Abstract
RNAs that undergo a rapid site-specific cleavage at low pH have been selected by in vitro selection (the SELEX process). The cleavage does not require the addition of any divalent metal ions, and is in fact inhibited by divalent metal ions, spermine, or high concentrations of monovalent metal ions. This low pH catalyzed cleavage results in a 2′,3′-cyclic phosphate at the 3′ end and a free hydroxyl at the 5′ end. The reaction proceeds with a calculated rate of 1.1 min−1 at room temperature in cacodylate buffer at pH 5.0. The rate of cleavage is dependent on the pH and shows an optimum around pH 4.0. The rate constant is independent of RNA concentration, indicating to an intramolecular reaction. Autocatalytic cleavage at low pH, in the absence of a metal ion requirement, adds to the reaction possibilities that may have existed on the prebiotic earth.
The discovery that certain RNAs are capable of carrying out catalytic functions that had once been attributed solely to protein enzymes (1, 2), has changed our view of nucleic acids as mere bearers of genetic information. Chemical reactions occurring in the presence of RNA within biological systems, such as RNA splicing or protein synthesis, may at least in some cases occur with RNA participating as a catalytic component. Evidence that 23S rRNA is at least partially responsible for the ribosomal peptidyl transferase activity has recently come to light (3). Several types of catalytic RNA molecules (ribozymes) that have the ability to undergo specific self-cleavage or self-ligation reactions within biological systems have been described (4–8). The SELEX§ (systematic evolution of ligands by exponential enrichment) protocol (reviewed in ref. 9), using a large number of random sequences, has brought to light a whole array of newly discovered reactions that RNA is capable of catalyzing outside the cell (10–12). Whether or not any of these reactions are catalyzed by RNA in any biological system remains to be seen. SELEX provides a useful approach to the generation of RNAs with novel catalytic activities. This report describes an unusual catalytic property of RNAs derived from a SELEX experiment in which the RNA undergoes a rapid, site-specific cleavage under low pH conditions, without a requirement for metal ions. To our knowledge, this is the first instance of RNA self-cleavage observed under these conditions, and highlights the expanding catalytic capacity of RNA.
MATERIALS AND METHOD
Enzymes and Chemicals.
T4 polynucleotide kinase, calf intestinal alkaline phosphatase, and T4 RNA ligase were from New England Biolabs; avian myeloblastosis virus reverse transcriptase from Promega; and Taq DNA polymerase from GIBCO/BRL. T7 RNA polymerase was generously provided by NeXstar Pharmaceuticals. Nuclease P, RNase T1, RNase inhibitor, and T4 polynucleotide kinase lacking the 3′ phosphatase activity were purchased from Boehringer Mannheim. Up (uridine 3′-monophosphate) and U>p (uridine 2′,3′-cyclic monophosphate) were from Sigma. Polyethyleneimine–cellulose thin-layer chromatography plates were obtained from Machery & Nagel.
Selection Protocol.
Selection and amplification followed a protocol described earlier (13, 14) and was, in fact, the control for our work on the T4 RegB endonuclease (14). Our aim was to select for RNA molecules that undergo spontaneous, site-specific phosphodiester bond cleavage. The RNA pool was comprised of T7 RNA polymerase transcripts (15) carrying a 30-residue contiguous tract of randomized nucleotides. Briefly, the RNA transcripts from a DNA library with a complexity of about 1014 molecules were ligated into intramolecular circles in 50 mM Hepes (pH 7.5), 3 mM DTT, 10 mM MgCl2, 10 μg/ml BSA, 10% dimethyl sulfoxide, 1 mM ATP, and 1 unit/μl T4 RNA ligase for 2 hr at 37°C. Circular products were isolated from polyacrylamide gels under denaturing conditions. The selection step has been carried out unintentionally during the extraction of circular RNA from the gel in 0.5 M NaOAc buffer (pH 5.2). Molecules that underwent a single cleavage event are now linearized and were isolated from unreacted circles by PAGE under denaturing conditions. Reacted RNAs were dephosphorylated and kinased to get the ends compatible for ligation and then re-ligated to generate the complete randomized region as described earlier (14). The selected RNAs were reverse transcribed, PCR amplified, and the selection was continued for 12 rounds. RNAs from the round 12 pool were cloned and sequences analyzed using standard protocols.
