Abstract
Subcellular localization directed by specific A kinase anchoring proteins (AKAPs) is a mechanism for compartmentalization of cAMP-dependent protein kinase (PKA). Using a two-hybrid screen, a novel AKAP was isolated. Because it interacts with both the type I and type II regulatory subunits, it was defined as a dual specific AKAP or D-AKAP1. Here we report the cloning and characterization of another novel cDNA isolated from that screen. This new member of the D-AKAP family, D-AKAP2, also binds both types of regulatory subunits. A message of 5 kb pairs was detected for D-AKAP2 in all embryonic stages and in all adult tissues tested. In brain, skeletal muscle, kidney, and testis, a 10-kb mRNA was identified. In testis, several small mRNAs were observed. Therefore, D-AKAP2 represents a novel family of proteins. cDNA cloning from a mouse testis library identified the full length D-AKAP2. It is composed of 372 amino acids which includes the R binding fragment, residues 333–372, at its C-terminus. Based on coprecipitation assays, the R binding domain interacts with the N-terminal dimerization domain of RIα and RIIα. A putative RGS domain was identified near the N-terminal region of D-AKAP2. The presence of this domain raises the intriguing possibility that D-AKAP2 may interact with a Gα protein thus providing a link between the signaling machinery at the plasma membrane and the downstream kinase.
Protein phosphorylation is one of the most important mechanisms for enzyme regulation and signal transduction in eukaryotic cells. cAMP-dependent protein kinase (PKA), one of the first protein kinases discovered, mediates a variety of hormonal and neurotransmitter responses by phosphorylating serine and threonine residues on various substrate proteins. The observation of preferential phosphorylation of specific target substrates by PKA, despite its broad specificity, may be explained by the discovery of the anchoring proteins of PKA. These A kinase anchoring proteins (AKAPs) function as targeting units and tether the kinase to specific subcellular localizations. Compartmentalization of PKA therefore serves as a potential regulatory mechanism that increase the selectivity and intensity of a cAMP-mediated hormonal response.
Recently, it was shown that anchoring proteins may also act as adapters for assembling multiprotein complexes. For example, Scott and coworkers (1, 2) showed that AKAP79, in addition to binding tightly to PKA, also interacts with calcineurin and protein kinase C. Therefore, the targeting of AKAP79 to neuronal postsynaptic densities brings enzymes with opposite catalytic activities together in a single transduction complex. This adds another level of intracellular organization for PKA and also facilitates the diversity of the cAMP-mediated signal transduction pathways (3).
In the absence of its activating ligand, cAMP, PKA exists as an inactive holoenzyme containing two regulatory (R) and two catalytic subunits. The two classes of R subunits, RI and RII, based on their elution from the DEAE cellulose, define the two types of holoenzymes. Because a significant proportion of the type II holoenzyme associates with the particulate fraction of cell homogenates, most previous studies focused on identification of AKAPs that anchor the kinase through RII (4, 5). Recently, using a yeast two-hybrid screen, a new member of the anchoring protein family, D-AKAP1, was identified that binds both RI and RII. This raises the possibility that like RII, RI may also serve to compartmentalize PKA. There are, in fact, indications that RI is also compartmentalized in some case in a dynamic way. For example, RI subunits in human erythrocytes are tightly bound to the plasma membrane (6). RIα is also enriched in the neuromuscular junctions (7). Type I holoenzyme is also depleted from the cytoplasm and accumulates at the “cap” site when lymphocytes are stimulated with anti-CD3 antibodies (8). RI was also found to associate with activated B cell receptors (9).
Here we report the identification and characterization of another potential A kinase anchoring protein, D-AKAP2. In addition to an R binding domain, D-AKAP2 contains a putative RGS domain characteristic of proteins that interact with the Gα subunits and possess GTPase activating protein-like activity (10–12). The presence of these two motifs in a single molecule provides a previously unrecognized direct link between two major components of the cAMP signaling pathway.
EXPERIMENTAL PROCEDURES
Materials.
