Abstract
Biological sensing of small molecules such as NO, O2, and CO is an important area of research; however, little is know about how CO is sensed biologically. The photosynthetic bacterium Rhodospirillum rubrum responds to CO by activating transcription of two operons that encode a CO-oxidizing system. A protein, CooA, has been identified as necessary for this response. CooA is a member of a family of transcriptional regulators similar to the cAMP receptor protein and fumavate nitrate reduction from Escherichia coli. In this study we report the purification of wild-type CooA from its native organism, R. rubrum, to greater than 95% purity. The purified protein is active in sequence-specific DNA binding in the presence of CO, but not in the absence of CO. Gel filtration experiments reveal the protein to be a dimer in the absence of CO. Purified CooA contains 1.6 mol heme per mol of dimer. Upon interacting with CO, the electronic spectrum of CooA is perturbed, indicating the direct binding of CO to the heme of CooA. A hypothesis for the mechanism of the protein’s response to CO is proposed.
The sensing of small molecules such as NO, CO, and O2 is a highly active area of research in biology. Although NO-sensing (soluble guanylyl cyclase) (1, 2) and O2-sensing (FixL) (3, 4) proteins have been studied, little data on CO-sensing proteins exists. The CO binding and CO inhibition of metalloproteins such as cytochromes has been extensively researched, but little is known of regulatory factors that have a clear biological role in the recognition of CO. In this paper, we report the isolation and characterization of a bacterial protein that binds CO and elicits a physiologically important response upon activation with CO.
The photosynthetic bacterium Rhodospirillum rubrum has the ability to respond to CO, and the CooA protein is required for this response. R. rubrum responds to CO by inducing a multicomponent CO-oxidation system that evolves H2 from CO, producing CO2 in the process (5). Two linked operons, cooMKLXUH and cooFSCTJ, encode these proteins (6–10). The biological role of the system is not completely clear; however, R. rubrum can grow on CO as a sole energy source (11). The system also may provide protection for certain CO-sensitive metabolic activities, such as nitrogen fixation. Mutations in cooA, which lies 3′ to cooFSCTJ, render the organism unable to respond to CO (8).
CooA is a member of the cAMP receptor protein (CRP)/fumavate nitrate reduction (FNR) family of single component regulatory proteins. The dimeric CRP protein of Escherichia coli binds its effector, cAMP, and responds by binding to specific DNA sequences (12). Similarly, the FNR protein of E. coli responds to changes in O2 levels and binds to DNA sites similar to CRP sites (13). FNR is dimeric when isolated anoxically and monomeric when exposed to O2, suggesting that its DNA-binding activity is modulated by this change in oligomeric state (14). Relative to CRP and CooA, FNR bears a N-terminal extension (8) that is believed to bind an Fe-S center (13, 14).
Whereas the putative effector-recognition region of CooA differs significantly from its homologs FNR and CRP (8), the DNA-binding region is similar and binding sites for CooA are highly reminiscent of CRP and FNR sites; one such site exists 5′ of each of the two CO-regulated operons (10, 15). In early experiments with crude extracts of a CooA-overexpressing strain, CO-dependent DNase protection of these sites was observed, suggesting that CooA was directly involved in the response of the organism to CO (15); however, it was not known if CooA was sufficient or simply necessary for this response, because other factors may be involved in the process.
While the current study was in progress, Aono et al. (19) reported the purification of R. rubrum CooA expressed in E. coli, finding that the protein incorporated heme and that CooA bound CO through its heme. This indicated the possibility that CO was the CooA effector, as suggested previously (8, 15). However, because a diverse array of hemoproteins that do not have a physiological role in interacting with CO will bind CO with high affinity, it was of interest to establish the biological relevance of the CO-CooA interaction. The demonstration of CO-dependent DNA binding would indicate physiological importance of CO as a CooA effector.
In this study, we have purified CooA from its native organism, R. rubrum. We demonstrate that the purified protein upon exposure to CO binds to the promoter region of cooF. The protein is a dimer in the presence and absence of its effector, similar to the homologous CRP protein. We find that CooA from R. rubrum uses a heme moiety to bind CO directly. The heme spectral characteristics are similar to cytochromes b and suggest that the heme in CooA exists in the low-spin state in the ferric (oxidized), ferrous (reduced), and ferrous-CO forms.