Cleavage Reactions.
RNA was transcribed in vitro from PCR-amplified DNA templates as described (14, 15) and purified on 8% polyacrylamide/7 M urea gels. For the cleavage reaction, RNA was internally labeled by including [α-32P]GTP in the transcription reaction. RNA was recovered by crushing the gel slice containing RNA in 100 mM Tris⋅HCl (pH 7.4)/1 mM EDTA and precipitating in ethanol. The RNA pellet was dried and resuspended in 10 mM Tris⋅HCl/1 mM EDTA (pH 7.5). The cleavage reaction was typically carried out in 50 mM sodium Mes (pH 5.5) or 50 mM sodium cacodylate (pH 5.0) buffer at room temperature at a final RNA concentration of 1 μM. For some experiments, other buffer systems were used for various time points as specified in the figure legends. Reactions were stopped by placing on ice and adding de-ionized formamide to a final 50%. Samples were heat denatured at 70°C for 3 min, chilled on ice, and run on denaturing polyacrylamide gels. Gels were dried and analyzed quantitatively using a Molecular Dynamics PhosphorImager.
For RNA sequencing reactions, RNA was radiolabeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase and purified by denaturing gel electrophoresis. Approximately 4 pmol of RNA was subjected to partial alkaline hydrolysis in 50 mM NaOH for 30 sec at 94°C to obtain a ladder. A partial RNase T1 ladder was obtained by treating RNA with 1 unit of RNase T1 in 50 mM Tris⋅HCl (pH 7.5), 4 mM EDTA, 3.5 M urea, and 5 μg/μl tRNA at 37°C for 15 min.
Determination of the End Products of Cleavage.
The ends produced by the cleavage were determined using established procedures (16) with some modifications. The truncated RNA dtr31 was internally radiolabeled using [α-32P]ATP during transcription, subjected to low pH-catalyzed self-cleavage, and the six nucleotide long 5′ product was isolated from a 20% polyacrylamide/7 M urea gel. This product was completely digested to mononucleotide 5′-phosphates by treating with 0.3 unit of nuclease P in 50 mM ammonium acetate (pH 7.0) buffer at 37°C for 30 min. The samples were spotted on polyethyleneimine–cellulose thin-layer plates and developed in 1 M ammonium acetate to a distance of 15 cm, along with nonradioactive markers. Because we were unable to obtain a commercial source of pUp (uridine 3′,5′-diphosphate) or pU>p (uridine 5′-monophosphate-2′,3′-cyclic monophosphate), these markers were synthesized by kinasing 100 nmol of Up or U>p with 20 units of T4 polynucleotide kinase, which lacks the 3′-phosphatase activity, in 50 mM Tris⋅HCl (pH 8.2), 10 mM MgCl2, 5 mM DTT, 0.1 mM EDTA, 0.1 mM spermidine, and 200 nmol ATP at 37°C for 30 min.
RESULTS AND DISCUSSION
Selection and amplification of RNA for a single cleavage event has been described (13, 14). The isolation of active RNAs is based on the ability of polyacrylamide gels to separate linear from circular molecules of the same length. A starting RNA repertoire of about 1014 unique sequences was subjected to low pH conditions in which RNA is normally stable. The RNAs that undergo a unique cleavage in low pH were selected and amplified for 12 rounds. The ligands obtained from this selection are listed in Fig. 1. Of a total of 22 clones sampled, 16 were found to be autocatalytic. Two of those sequences undergo self-cleavage at neutral pH and are not shown here. Clones 8, 9, 13, 22, and 5 and 17 have almost identical sequences at the 3′ end of the randomized region. The other clones do not share extensive sequence similarities, suggesting that low pH-catalyzed site-specific cleavage is probably not a rarity.
Figure 1.
Sequences obtained by cloning and sequencing after 12 rounds of selection for self-cleavage at low pH. The 5′ and 3′ fixed regions are in lowercase letters and the randomized region is in uppercase letters. The site of cleavage for each sequence is indicated by an arrow. The sequence that was studied in detail for this work appears within the bracketed area.
Cleavage Is Site Specific and Requires Low pH.