All vectors for the yeast two-hybrid system were from S. Hollenberg (Vollum Institute, Oregon). The following reagents were purchased as indicated: Mouse testis cDNA library (CLONTECH), Mouse Multiple Tissue Northern Blot and Mouse Embryonic Northern Blot (CLONTECH), Ready-to-Go DNA labeling kit (Pharmacia), Genius labeling system (Boehringer Mannheim), ATP, phenylmethylsulfonyl fluoride, benzamidine, Triton X-100 and glutathione S-transferase (GST) agarose resin (Sigma), standard saline citrate (SSC) buffer (5 Prime–3 Prime Inc.), nickel-nitrilotriacetic acid resin (Qiagen, Chatsworth, CA), 5-bromo-4-chloro-3-indoyl β-d-galactoside and enzymes used for DNA manipulations (Life Technologies, Grand Island, NY), DNA sequencing kit (United States Biochemical). All oligonucleotides were synthesized at the Peptide and Oligonucleotide Facility at the University of California, San Diego.
Two-Hybrid Screen.
A yeast two-hybrid screen was used to isolate proteins that associate with the Ret/ptc2 oncoprotein, where the C-terminal domain of the Ret receptor tyrosine kinase, is fused to the N terminus of the RIα subunit of PKA (13). Using Ret/ptc2 to screen a mouse embryonic cDNA library, several interacting clones were isolated. Eight of these cDNA clones coded for three novel protein fragments, designated RPP7, RPP8, and RPP9, that associated specifically with the RI portion of Ret/ptc2. RPP7 was later identified to be part of a novel anchoring protein of PKA, D-AKAP1 (14). Here we report the cloning of full-length RPP8.
Expression and Purification of Recombinant R Binding Proteins.
To determine whether the protein coded by RPP8 binds to the RI-subunit in vitro, the RPP8 cDNA was subcloned from the pVP16 plasmid into pGEX-KG to make a GST-fusion protein. This RPP8 fusion protein, designated GST-RPP8, was expressed in Escherichia coli BL21(DE3) at 37°C and purified to near homogeneity using glutathione resin as described earlier (15). The BL21(DE3) cell strain was a gift from Bill Studier (Brookhaven National Laboratories, Brookhaven, NY).
Construction of Mutants of R Subunits.
Site-directed mutations of the RIα-subunit were generated according to Kunkel et al. (16). The RIα mutant with Cys-16 changed to His was designated RI(C16H), and mutant with Cys-37 changed to His or Ala were designated RI(C37H) and RI(C37A), respectively. A double mutant with both Cys-16 and Cys-37 changed to Ala was designated as RI(C16A-C37A). In addition, Val-20 and Ile-25 were mutated to Ala on RIα, respectively, to make RI(V20A) and RI(I25A).
Expression and Purification of R Subunits.
RI and the various RI mutants were either expressed in BL21(DE3) cells and purified on a DE52 ion exchange column (17) or by cAMP resin (18). His6RII, His6RI(Δ63–379), and RII(Δ46–400) were purified as described (14).
In Vitro Binding Assay.
Bacterial cell lysates containing GST-RPP8 were incubated with glutathione resin for 2 hr at 4°C in PBS (10 mM potassium phosphate, 150 mM NaCl, pH 7.4) with 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 5 mM benzamidine and 5 mM 2-mercaptoethanol, and then washed extensively with the same buffer. Wild type and mutants (100–200 μg) of RI and/or RII were added to the resin and incubated for 2 hr at 4°C. After washing the resin extensively with PBS, proteins associated with the GST-RPP8 were eluted by boiling in SDS gel-loading buffer and analyzed by SDS/PAGE. All electrophoresis was performed using Mini-Protean II electrophoresis system (Bio-Rad). SDS/PAGE reagents were prepared according to Laemmli (19). Proteins were visualized by Coomasie blue staining.
Northern Blot Analysis.
Blots containing 2 μg immobilized samples of mRNAs from selected adult tissues or total mRNA at different embryonic stages were probed with 32P-radiolabeled RPP8 cDNA. Nitrocellulose filters were prehybridized in 5× SSC (750 mM sodium chloride, 75 mM sodium citrate, pH 7.0), 5× Denhardt’s reagent (0.1% Ficoll/0.1% polyvinylpyrrolidone/0.1% acetylated bovine serum albumin), 0.5% SDS, and 50% formamide for 6 hr at 42°C, and then hybridized to 1.5 × 106 cpm/ml of denatured radiolabeled cDNA probe in the same buffer. Hybridization was performed at 42°C for 16 hr and nonhybridized probe was removed with 0.1× SSC, 0.1% SDS at 68°C. Hybridizing mRNA signals were detected by autoradiography.
Screening of cDNA Libraries.