MATERIALS AND METHODS
Production of Antibody for CooA Using a Maltose-Binding Protein Fusion.
The fusion between the maltose-binding protein and a fragment of CooA (residues 22–222) was generated in the following way: the 3′ overhang of a 880-bp SacII–HincII fragment of cooA was removed by Klenow polymerase and cloned into the XmnI site of pMAL-c2 (New England Biolabs), yielding a malE::′cooA fusion. The fusion junction was verified by sequencing, and the fusion protein was expressed in E. coli, isolated, and cleaved with factor Xa. The truncated CooA was excised from an acrylamide gel and purified. Rabbit serum antibodies to the CooA fragment were prepared by the University of Wisconsin-Madison Medical School Animal Care Unit.
Purification of CooA from R. rubrum.
A R. rubrum strain (UR459) (15), in which CooA is under the control of a nif promoter, was grown in 22-l carboys under nitrogen-fixing conditions. Cells were harvested at an OD680 of 1.6, and approximately 160 g cells were obtained from two 22-l carboys. After harvesting, the cells were frozen in liquid nitrogen and stored at −80°C.
All protein purification procedures were conducted anoxically; solutions were sparged with N2 or degassed before use. After the solutions were rendered anoxic, they received additions (unless otherwise noted) to the following concentrations: 1 mM DTT, 1.7 mM dithionite, 200 μM phenylmethylsulfonyl fluoride, and 1 μg/ml leupeptin. Phosphate buffer (PO4) was prepared with KH2PO4 and K2HPO4 to attain a pH of 7.4. All manipulations were carried out at 4°C. All centrifugation was carried out in 45-ml centrifuge tubes fitted with septa.
Ninety-six grams of frozen cell pellets were resuspended in 240 ml of 0.2 M NaCl/10% glycerol/25 mM Mops (3-[N-morpholino]propanesulfonic acid), pH 7.4 and broken using a French press at 16,000 psi. Unbroken cells were removed by centrifugation at 11,200 × g for 40 min. The resulting supernatant was distributed to 45-ml centrifuge tubes, and polyethylenimine (5% stock, pH 7.9) was slowly added with mixing to a final concentration of 0.1%. This solution was allowed to incubate 30 min before centrifuging at 11,200 × g for 20 min. The resulting pellets were resuspended in 100-ml final volume of 0.8 M NaCl/10% glycerol/25 mM Mops, pH 7.4 using a homogenizer and allowed to incubate for 2 hr on ice. The resulting suspension was centrifuged at 11,200 × g for 20 min, and the supernatant was recovered. To the supernatant, ammonium sulfate (85% saturated stock, containing 25 mM Mops, pH 7.4 and 10% glycerol) was added to a final concentration of 50% saturation, and the mixture was allowed to precipitate overnight. The precipitate was collected by centrifugation at 11,200 × g for 20 min and resuspended in 125-ml final volume of 25 mM Mops, pH 7.4. The remaining chromatophores were removed by ultracentrifugation in 25-ml centrifuge tubes fitted with septa at 148,000 × g for 2 hr.
The ultracentrifuge supernatant was diluted 1:3 in 0.2 M KCl/10 mM PO4/5% glycerol/25 mM Mops, pH 7.4 and applied to a 4 × 6-cm Bio-Gel HTP (Bio-Rad) hydroxylapatite column equilibrated with 0.2 M KCl/10 mM PO4/5% glycerol/25 mM Mops, pH 7.4. The column was washed with 160 ml of 1.2 M KCl/10 mM PO4/5% glycerol/25 mM Mops, pH 7.4 followed with 80 ml of 40 mM PO4/50 mM NaCl/5% glycerol/25 mM Mops, pH 7.4. CooA was eluted from the column with a 800-ml linear gradient (40–160 mM PO4/50 mM NaCl/5% glycerol/25 mM Mops, pH 7.4). During the gradient, 30-ml fractions were collected, and those containing CooA were pooled. CooA eluted at approximately 110 mM PO4.