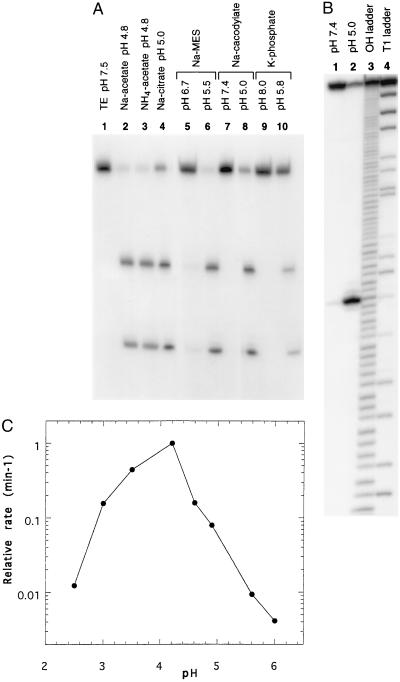
The cleavage pattern of one RNA, namely clone 8 RNA, in several different buffer systems is shown in Fig. 2A. There is no significant amount of cleavage with any of the buffers at neutral pH; however, the RNA undergoes cleavage efficiently and site specifically when the pH is lowered in the apparent absence of any divalent metal ions. Cleavage is not dependent on a particular cation or anion component in the buffer, although the extent of cleavage may be affected considerably. Ammonium acetate buffer is just as effective in cleaving the RNA as sodium acetate buffer at the same pH (compare lanes 2 and 3). The same result is observed with sodium and potassium phosphate buffers (data not shown). Sodium Mes buffer is found to be more effective than sodium citrate or sodium cacodylate buffers of the same pH in catalyzing the site-specific cleavage (compare lane 6 with lanes 4 and 8).
Figure 2.
(A) Self-cleavage of clone 8 RNA in the different buffer systems studied. Internally labeled RNA was incubated at room temperature in TE buffer at pH 7.5 (lane 1), 50 mM sodium acetate buffer at pH 4.8 (lane 2), 50 mM ammonium acetate buffer at pH 4.8 (lane 3), 50 mM sodium citrate at pH 5.0 (lane 4), 50 mM sodium Mes buffer at pH 6.7 (lane 5) or pH 5.5 (lane 6), sodium cacodylate buffer at pH 7.4 (lane 7) or pH 5.0 (lane 8), potassium phosphate buffer at pH 8.0 (lane 9) or pH 5.8 (lane 10) at a final RNA concentration of 1 μM (15). (B) Site of self-cleavage for clone 8 RNA. RNA labeled at the 5′ end was treated with 50 mM sodium cacodylate buffer at pH 7.4 (lane 1) or pH 5.0 (lane 2). To determine the cleavage site, samples were run on a 10% polyacrylamide/7 M urea gel along with partial alkaline hydrolysis ladder (lane 3) and partial RNase T1 digestion samples (lane 4). (C) Effect of pH on cleavage of clone 8 RNA in 50 mM sodium citrate buffer. The rate of cleavage obtained at each pH value was normalized to the maximum rate measured at pH 4.2.
The cleavage site was mapped to be at the U-A phosphodiester bond 10 nucleotides downstream from the 5′ end of the randomized region (Fig. 2B). The cleavage sites of the other clones have been similarly determined and are shown in Fig. 1. Although low pH is required for the reaction to occur efficiently, all the RNAs exhibit a low level of cleavage at the same site in neutral pH (Fig. 2B, compare lanes 1 and 2). With the single exception in clone 19, low pH-induced site-specific cleavage occurs predominantly at a pyrimidine-A dinucleotide. Unusual susceptibility of pyrimidine-A phosphodiester bonds to undergo site-specific self-cleavage in the presence of detergents, high molecular weight polymers or various protein factors at neutral pH have been reported (17–20). However, site-specific cleavage of RNA at low pH has not been reported previously. Furthermore, the reactions referenced above occur at rates several orders of magnitude slower than the rates obtained for the low pH catalyzed cleavage reaction described in this study (see Fig. 4 A and B). Interestingly, all of our selected clones carry two or more pyrimidine-A phosphodiester bonds in the randomized region alone, but only a unique pyrimidine-A phosphodiester bond is preferentially cleaved in each case, indicating that the mere existence of a pyrimidine-A phosphodiester bond is not sufficient for the rate enhancement observed here. It has been proposed that pyrimidine-A phosphodiester bonds are inherently more susceptible to cleavage and can be further destabilized by the structures adopted by the RNA, such that spontaneous cleavage is greatly facilitated (17, 18).