An adult mouse testis cDNA library in λgt10 was screened with 32P-labeled RPP8 cDNA. The cDNA fragment RPP8 was excised from the two-hybrid library plasmid pVP16 using the NotI restriction endonuclease, and purified on agarose gel. This purified cDNA was then labeled with [32P]αdCTP using random-prime labeling. Approximately 1.6 million plaques were screened, and positive clones were plaque-purified. Positive phage cDNAs were then isolated and digested with EcoRI, then the inserts were subcloned into the EcoRI site of pBluescriptII KS(+). All DNA sequences were determined by the dideoxynucleotide chain termination procedure (20). All sequences were analyzed using pcgene software (IntelliGenetics) and the blast program provided by the National Center for Biotechnology Information server at the National Library of Medicine, National Institute of Health.
In Situ Hybridization.
RNA probes of D-AKAP2 were derived from cDNA sequence 1–1469. RIα probes were derived from residues 18 to 169. These cDNA fragments were first subcloned into pBluescriptII KS(+), and digoxigenin-labeled riboprobes were transcribed in the sense and antisense orientations following template linearization. Probe synthesis employed the Genius labeling system, and hybridization was carried out on 20-μm cryostat sections of the testis of adult BALB/c mice as previously described by Chun and coworkers (21, 22). Hybridized probe was visualized by alkaline phosphatase histochemistry.
RESULTS
Identification of a Novel RI Binding Protein.
Using Ret/ptc2 to screen a mouse embryonic library, three novel cDNAs were isolated that coded for proteins designated RPP7, RPP8, and RPP9, which interacted specifically with the RIα portion of the bait. RPP7 was further characterized and identified as a fragment of a novel PKA anchoring protein, D-AKAP1 (14). Here we describe the characterization of RPP8 and the cloning of the full length cDNA from which it was derived. RPP8 coded for a 40-amino acid protein fragment.
To determine whether RPP8 binds RIα in vitro, the fragment corresponding to RPP8 was expressed as a fusion protein to GST in E. coli from a pGEX vector encoding GST fused to the RPP8 cDNA. GST-RPP8 was then expressed and tested for its ability to bind RIα in an affinity precipitation assay as described earlier (14). Cell lysates containing either GST or GST-RPP8 were incubated with glutathione resin. Bacterially expressed RIα-subunit was then added. Proteins associated with the resin after stringent washing were analyzed by SDS/PAGE. As seen in Fig. 1, this construct codes for a stable protein that can specifically pull down near stoichiometric amounts of the RIα-subunit. GST alone does not interact with RIα. These results confirmed the results of the two-hybrid screen, and established furthermore that no other factors are required for the interaction between RIα and RPP8 in vitro and in vivo.
Figure 1.
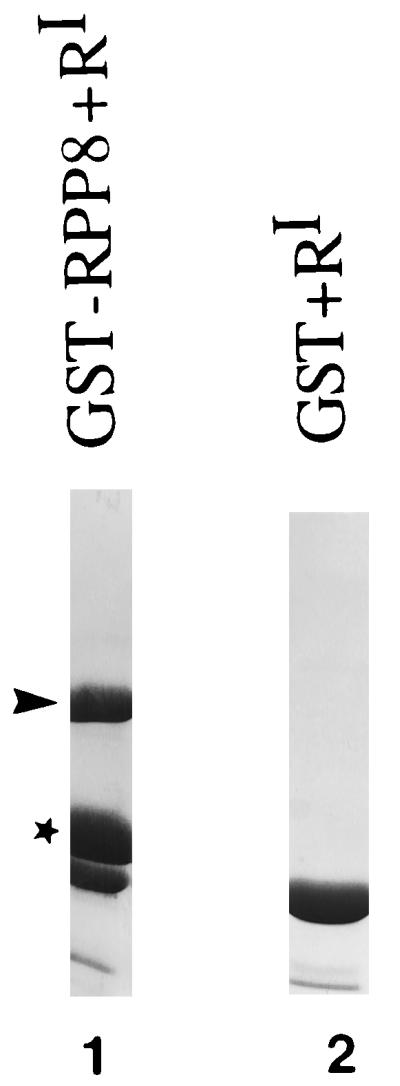
GST-RPP8 binds to RIα. Lane 1 shows purified GST-RPP8. GST-RPP8 or GST were immobilized on glutathione resin as described in Experimental Procedures. After removal of nonspecific proteins, 200 μg of RIα-subunit were added to the resin and incubated for 2 hr at 4°C. After washing the resin extensively, proteins associated with either GST-RPP7 or GST were eluted from the glutathione resin by boiling in SDS gel-loading buffer and analyzed on SDS/PAGE. As shown in lanes 1 and 2 RIα only associates with GST-RPP8 but not GST. ∗, GST-RPP8; ➤, RIα.