Fractions containing CooA were diluted 1:6 in 25 mM Mops, pH 7.4/10% glycerol, concentrated on a Q-Sepharose (Pharmacia) column, and eluted with 0.3 M NaCl. CooA then was purified with a 5-ml QMA MemSep HP101 column (Millipore) using a System Gold HPLC (Beckman). Fractions containing CooA were diluted 1:6 in 25 mM Mops, pH 7.4/10% glycerol and applied to the QMA column. The column was washed with 10 ml of 25 mM Mops, pH 7.4/25 mM NaCl and eluted with a 50-ml linear gradient of 25–300 mM NaCl/25 mM Mops, pH 7.4. Two-milliliter fractions were collected with CooA eluted at approximately 120 mM NaCl. Pooled CooA fractions were adjusted to 25% glycerol by addition of an equal volume of 50% glycerol/25 mM Mops, pH 7.4 and stored at −80°C for use in the experiments described below.
Gel Filtration Chromatography and Determination of Molecular Weight.
Gel filtration experiments were performed using a Superose 12 FPLC column (Pharmacia), using a flow rate of 0.25 ml/min. Chromatography was performed in anoxic, reduced column buffer (0.1 M NaCl/25 mM Mops, pH 7.4/1 μg/ml leupeptin/0.2 mM phenylmethylsulfonyl fluoride/1 mM DTT/1.7 mM dithionite). A calibration curve was constructed using BSA, ovalbumin, and carbonic anhydrase. Purified CooA was diluted 1:2 with anoxic, reduced column buffer, attaining a final concentration of 3.7 μM CooA dimer, and 300 μl was loaded onto the column. For determination of molecular weight in the presence of CO, the headspace of the vial containing diluted CooA was flushed with 100% CO for 10 min, and the sample was mixed gently. The column buffer was sparged with 100% CO for 30 min before use and maintained under a CO headspace. Column fractions (0.125 ml) were collected and analyzed for CooA by SDS/PAGE and Western blotting of similar gels. The percentage of CooA present in each fraction was determined using a densitometric scan of the Coomassie-stained SDS gels.
Spectrophotometric Methods and CO-Binding Assay.
Spectrophotometric measurements were recorded using a Shimadzu UV-1601PC (slit width of 2 nm). Purified, reduced CooA (370 μl) was diluted with 863 μl of anoxic buffer (25 mM Mops/1.7 mM dithionite/1 mM DTT) to attain a final concentration of 2.2 μM CooA dimer. The purified protein then was transferred to a stoppered, degassed cuvette, and a spectrum was recorded. To generate CO-bound CooA, the solution containing CooA was equilibrated with 100% CO by flushing the headspace of the sample for 10 min and mixing the sample gently; a spectrum then was recorded. Extinction coefficients were calculated by dividing the absorbances at the peak maxima by the concentration of heme determined by the pyridine hemochrome assay (see below).
The oxidized form of CooA was generated by removal of dithionite using a small hydroxylapatite column and subsequent exposure of the sample to air for 1 hr with gentle shaking. The oxidized form of CooA also was generated by desalting the dithionite-containing sample with Sephadex G-25 (Pharmacia) and subsequent exposure to air or potassium ferricyanide under argon.
Preparation and Electronic Absorption Spectrum of the Reduced Pyridine Hemochrome of CooA.
Purified, reduced CooA (360 μl) was added to 140 μl of anoxic buffer (25 mM Mops, pH 7.4/1.7 mM dithionite) and 500 μl of 4.4 M pyridine/0.2 M NaOH, obtaining a final CooA concentration of 2.7 μM CooA dimer. This solution was transferred to a stoppered, degassed cuvette, and a spectrum was recorded.
Determination of Amino Acid Composition, Protein Concentration, and Metal Content.
The amino acid composition determination was performed at the W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University, New Haven, CT. Metal analysis was performed on CooA at the Department of Chemistry, University of Wisconsin-Madison, using atomic absorption spectrophotometry. Protein concentrations reported were determined by the method of Minamide and Bamburg (16). For determination of protein concentration for heme and iron stoichiometry, the bicinchoninic acid assay (17) was used (see below).
Determination of Heme Stoichiometry.