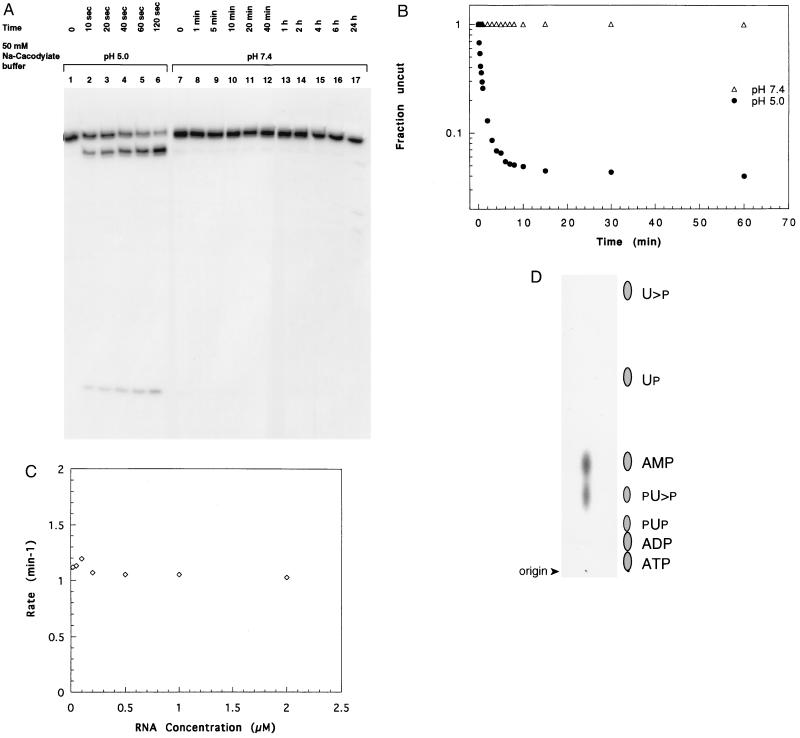
Figure 4.
(A) Time course of cleavage for truncated dtr31. Reactions were carried out in the presence of 50 mM sodium cacodylate buffer at pH 5.0 (lanes 1–6) or pH 7.4 (lanes 7–17) for the times indicated, and the samples were separated on a 20% polyacrylamide/8 M urea gel. (B) The fraction of the RNA remaining uncut was plotted against time using data from an experiment similar to A. The fraction of molecules remaining at each time point was calculated and the background at zero time was subtracted from each value. Solid and open symbols indicate the reactions at pH 5.0 and pH 7.4, respectively. (C) The rate of cleavage reaction was plotted as a function of RNA concentration. (D) Thin-layer chromatography of nuclease P-digested 5′ reaction product of dtr31 on polyethyleneimine–cellulose plates to determine the ends of cleavage. The positions of nonradioactive markers are indicated alongside.
The origin of the faint band immediately 3′ to the major cleavage site was not investigated. It could have resulted from the opening of the 2′,3′-cyclic ring of the 5′ reaction product (21) or from the possible heterogeneity of the full-length RNA at the 5′ end during transcription (22). The fact that the band is not observed in the 5′ cleavage product of the truncated RNA dtr31 favors the second possibility.
Cleavage was studied at various pH values using two different buffers to determine the optimum pH for the cleavage. The rate of reaction increases with decreasing pH until an optimum value is reached at around pH 4.2 in sodium citrate buffer, as shown in Fig. 2C. Similarly, sodium acetate buffer shows a pH optimum at about 4.0. Interestingly, the pKa values for N1 of adenosine and N3 of cytidine residues fall in this range, suggesting that a protonated base may play a structural or a catalytic role (23). At low pH values, non-Watson–Crick interactions may play important roles in determining the structure of the self-cleaving RNA. The decrease in the activity below the pH optimum may be due to changes in RNA conformation upon protonation at alternate sites, or denaturation of RNA. The pH profile of clone 8 RNA can be compared with that of Neurospora VS RNA, the most active natural ribozyme at low pH. VS RNA exhibits no change in reaction rate in the pH range between 8.9 to about 5.5; however, at pH 5.0, the reaction proceeds much slower (24). At this pH, the hammerhead ribozyme shows no activity (16).