Specificity of RPP8.
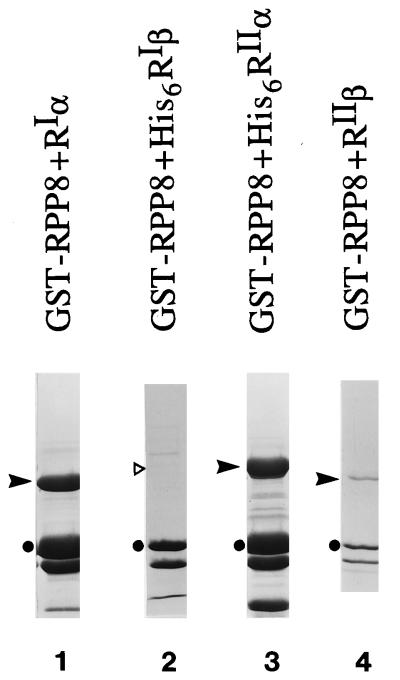
All AKAPs previously identified bind only to RII with the exception of D-AKAP1 that was isolated from the same screen and was shown to be a D-AKAP based on its ability to bind both RIα and RIIα. Therefore, the binding specificity of RPP8 toward the various isoforms of the R subunits was examined. As seen in Fig. 2, in addition to RIα, RPP8 also interacted with His6RIIα and RIIβ in the assay. However, GST-RPP8 was not capable of binding His6RIβ. Thus, RPP8 also belongs to the AKAP family that binds both RI and RII with high affinity. For this reason, we designate RPP8 as an active fragment of D-AKAP2.
Figure 2.
Specificity of GST-RPP8. Bacterial cell lysates containing GST-RPP8 were incubated with glutathione agarose resin for 2 hr at 4°C in binding buffer as described. One to two hundred micrograms of different forms of RI or RII were then added to the resin and incubated for 2 hr at 4°C. After washing the resin extensively, proteins associated with the GST-RPP8 were eluted from the GST resin by boiling in SDS gel-loading buffer and analyzed on SDS/PAGE. ➤, The different R subunits in each sample. In lane 2, ▹ indicates where His6RIβ should run on this gel. Lanes: 1, RIα; 2, His6RIβ; 3, His6RIIα; 4, RIIβ.
Localization of D-AKAP2 Binding Site.
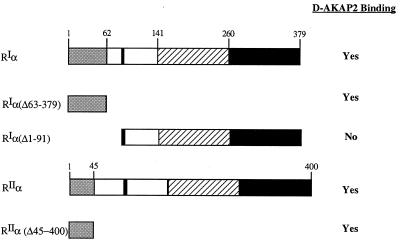
To localize more precisely the RPP8 binding site on RIα and RIIα, a series of deletion mutants, summarized in Fig. 3, were used. As seen in Fig. 3, GST-RPP8 associated with His6RIα(Δ63–379), which contains only the first 62 residues of RI. In contrast, RIα(Δ1–91), which lacks the N-terminal 91 residues of RI, was not precipitated with GST-RPP8. All AKAPs for RII, including D-AKAP1, bound the N terminus of RIIα (13, 23, 24). To determine whether RPP8 also bound RIIα through the N terminus, several deletion mutants of RIIα were tested for their ability to bind GST-RPP8. As shown in Fig. 3, GST-RPP8 interacted with RIIα(Δ46–400), a construct containing only the N-terminal 45 residues of RIIα. GST alone does not interact with any of the R subunits we have tested. Thus, the N terminus of RIα or RIIα is sufficient for the interaction with D-AKAP2.
Figure 3.
RPP8 interacts with the N terminus of the RI-subunit. Schematic diagram indicating the binding capacity of various truncation mutants of RI to GST-RPP8. The N-terminal dimerization domain, inhibitory site, and two cAMP-binding domains are indicated on the wild-type RIα.
Specific Residues on RIα That Are Necessary to Interact with D-AKAP2.