To determine the heme content of purified CooA, dithionite and DTT were removed anoxically by using a small hydroxylapatite column, and the protein concentration was determined by using the bicinchoninic acid method (17). The protein solution was degassed, and dithionite was added to a final concentration of 1.7 mM. The pyridine hemochrome was generated as described above. The known extinction coefficient [34.4 mM−1 cm−1] (18) at the absorbance maximum of the alpha peak of the reduced pyridine hemochrome of protoheme was used to calculate the concentration of heme in purified CooA.
DNase I Protection Assays for CooA Activity.
DNase I protection assays were performed as described (15). Briefly, a 32P- labeled 294-bp EcoRV–EagI fragment bearing the CooA-binding site located upstream of cooF was used as the substrate for the reactions. The reaction mixtures contained anoxic buffer [20 mM Tris⋅HCl, pH 7.6/7 mM MgCl2/50 mM KCl/7 mM DTT/50 μg/ml BSA/5% (vol/vol) glycerol/1.7 mM dithionite], and purified CooA was added in increasing concentration, either in the presence or absence of CO. The binding mixture was allowed to equilibrate for 30 min at 25°C, and the mixture was subjected to DNase I cleavage. The cleavage products were phenol-chloroform-extracted, ethanol-precipitated, and separated on a 6% polyacrylamide-urea sequencing gel.
RESULTS AND DISCUSSION
CooA Purified from Its Native Organism, R rubrum.
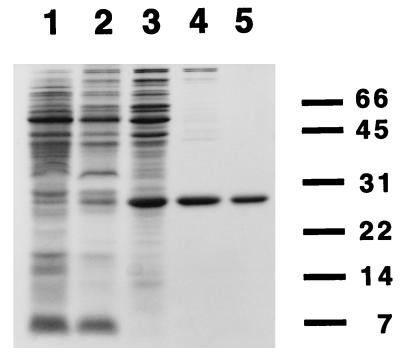
CooA was purified from an overexpressing R. rubrum strain (UR459) (15) to greater than 95% purity, as revealed by densitometric scanning of a Coomassie-stained SDS/PAGE gel (Fig. 1). The purification was performed anoxically under reducing conditions to prevent loss of any oxygen-labile activities that could potentially be associated with CooA (15, 5). The purification used, in the following order, polyethylenimine precipitation, ammonium sulfate precipitation, ultracentrifugation, chromatography on hydroxylapatite, and HPLC anion ion exchange chromatography. The progress of the purification was monitored by SDS/PAGE (Fig. 1), and CooA was detected using an antibody produced to a CooA-maltose-binding protein fusion in E. coli. The yield of protein was approximately 8 mg from 96 g of R. rubrum cells. When the amino acid composition was determined on the purified material, it matched the composition predicted for CooA (data not shown), indicating that the cooA gene product was indeed purified. As described below, the purified protein had the activities previously predicted for CooA (8, 15).
Figure 1.
Purity of CooA at different stages of the isolation procedure as revealed by SDS/PAGE. Samples at different stages of the purification of CooA from R. rubrum were subjected to electrophoresis on a 13.5% SDS gel and Coomassie-stained. The stages in the purification are: Lane 1, crude extract of R. rubrum strain UR459 (CooA-overexpressing strain); lane 2, material extracted with 0.8 M NaCl after polyethylenimine precipitation; lane 3, ultracentrifuge supernatant; lane 4, pooled hydroxylapatite fractions; and lane 5, pooled QMA (HPLC anion exchange) fractions. The dashes and numbers to the right of the gel indicate the migration position and molecular mass in kilodaltons, respectively, of electrophoresis standards.
Purified CooA Is Active in CO-Dependent, Sequence-Specific DNA-Binding.