The pattern of cleavage 3′ to a pyrimidine nucleotide raises the doubt of whether this observation results from a trace contamination of ribonuclease A, although that nuclease is less active at low pH than at neutral pH (25). Furthermore, prolonged incubation of RNA at neutral pH for up to several hours did not cause further degradation (see Fig. 4A). However, to eliminate the possibility of a trace amount of nuclease contamination, various reagents that degrade proteins were included in the reaction. Cleavage was not abolished or even diminished by extracting RNA with phenol/chloroform or including proteinase K, SDS, or RNase inhibitor in the reaction (data not shown). At the concentrations used in the reaction, proteinase K, SDS, and RNase inhibitor were sufficient to completely protect RNA from ribonuclease A degradation.
Effect of Metal Ions on Cleavage.
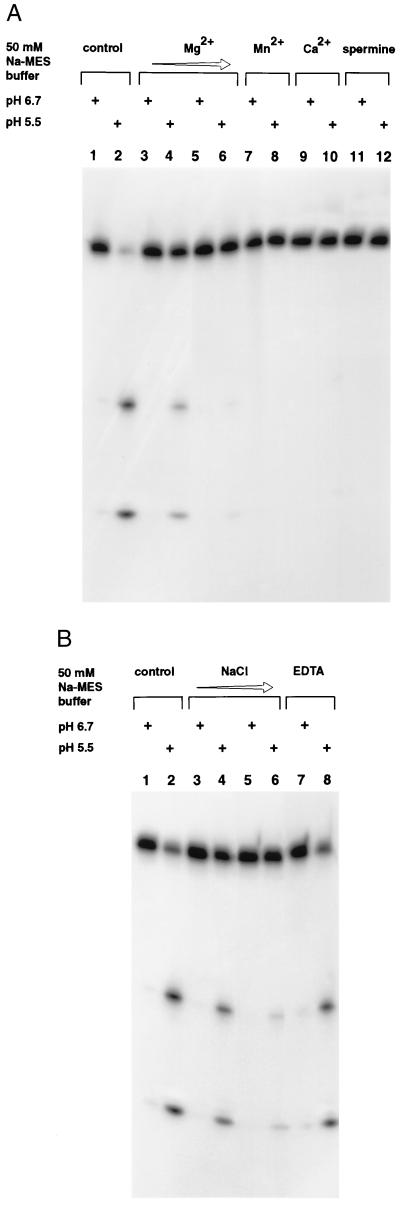
Cleavage reaction does not require the addition of any divalent metal ions, although this does not exclude the possibility of a trace quantity of a contaminating metal ion being responsible for the reaction. The addition of EDTA up to a final concentration of 5 mM does not inhibit the reaction (Fig. 3B, lanes 7 and 8), suggesting that a trace metal ion is probably not involved in the cleavage reaction. Furthermore, metal ion-catalyzed RNA cleavage proceeds faster with increasing pH (16, 26). The effect of including various divalent metal ions in the reaction is shown in Fig. 3A. The Mg2+ ion inhibits the cleavage reaction in a concentration-dependent manner, allowing the reaction to occur to a lesser degree at 2 mM and completely inhibiting at 10 mM (Fig. 3A, compare lanes 3–6 with lanes 1 and 2). Other divalent metal ions such as Mn2+ and Ca2+ (lanes 7–10), and Co2+ and Cu2+ (data not shown), exhibit the same inhibitory effect on cleavage.
Figure 3.
(A) The effect of divalent cations and spermine on self-cleavage of clone 8 RNA. The RNA was treated with 50 mM sodium Mes buffer at pH 6.7 or pH 5.5, as indicated by + signs in the absence of any added metal ions (lanes 1 and 2) or in the presence of 2 mM MgCl2 (lanes 3 and 4), 10 mM MgCl2 (lanes 5 and 6), 10 mM MnCl2 (lanes 7 and 8), 10 mM CaCl2 (lanes 9 and 10), or in the presence of 0.5 mM spermine (lanes 11 and 12). (B) The effect of EDTA or increasing concentration of NaCl on self-cleavage. Reactions were carried out as described above in the absence of added metal ions (lanes 1 and 2) or in the presence of 50 mM NaCl (lanes 3 and 4), 250 mM NaCl (lanes 5 and 6), or 5 mM EDTA (lanes 7 and 8).