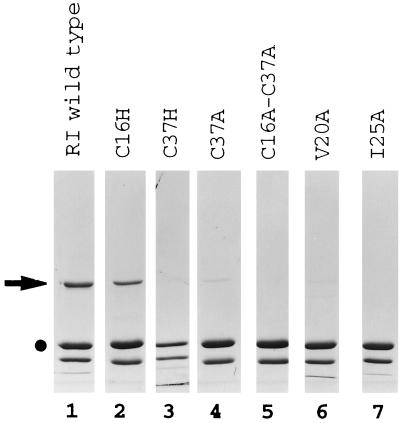
To determine specific residues on RIα that are necessary to interact with D-AKAP2, a series of site-directed mutants of RIα were also tested to identify specific amino acids on the N-terminal dimerization domain that are essential for interaction with RIα and D-AKAP2. As seen in Fig. 4, a mutant of RIα where Cys-16 was mutated to His, RI(C16H), interacted with GST-RPP8 like wild-type RIα in this assay. Mutants of Cys-37 to Ala, RI(C37A), and Val-20 to Ala, RI(V20A), had dramatic decreased affinity for GST-RPP8. Mutation of Cys-37 to His, RI(C37H), Ile-25 to Ala, RI(I25A), or both Cys-16 and Cys-37 to Ala, RI(C16A-C37A), completely lost the ability to interact with GST-RPP8 although all of these proteins were functional dimers (unpublished data).
Figure 4.
Specific residues on RIα that are necessary to interact with D-AKAP2. Equal amounts of the various RIα-subunit mutants were tested in the binding assay. •, GST-RPP8; ➤, Various RIα mutants.
Tissue Distribution of D-AKAP2 mRNA.
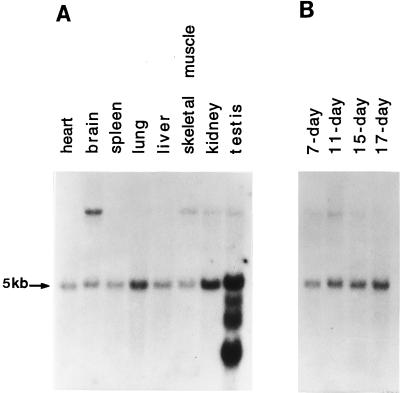
To investigate the tissue and developmental expression patterns of D-AKAP2, Northern blots containing 2 μg of poly(A+)RNA from different adult mouse tissues or different embryonic stages were probed with 32P-labeled RPP8 cDNA. As shown in Fig. 5A, a 5kb mRNA was detected in all tissues with equal intensity. A weak signal at 10 kb was detected in brain, skeletal muscle, kidney, and testis. In addition, mRNA of 3.4 kb, 2.8 kb, and 1.8 kb were detected only in testis. The 5-kb transcript was detected in all embryonic stages with comparable intensity (Fig. 5B), indicating that the mRNA expression level in the embryo was the same throughout different developmental stages. The other mRNAs were not detected in the embryonic samples.
Figure 5.
Size and abundance of D-AKAP2 mRNA in mouse tissues and various embryonic stages. Blots containing 2 μg of mRNAs from selected adult tissues (A) or total mRNA at different embryonic stages (B) were probed with 32P-radiolabeled RPP8 cDNA for 16 hr at 42°C in 5× SSC, 5× Denhardt’s reagent, 0.5% SDS, and 50% formamide. After washing with 0.1× SSC and 0.1% SDS at 68°C, hybridizing signals were detected by autoradiography.
Cloning of D-AKAP2.