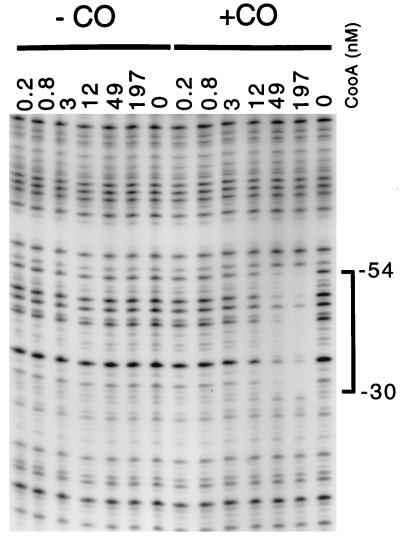
To determine if CooA was active in DNA binding in the absence of other factors, DNase I footprinting under anoxic, reducing conditions (15) was used. CooA exhibited sequence-specific DNA-binding and bound DNA only in the presence of CO (Fig. 2). This observation is consistent with results obtained using a crude extract (15) and indicates that no factors essential for either CO-sensing or DNA-binding were lost during the purification; CooA alone is competent in these coordinated activities. Purified, CO-activated CooA bound its DNA target site (15), which is similar to the CRP and FNR target sites (12). Because the protected portion includes a region of 2-fold rotational symmetry, this suggests the protein is bound to its site as a dimer.
Figure 2.
CO-dependent footprinting of the cooF promoter region with purified CooA. A 294-bp fragment bearing the CooA binding site upstream of cooF was used as a substrate for a DNase I protection experiment, either in the presence of CO or in the absence of CO. All reactions were performed anoxically under reducing conditions. The numbers at the top reflect the nM concentration of CooA in each reaction. The bracket on the right indicates the protected region, which encompasses the CooA-binding site identified previously (15). The numbers next to the bracket indicate the position of the protected bases relative to the CO-regulated cooF transcription start site (15).
Purified CooA Exists in a Dimeric State in Both the Presence and Absence of CO.
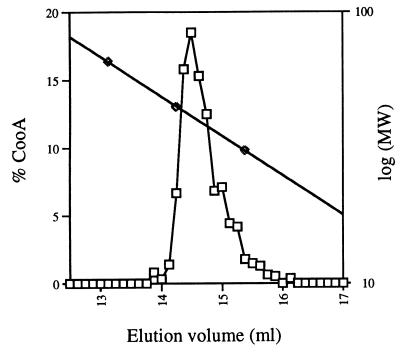
To determine the oligomeric state of CooA, we performed gel filtration experiments on the purified protein. These experiments were performed under reducing, anoxic conditions because the protein is known to be active under these conditions. In the absence of CO, all detectable CooA eluted from the gel filtration column as a single peak (Fig. 3). The calculated molecular mass from these experiments was 41 kDa. The theoretical molecular mass of the monomeric protein predicted from its DNA sequence (8) is 24.6 kDa, indicating that the protein is a dimer under the experimental conditions used. Experiments conducted similarly in the presence of CO under anoxic, reducing conditions gave a similar molecular mass value, indicating the protein is a dimer in the presence of CO (data not shown).
Figure 3.
Molecular mass determination of CooA using gel filtration chromatography. Reduced, purified CooA was loaded onto a Superose 12 (Pharmacia) column equilibrated with anoxic, dithionite-containing buffer in the absence of CO. The squares represent the percentage of CooA relative to the total amount of CooA eluting from the column. The diamonds represent the molecular markers used to calibrate the column: BSA (66 kDa), carbonic anhydrase (45 kDa), and ovalbumin (31 kDa).
The oligomeric state of CRP does not change in response to cAMP; the protein is a dimer regardless of the presence of its effector. In contrast, the FNR protein is monomeric in its inactive, oxygen-exposed state. FNR, when isolated anoxically, is dimeric and active in DNA binding; the oligomeric state of FNR reflects its activity. This change is believed to be effected through an oxygen-labile iron-sulfur center. The absence of a detectable change in CooA oligomerization upon exposure to CO indicates that CooA is more similar to CRP than FNR and suggests CooA undergoes a conformational change in the presence of its effector that allows it to bind DNA.
Purified CooA from R. rubrum Contains Protoheme and Binds CO.
