis (azole) but not allow the cell to utilize lanosterol (hemin reversal of slu) as a sterol. Thus, growth would not be initiated and the existing 4,4-dimethylzymosterol would not be depleted. In the case of hemin reversal of the phenotype of the triple mutant, erg11 erg25 slu1 (or slu2), the starting strain makes no 4,4-dimethylzymosterol, is capable of utilizing lanosterol, and is fully viable. Hemin will adversely effect growth by eliminating the slu cosuppressor function. The accommodation for growth on lanosterol afforded by slu1 and slu2 is not immediately or completely reversed by hemin and slow growth is noted. The mechanism responsible could be related to the fact that the cells are initially able to grow with lanosterol or it could be a function of the permeability of the triple mutants to exogenous hemin (intracellular hemin concentration). In both cases, the results of hemin addition are consistent with a critical role for lowered heme concentration in the suppression of erg25.
From the data presented it is clear that yeast cannot utilize 4,4-dimethylzymosterol as a membrane sterol. To survive under conditions in which this sterol accumulates, as in an erg25 mutant, a mutation in a second required gene, ERG11, is necessary. The combination of erg25 and erg11 mutations does not result in downstream interconversions of sterols (with the exception of low levels of 24-methylenelanosterol) as is seen when erg11 is present alone (Fig. 1). The requirement for a leaky heme cosuppressor represents a novel suppression mechanism in yeast. Heme mutations resulting in a complete loss of heme could not act as suppressors in this system because such mutations themselves would result in auxotrophy not only for sterols but for unsaturated fatty acids and methionine (18, 19). Because heme mutations allow uptake of sterols aerobically, it is tempting to speculate that these leaky heme mutations (slu) allow uptake of “sparking levels” of ergosterol (27) present in the YPD growth medium. Such sparking levels of ergosterol although undetectable upon GC/MS analyses of the triple mutants may be present in YPD medium. The fact that both triple mutants respond to sparking levels of ergosterol added to CSM medium compels us to conclude that slu suppression at least in YPD medium may result from uptake of sparking levels of sterol. However, growth of erg11erg25slu2 on CSM (with Difco Noble agar) must be explained by a mechanism other than ergosterol uptake. This mechanism permits growth on lanosterol-type sterols.
The cosuppressor activity of azoles in place of erg11 in an erg25 slu1 (or slu2) double mutant presents a novel situation where an antifungal drug actually promotes rather than inhibits growth. In the event that an antifungal inhibitor of the ERG25 gene product is developed, treatment with azoles would negate the inhibition of the sterol methyl oxidase (ERG25) and result in restored growth under low heme conditions. Thus, resistance to the inhibitor would require only a leaky mutation in any one of the heme pathway genes (slu) if azole is present.
Table 1.
Sterol accumulation profiles of wild-type and ergosterol-deficient strains
| Yeast genotype | Sterols accumulated |
|---|---|
| WA1 (ERG11 ERG25 SLU1) | Ergosterol (65), zymosterol (6), fecosterol (9), episterol (9), lanosterol (5), 4,4-dimethylzymosterol (3) |
| erg25(*) | 4,4-Dimethylzymosterol (100) |
| erg11(*) | Lanosterol (86), 14-methylfecosterol (7), obtusifoliol (7) |
| erg11 erg25(*) | Lanosterol (94), 24-methylenelanosterol (6) |
| slu1 | Ergosta 5,7-dien-3β-ol (65), ergosterol (18), lanosterol (8), zymosterol (5), 14-methylfecosterol (3), fecosterol (1) |
| slu2 | Lanosterol (45), fecosterol (or ergosta-5,7-dien-3β-ol) (32), obtusifoliol (5), 14-methylfecosterol (15), ergosterol (3) |
| erg25 slu1(*) | Lanosterol (22), 4,4-dimethylzymosterol (78) |
| erg25 slu2 (*) | Lanosterol (42), 4,4-dimethylzymosterol (58) |
| erg11 erg25 slu1 | Lanosterol (92), 24-methylenelanosterol (8) |
| erg11 erg25 slu2 | Lanosterol (93), 24-methylenelanosterol (7) |
Sterols accumulating in each strain are listed with percentage of each sterol in parentheses. Only the wild-type slu1, slu2, and erg11 erg25 slu1 (slu2) strains were grown aerobically in YPD cultures. Sterol requiring strains (*) were grown in anaerobic jars on solid medium containing exogenous sterol.