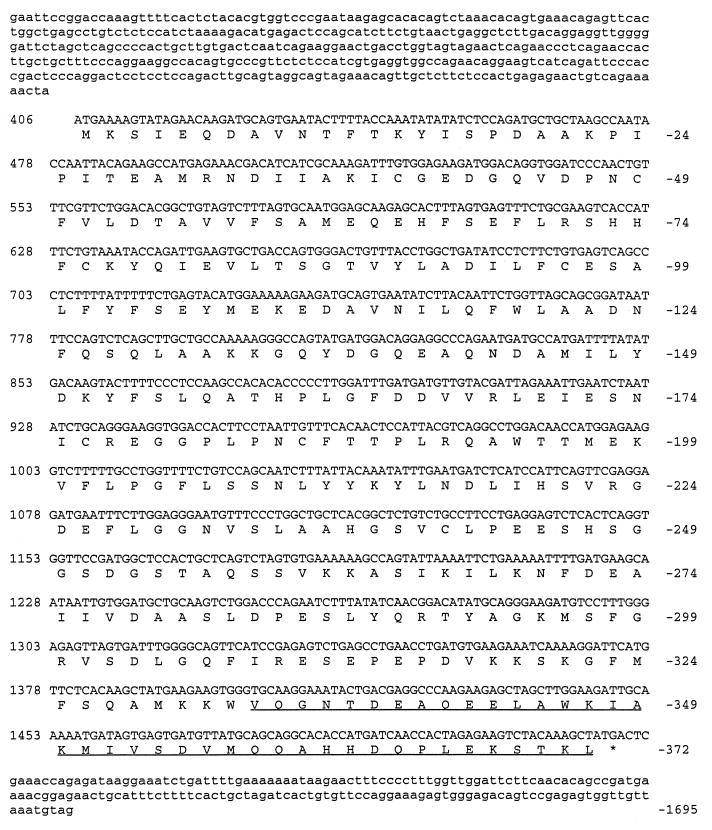
To obtain a full-length clone of D-AKAP2, a mouse testis cDNA library was screened using RPP8 cDNA as a probe. After radiolabeling of the RPP8 probe, positive clones were isolated from ≈1.6 million recombinants. After sequence analysis, one cDNA clone was identified that contained a full-length cDNA of D-AKAP2. The DNA sequence and the deduced amino acid are shown in Fig. 6. This cDNA fragment is likely to represent a full-length clone of D-AKAP2 because it contained a proper initiation sequence and stop codons followed by the polyA tail (25). Stop codons were also identified in frame upstream from the initiation site, precluding any possible use of another ATG upstream.
Figure 6.
cDNA and deduced amino acid sequences for D-AKAP2 core. Numbers on the left are for cDNA sequence, and numbers on the right indicate amino acid position. RPP8 sequence is underlined.
D-AKAP2 contains 372 amino acids. The D-AKAP2 protein sequence was compared with the GenBank database and there was no overall homology to known protein sequences. Surprisingly, however, residues 70–215 showed homology to the RGS domain found in RGS proteins.
In Situ Hybridization.
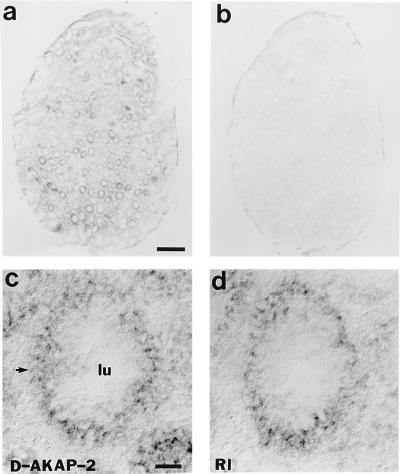
To determine the expression pattern of D-AKAP2 in comparison to RIα, in situ hybridization was performed using probes for both D-AKAP2 and RIα in mouse testicle sections. As shown in Fig. 7, D-AKAP2 and RIα were expressed in many of the same regions in testis.
Figure 7.
In situ hybridization pattern of D-AKAP2 that shows overlapping regions for RIα. RNA probes of D-AKAP2 were derived from the cDNA sequence of 1–1469. RIα probes were derived from cDNA coding for residues 18–169. Hybridization was carried out on adjacent 20-μm cryostat sections of adult mouse testis and signals were visualized by alkaline phosphatase histochemistry. (a) D-AKAP1 mRNA is present in various regions of testis. (b) Signal of the sense strand as negative control. (c) High magnification of signal for D-AKAP2 on one of the testicular tube cross sections. (d) High magnification of signal for RIα on one of the testicular tube cross sections. (Bar = 100 μm.)
DISCUSSION
D-AKAP2, named for its potential dual specificity, was first identified as a fragment from a yeast two-hybrid screen based on specific interaction with the RI portion of Ret/ptc2. In that screen, another anchoring protein, D-AKAP1, was also identified and was found to bind both RI and RII. As demonstrated in an affinity precipitation assay, this fragment of D-AKAP2, RPP8, includes most, if not all, of the RI/RII binding domain. Secondary structure predictions indicate that RPP8 has an amphipathic α-helix at its N terminus, and this is consistent with the proposed model for other AKAPs where predicted amphipathic helices are hallmarks for R binding domains (24, 26–28). RPP8 was shown to bind RIα, RIIα, and RIIβ, but not RIβ. This isoform selectivity of AKAPs is consistent with results published earlier. Leiser et al. (5) showed that RIIα had a 6-fold preference for microtubule-associated protein-2 (MAP2) over RIIβ, whereas RIIβ had a two fold preference for AKAP75 over RIIα. Follicle stimulating hormone treatment of rat granulosa cells also induces an 80-kDa RIIα selective AKAP (29).