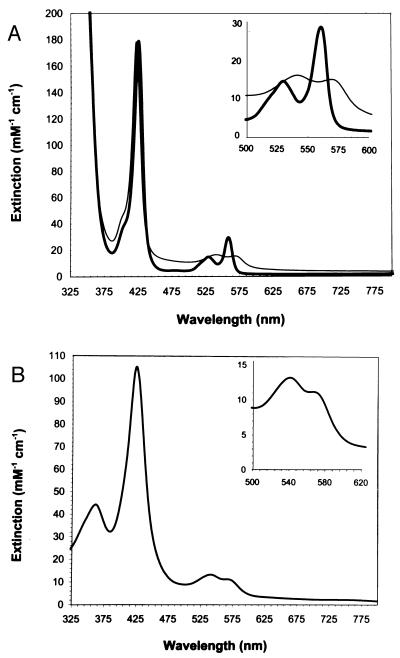
Purified CooA was orange-red in appearance, and a spectrum (Fig. 4A) of the reduced, anoxic protein appeared very similar to cytochromes b, which incorporate protoheme. The spectrum was consistent with a low-spin, six-coordinate ferrous iron in the resting (CO-unbound) state, as revealed by the presence of two distinct peaks in the 500- to 600-nm range. To determine if the protein contained protoheme, the pyridine hemochrome of CooA was prepared, and a spectrum consistent with the pyridine hemochrome of protoheme (absorbance maxima at 556, 524, and 419 nm) was obtained (data not shown), consistent with the observations of Aono et al. (19) on CooA purified from E. coli.
Figure 4.
Electronic absorption spectra of purified CooA. (Inset) The alpha-beta region of the spectra. (A) CooA in the presence and absence of CO. The heavy trace represents the spectrum of reduced CooA in the absence of CO. The light trace represents the spectrum of reduced CooA after exposure to CO. (B) CooA oxidized by air in the absence of dithionite. Extinction coefficients and peak maxima are reported in the text.
The heme content of purified CooA was 1.6 mol heme per mol of dimeric CooA, as determined by the pyridine hemochrome method (see Materials and Methods). The iron to heme ratio (iron content determined by atomic absorption spectrophotometry) was 1.1:1, suggesting that the iron in the protein is incorporated only in the heme and that nonheme iron is not present. The heme content is consistent with a model in which each monomer of CooA binds a single molecule of heme; it is possible that the experimentally determined heme content is lower than 2.0 because the protein assay used (17) may be overestimating the protein concentration. Alternatively, some heme may be lost from the protein during purification.
The oxidized (ferric) form of CooA was prepared by the removal of dithionite from the protein sample (see Materials and Methods) followed by exposure to air. CooA rapidly oxidized in the presence of air as indicated by a change in the spectral features of the protein (Fig. 4B). These results were consistent with the observations of Aono et al. (19) on CooA isolated from E. coli (however, see below for spectral differences). When air-exposed CooA was anoxically exposed to the chemical oxidant potassium ferricyanide, the spectrum did not change (data not shown), further suggesting that the spectrum was that of the oxidized CooA. The positions of the bands (extinction coefficients in mM−1 cm−1 in brackets) were 361 nm [44], 424 nm [105], 541 nm [13], and 566 nm (shoulder) [11].
The spectrum of oxidized CooA is consistent with a low-spin ferric species. The position of the Soret band of oxidized CooA (424 nm) is similar to that of low-spin, bis histidine-ligated ferric cytochromes b such as cytochrome b5 (20) and is red-shifted relative to other high-spin ferric hemoproteins. High-spin species usually give Soret bands in the vicinity of 400–410 nm, whereas low-spin species have Soret bands above 410 nm. In the spectrum we obtained of oxidized CooA, two maxima are present in the alpha-beta region (500–600 nm). The features of this region are also similar to the low-spin ferric cytochromes b, such as cytochrome b5. Low-spin ferric species give two prominent bands in the alpha-beta region, whereas high-spin species have more complex spectral features in this region (21). Additionally, a charge-transfer band in the vicinity of 630 nm, an indication of a high-spin species, was not detectable for CooA. Taken together, these results suggest the iron of CooA is low spin in the oxidized form and should have two strong-field ligands to its heme. Amino acids that bear side-chains capable of generating such low-spin species include His, Met, Cys, and Lys (22); the most commonly observed liganding arrangement observed in cytochromes b is bis-histidine ligation.
Our oxidized CooA spectrum differs somewhat from the results of Aono et al. (19), who reported a Soret band for oxidized CooA at 418.5 nm, different from the 424-nm position that we measured. Additionally, the authors reported a “broad band” in the alpha-beta region of the spectrum; in our spectra, two maxima are clearly observable in this region. To determine if the differences in oxidized spectra between our samples and those of Aono et al. were a result of expression in E. coli versus R. rubrum, we purified CooA from E. coli aerobically in the absence of dithionite using our protocol and obtained identical oxidized and reduced spectra as for the R. rubrum-isolated protein (data not shown). Therefore, the differences in the spectral features and observed maxima most likely result from differences in purification or sample preparation for spectroscopy.