Acknowledgments
This work was supported by National Institutes of Health Grant 1R01 AI38598-01 to M.B. and a Johnson & Johnson Focused Giving Award to M.B. and N.D.L.
ABBREVIATIONS
- CSM
complete synthetic medium
- GC/MS
gas chromatography/mass spectrometry
- YPD
yeast extract peptone glucose
- YPE
yeast extract peptone ethanol
- slu
suppressor of lanosterol utilization
References
- 1.Lees N D, Bard M, Kirsch D R. In: Biochemistry and Function of Sterols. Parish E J, Nes W D, editors; Parish E J, Nes W D, editors. Boca Raton, FL: CRC; 1997. pp. 85–99. [Google Scholar]
- 2.Lees N D, Skaggs B, Kirsch D R, Bard M. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- 3.Vanden Bossche H, Willemsens G, Marichal P. Crit Rev Microbiol. 1987;15:57–72. doi: 10.3109/10408418709104448. [DOI] [PubMed] [Google Scholar]
- 4.Georgopapadakou N H, Walsh T J. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 5.Hitchcock C A. Biochem Soc Trans. 1993;21:1039–1047. doi: 10.1042/bst0211039. [DOI] [PubMed] [Google Scholar]
- 6.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. FEMS Microbiol Lett. 1995;131:337–341. doi: 10.1111/j.1574-6968.1995.tb07797.x. [DOI] [PubMed] [Google Scholar]
- 7.Bard M, Bruner D A, Pierson C A, Lees N D, Biermann B, Frye L, Koegel C, Barbuch R. Proc Natl Acad Sci USA. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trocha P J, Jasne S J, Sprinson D B. Biochemistry. 1977;16:4721–4726. doi: 10.1021/bi00640a029. [DOI] [PubMed] [Google Scholar]
- 9.Kalb V, Woods C W, Turi T G, Dey C R, Sutter T R, Loper J C. DNA. 1987;6:529–537. doi: 10.1089/dna.1987.6.529. [DOI] [PubMed] [Google Scholar]
- 10.Ladeveze V, Marcireau C, Delourme D, Karst F. Lipids. 1993;28:907–912. doi: 10.1007/BF02537499. [DOI] [PubMed] [Google Scholar]
- 11.Taylor F R, Rodriguez F J, Parks L W. J Bacteriol. 1983;155:64–68. doi: 10.1128/jb.155.1.64-68.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz R T, Parks L W. DNA Cell Biol. 1992;11:685–692. doi: 10.1089/dna.1992.11.685. [DOI] [PubMed] [Google Scholar]
- 14.Berben G, Dumont J, Gilliquet P-A, Hilger F. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 15.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 16.Sherman F, Fink G R, Hicks J. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab Press; 1986. [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning. Plainview, NY: Cold Spring Harbor Lab Press; 1982. [Google Scholar]
- 18.Bard M, Woods R A, Haslam J M. Biochem Biophys Res Commun. 1974;56:324–330. doi: 10.1016/0006-291x(74)90845-6. [DOI] [PubMed] [Google Scholar]
- 19.Gollub E G, Liu K, Dayan J, Adlersberg M, Sprinson D B. J Biol Chem. 1977;252:2846–2854. [PubMed] [Google Scholar]
- 20.Lewis T L, Keesler G A, Fenner G P, Parks L W. Yeast. 1988;4:93–106. doi: 10.1002/yea.320040203. [DOI] [PubMed] [Google Scholar]
- 21.Bard M, Lees N D, Turi T, Craft D, Cofrin L, Barbuch R, Koegel C, Loper J C. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 22.Arthington-Skaggs B A, Crowell D N, Yang H, Sturley S L, Bard M. FEBS Lett. 1996;392:161–165. doi: 10.1016/0014-5793(96)00807-1. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nes W D, Janssen G G, Crumley F G, Kalinowska M, Akihisa T. Arch Biochem Biophys. 1993;300:724–733. doi: 10.1006/abbi.1993.1100. [DOI] [PubMed] [Google Scholar]
- 25.Buttke T M, Bloch K. Biochem Biophys Res Commun. 1980;92:229–236. doi: 10.1016/0006-291x(80)91543-0. [DOI] [PubMed] [Google Scholar]
- 26.Buttke T M, Van Cleave S. Biochem Biophys Res Commun. 1994;200:206–212. doi: 10.1006/bbrc.1994.1435. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez R J, Low C, Bottema C D K, Parks L W. Biochim Biophys Acta. 1985;837:336–343. doi: 10.1016/0005-2760(85)90057-8. [DOI] [PubMed] [Google Scholar]