Using this functional R binding fragment, RPP8, the interaction regions on both RIα and RIIα were localized to their N termini. This N-terminal region corresponds to the first 62 amino acids in RI and the first 45 amino acids in RIIα. The actual microenvironment for D-AKAP2 with respect to RIα and/or RIIα is not understood at this point. Further characterization will be required to predict the interaction patterns between D-AKAP2 and both types of R subunits in vivo.
To further characterize the novel interaction between D-AKAP2 and RIα, various mutations were made in the N-terminal region of RIα and tested for their effects on D-AKAP2 binding. Mutating Cys-16 to His had no effect, however, changing Cys-37 to His totally abolished binding. Therefore, although Cys-16 is disulfide bonded to Cys-37 in the RIα dimer, they exhibited different roles in contributing to the binding interface for D-AKAP2. Mutation at Val-20 had dramatically decreased binding, whereas mutation at Ile-25 totally abolished binding, indicating that these residues are important, either directly or indirectly, for binding D-AKAP2. Mutation of Cys-37 to Ala had dramatically decreased binding and a double mutant with both cysteines changed to alanines totally abolished binding to GST-RPP8. Whether the better tolerance of an Ala than a His at position 37 is due to disruption of a hydrophobic interaction surface or due to the disruption of a specific conformation is still unclear.
A complicated mRNA expression pattern was detected for D-AKAP2, therefore it could represent a novel family of proteins. In addition to the 5-kb mRNA that was expressed in all tissues tested, and also throughout all embryonic stages, brain, skeletal muscle, and kidney had an additional 10-kb message. Moreover, in testis, there are total of five mRNA species. The expression of D-AKAP2 in testis was further confirmed by in situ hybridization. D-AKAP2 and RIα appeared to be expressed in many of the same regions in adult mouse testicle sections.
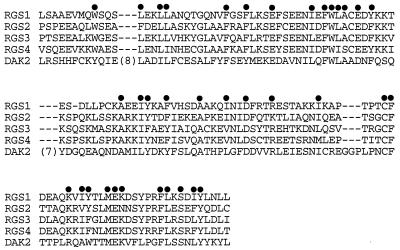
Sequence comparison revealed that D-AKAP2 contained a potential RGS domain in its N-terminal region. RGS proteins are a new family of “regulator of G-protein signaling” proteins, which down-regulate signaling by heterotrimeric G proteins. The first biochemical studies of RGS proteins have shown that they accelerate the GTPase activities of G-protein α subunits, thus driving G proteins into their inactive GDP-bound forms. These RGS domains therefore function as GTPase-activating proteins of the heterotrimeric G proteins (10–12). Alignment of this region of D-AKAP2 to some of the known RGS domains from RGS proteins showed striking similarity (Fig. 8). All conserved hydrophobic residues proposed to be important for maintaining the folding of the domain were highly conserved. Surprisingly, not all of the charged residues conserved in RGS proteins reported so far were conserved in D-AKAP2. In contrast to the hydrophobic residues, these charged amino acids lie on the surface. The lack of conservation of these charge residues suggests that if D-AKAP2 is a functional RGS, it may not interact with Giα as most of the known RGSs do, but with other Gα isoforms.
Figure 8.
D-AKAP2 contains a potential RGS domain. Sequence alignment of the putative RGS domain in D-AKAP2 with other RGS proteins. •, Conserved residues.
We report here the identification of another novel anchoring protein family of PKA, D-AKAP2, that binds to both RI and RII. In addition to an R binding domain, D-AKAP2 contains a potential RGS domain that was recently shown to interact with Gα subunits and possess GAP-like activity. The presence of these two motifs in a single molecule provides a previously unrecognized direct link between two major components of the cAMP signaling pathway.
Acknowledgments
We thank Dr. Taroh Iiri at the University of California, San Francisco, for pointing out a sequencing error in our original cloning. This research was supported in part by American Cancer Society Grant BE-48L (to S.S.T.) and National Institutes of Health Grant R29MH51699 (to J.C.). S.S.T. is a Howard Hughes Medical Investigator. L.J.-s.H. was supported by University of California, San Diego, Cell Molecular and Genetics Training Grant 2T32GM07240–21A1. K.D. was supported by the Markey Charitable Trust as a Fellow and is currently supported by National Institutes of Health Training Grant National Cancer Institute T32 CA09523. J.A.W. was supported by a National Science Foundation Graduate Fellowship.