When purified, reduced CooA was exposed to CO, a shift in the absorbance maxima occurred. This result indicates that a change occurred in the electronic environment of the heme and suggests that CO was binding directly to the heme moiety of CooA (Fig. 4A). The absorbance maxima (extinction coefficients in mM−1 cm−1 in brackets) shifted from 559 nm [29], 529 nm [15], and 426 nm [179] (reduced, −CO) to 568 nm [15], 540 nm [16], and 422 nm [179] (reduced, +CO). As expected, the spectrum in the presence of CO displayed features that were also characteristic of six-coordinate, low-spin heme and similar to CO adducts of hemoglobin, myoglobin, and a variety of other histidine-ligated heme proteins. The characteristic spectrum of the cytochrome P450-CO complex (23), a red-shifted Soret band at approximately 450 nm, was not observed, suggesting that cysteine is not a ligand to the heme of CooA in the CO-bound state.
The activation of CooA by CO may be mediated by replacement of a protein-bound heme ligand by CO. Because the resting state of the heme of CooA appears to be low spin, a spin transition does not seem to be involved in the activation of CooA. This is in contrast to the heme of the O2-sensing protein FixL, which undergoes a transition from high spin in the resting state to low spin upon binding O2 (4) and is also in contrast to hemoglobin, for which a spin-state transition is critical for the conformational change that the protein undergoes upon binding O2. The activation of CooA by CO instead may occur through the replacement of a heme ligand by CO, which could trigger a conformational change in the protein. In that it involves replacement of a protein-bound ligand, this mechanism may have similarities to the proposed activation mechanism of soluble guanylyl cyclase by NO (24–26). Future spectroscopic and mutational analysis will be needed to delineate the mechanism of activation of CooA.
The spectral features of CooA resemble those of cytochromes b, which commonly bear two histidine ligands to the heme. A His residue (His-77) is in the region of CooA (8) that is homologous to the cAMP-binding site of CRP (27); to test if this was a heme ligand we created a His to Tyr substitution at this position and expressed this mutant CooA in E. coli. In experiments done in crude extracts, the heme in this mutant CooA could not be stably reduced by a large excess of dithionite, consistent with the notion that an axial ligand has been changed to a Tyr (data not shown). It is possible, however, that His-77 is not an axial ligand to the heme but is in the general vicinity of the heme, and its mutation has simply perturbed the environment of the heme. Regardless, this observation suggests that for CooA, effector binding occurs in a region homologous to the cAMP-binding site in CRP.
To summarize, in this study we have shown that purified CooA from R. rubrum interacts with CO through the heme, and upon binding CO becomes competent to bind to its DNA target site. We also have shown that the protein is dimeric regardless of the presence of CO, similar to CRP. We have described a heme protein that responds to CO by binding to DNA in a sequence-specific manner and a protein definitively shown to be a biological sensor of CO.
Though many genes encoding CRP homologs have been discovered, only in the case of CRP is the effector definitively demonstrated. The structural dissimilarity between CO and cAMP indicates that this family of transcriptional regulators is versatile in the recognition of effector molecules.
Acknowledgments
We thank Judith Burstyn and Mark Reynolds for numerous helpful conversations, suggestions on the manuscript, and assistance with the iron analysis. We thank Vinod Shah, Paul Ludden, Mary Homer, and Jon Roll for assistance with protein purification. This research was supported by the College of Agricultural and Life Sciences, University of Wisconsin-Madison, and by National Institutes of Health Grant GM53228.