ABBREVIATIONS
- PKA
cAMP-dependent protein kinase
- AKAP
A kinase anchoring protein
- R
PKA regulatory subunit
- RI and RII
type I and II regulatory subunit
- GST
glutathione S transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (assession no. AF021833).
References
- 1.Coghlan V, Perrino B A, Howard M, Langeberg L K, Hicks J B, Gallatin W H, Scott J D. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 2.Klauck T M, Faux M C, Labudda K, Langeberg L K, Jaken S, Scott J D. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 3.Faux M C, Scott J D. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- 4.Scott J D, McCartney S. Mol Endocrinol. 1994;8:5–11. doi: 10.1210/mend.8.1.8152430. [DOI] [PubMed] [Google Scholar]
- 5.Leiser M, Rubin C S, Erlichman J. J Biol Chem. 1986;261:1904–1908. [PubMed] [Google Scholar]
- 6.Rubin C S, Erlichman J, Rosen O M. J Biol Chem. 1972;247:6135–6139. [PubMed] [Google Scholar]
- 7.Imaizumi-Scherrer T, Faust D M, Benichou J-C, Weiss M C. J Cell Biol. 1996;134:1241–1254. doi: 10.1083/jcb.134.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skalhegg B S, Tasken K, Hansson V, Huitfeldt H S, Jahnsen T, Lea T. Science. 1994;263:84–87. doi: 10.1126/science.8272870. [DOI] [PubMed] [Google Scholar]
- 9.Levy F O, Rasmussen A M, Tasken K, Skalhegg B S, Huitfeldt H S, Smeland E B, Hansson V. Eur J Immunol. 1996;26:1290–1296. doi: 10.1002/eji.1830260617. [DOI] [PubMed] [Google Scholar]
- 10.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. Nature (London) 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 11.Berman D M, Wilkie T M, Gilman A G. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 12.Loelle M R. Curr Opin Cell Biol. 1997;9:143–147. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 13.Luo Z, Shafit-Zagardo B, Erlichman J. J Biol Chem. 1990;265:21804–21810. [PubMed] [Google Scholar]
- 14.Huang L J, Durick K, Weiner J A, Chun J, Taylor S S. J Biol Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 15.Adams J A, McGlone M L, Gibson R, Taylor S S. Biochemistry. 1995;34:2447–2454. doi: 10.1021/bi00008a007. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraswat L D, Filutowics M, Taylor S. Methods Enzymol. 1988;159:325–336. doi: 10.1016/0076-6879(88)59033-x. [DOI] [PubMed] [Google Scholar]
- 18.Ringheim G E, Taylor S S. J Biol Chem. 1990;265:4800–4808. [PubMed] [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci, USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chun J M, Schatz D G, Oettinger M A, Jaenisch R, Baltimore D. Cell. 1991;64:189–200. doi: 10.1016/0092-8674(91)90220-s. [DOI] [PubMed] [Google Scholar]
- 22.Weiner J A, Chun J. J Comp Neurol. 1997;381:130–142. doi: 10.1002/(sici)1096-9861(19970505)381:2<130::aid-cne2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Scott J D, Stofko R E, McDonald J R, Comer J D, Vitalis E A, Mangili J A. J Biol Chem. 1990;265:21561–21566. [PubMed] [Google Scholar]
- 24.Carr D W, Stofko-Hahn R E, Fraser I D, Bishop S M, Acott T S, Brennan R G, Scott J D. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 25.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carr D W, Stofko-Hahn R E, Fraser I D C, Cone R D, Scott J D. J Biol Chem. 1992;267:16816–16823. [PubMed] [Google Scholar]
- 27.Carr D W, Hausken Z E, D C F, I, Stofko-Hahn R E, Scott J D. J Biol Chem. 1992;267:13376–13382. [PubMed] [Google Scholar]
- 28.Coghlan V M, Langeberg L K, Fernandez A, Lamb N J C, Scott J D. J Biol Chem. 1994;269:7658–7665. [PubMed] [Google Scholar]
- 29.Carr D W, DeManno D A, Atwood A, Hunzicker-Dunn M, Scott J D. J Biol Chem. 1993;268:20729–20732. [PubMed] [Google Scholar]