ABBREVIATIONS
- CRP
cAMP receptor protein
- FNR
fumavate nitrate reduction
References
- 1.Ignarro L J, Degnan J N, Baricos W H, Kadowitz P J, Wolin M S. Biochim Biophys Acta. 1982;718:49–59. doi: 10.1016/0304-4165(82)90008-3. [DOI] [PubMed] [Google Scholar]
- 2.Craven P A, DeRubertis F R. Biochim Biophys Acta. 1983;745:310–321. doi: 10.1016/0167-4838(83)90063-8. [DOI] [PubMed] [Google Scholar]
- 3.Gilles-Gonzalez M A, Ditta G S, Helinski D R. Nature (London) 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 4.Gilles-Gonzalez M A, Gonzalez G, Perutz M P. Biochemistry. 1995;34:232–236. doi: 10.1021/bi00001a027. [DOI] [PubMed] [Google Scholar]
- 5.Bonam D, Lehman L, Roberts G P, Ludden P W. J Bacteriol. 1989;171:3102–3107. doi: 10.1128/jb.171.6.3102-3107.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerby R L, Hong S S, Ensign S A, Coppoc L J, Ludden P W, Roberts G P. J Bacteriol. 1992;174:5284–5294. doi: 10.1128/jb.174.16.5284-5294.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerby R L, Ludden P W, Roberts G P. J Bacteriol. 1997;179:2259–2266. doi: 10.1128/jb.179.7.2259-2266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shelver D, Kerby R L, He Y-P, Roberts G P. J Bacteriol. 1995;177:2157–2163. doi: 10.1128/jb.177.8.2157-2163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox J D, Kerby R L, Roberts G P, Ludden P W. J Bacteriol. 1996;178:1515–1524. doi: 10.1128/jb.178.6.1515-1524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox J D, He Y, Shelver D, Roberts G P, Ludden P W. J Bacteriol. 1996;178:6200–6208. doi: 10.1128/jb.178.21.6200-6208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerby R L, Ludden P W, Roberts G P. J Bacteriol. 1995;177:2241–2244. doi: 10.1128/jb.177.8.2241-2244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb A, Busby S, Buc H, Garges S, Adhya S. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 13.Spiro S, Guest J R. FEMS Microbiol Rev. 1990;6:399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- 14.Lazazzera B A, Beinert H, Khoroshilova N, Kennedy M C, Kiley P J. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 15.He Y-P, Shelver D, Kerby R L, Roberts G P. J Biol Chem. 1996;271:120–123. doi: 10.1074/jbc.271.1.120. [DOI] [PubMed] [Google Scholar]
- 16.Minamide L S, Bamburg J R. Anal Biochem. 1990;190:66–70. doi: 10.1016/0003-2697(90)90134-u. [DOI] [PubMed] [Google Scholar]
- 17.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 18.de Duve C. Acta Chem Scand. 1948;2:264–289. doi: 10.3891/acta.chem.scand.02-0264. [DOI] [PubMed] [Google Scholar]
- 19.Aono S, Nakajima H, Saito K, Okada M. Biochem Biophys Res Comm. 1996;228:752–756. doi: 10.1006/bbrc.1996.1727. [DOI] [PubMed] [Google Scholar]
- 20.Bois-Poltoratsky R, Ehrenberg A. Eur J Biochem. 1967;2:361–365. doi: 10.1111/j.1432-1033.1967.tb00146.x. [DOI] [PubMed] [Google Scholar]
- 21.Tsai A-L, Kulmacz J-S W, Wang Y, Van Wart H E, Palmer G. J Biol Chem. 1993;268:8554–8563. [PubMed] [Google Scholar]
- 22.Cheesman M R, Thomson A J, Greenwood C, Moore G R, Kadir F. Nature (London) 1990;346:771–773. doi: 10.1038/346771a0. [DOI] [PubMed] [Google Scholar]
- 23.Dawson J H, Sono M. Chem Rev. 1987;87:1255–1276. [Google Scholar]
- 24.Traylor T G, Sharma V S. Biochemistry. 1992;31:2847–2849. doi: 10.1021/bi00126a001. [DOI] [PubMed] [Google Scholar]
- 25.Yu A E, Hu S, Spiro T G, Burstyn J N. J Am Chem Soc. 1994;116:4117–4118. [Google Scholar]
- 26.Burstyn J N, Yu A E, Dierks E A, Hawkins B K, Dawson J H. Biochemistry. 1995;34:5896–5903. doi: 10.1021/bi00017a019. [DOI] [PubMed] [Google Scholar]
- 27.Weber I T, Steitz T A. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]