Divalent metal ions play a dual role in promoting RNA folding and, in certain instances, catalyzing chemical reactions (27–29). Polycations such as spermine and spermidine are also capable of stabilizing RNA folding by neutralizing the charges on the backbone. Inclusion of spermine in the reaction results in the inhibition of cleavage similar to the effect of divalent metal ions (Fig. 3A, lanes 11 and 12). The presence of monovalent cation Na+ shows the same concentration-dependent inhibitory effect as the divalent ions, but at a much higher concentration (Fig. 3B, compare lanes 1 and 2 with lanes 3–6). This observation is consistent with the fact that divalent metal ions are much more effective in stabilizing RNA structure compared with monovalent ions. At about 0.5 M NaCl, the reaction is mostly inhibited (data not shown); this concentration range is about equally effective in stabilizing RNA structure as 10–20 mM MgCl2 (16). Collectively, these data suggest that the structure that is responsible for the cleavage reaction is accessible in low salt, and agents such as spermine, divalent and monovalent metal ions at higher concentration probably stabilize the RNA in a different conformation that is not susceptible to cleavage.
Implications for a Secondary Structure.
Attempts were made to truncate the RNA molecule to assess the minimal length sufficient and necessary to catalyze the reaction. The sequence of dtr31, the shortest truncate (a 31-mer) able to catalyze the reaction at the highest level is given in Fig. 1. It lacks a 28-nucleotide region from the 5′ end, leaving only a 6-nucleotide stretch 5′ to the cleavage site and a 21-nucleotide region from the 3′ end of the full-length RNA molecule (an 80-mer). Deleting five nucleotides from the 3′ end of dtr31 results in a drastic reduction of activity (data not shown). Deleting a further five nucleotides completely abolishes the reaction. The addition of two guanosine residues to the 5′ end of dtr31 for better transcription efficiency results in a very significant reduction of catalytic activity (data not shown). The reaction is completely inhibited in denaturants such as 50% formamide or 6 M urea (not shown). These results imply the involvement of a secondary structure that is important for the reaction to occur.
Kinetics of Cleavage Reaction.
Time course of reaction for dtr31 in cacodylate buffer at neutral and low pH at room temperature is shown in Fig. 4A. At pH 5.0, a significant amount of product is detectable as early as 10 sec and the reaction is mostly completed at about 2 min. In contrast, at pH 7.4, RNA does not undergo any significant level of cleavage, up to 6 hr, but by 24 hr RNA has begun to show degradation products (lanes 16 and 17).
Using data from a similar gel, the fraction of molecules remaining was plotted against time (Fig. 4B). About 4% of the RNA population does not undergo cleavage even after prolonged incubation for 20 hr at pH 5.0 (data not shown). This may represent molecules that carry misincorporated nucleotides and are thus unable to cleave or may simply represent molecules trapped in an alternate conformation(s) (30), which does not support cleavage. The cleavage reaction proceeds with a half-life (t1/2) of about 0.6 min, corresponding to a kobs value of 1.1 min−1 under the conditions specified above. The rate of cleavage shows a pH optimum around 4.0 as shown in Fig. 2C. We have not measured the rate of the truncated RNA at this pH and, thus, the rate obtained here has been measured under suboptimal reaction conditions. Although the rates of cleavage for the several different hammerheads studied so far vary by more than a thousand-fold, this t1/2 value compares with those of the faster cleaving hammerheads (31). Similar values for rates have been reported for the hairpin (26) and the hepatitis delta (32) ribozymes. The data points are linear at the earliest time points but adopt a definite curvature as the reaction proceeds, suggesting that another factor influences the reaction rate with time. This could be a conversion of an alternate conformation into the one that is susceptible to cleavage. The rate of cleavage was studied as a function of RNA concentration, and as shown in Fig. 4C, the rate remains unchanged over a hundred-fold difference in concentration (between 0.02 and 2 μM), indicating that the reaction is intramolecular.
The End Products of Cleavage.
The ends produced by low pH-catalyzed cleavage were determined using the truncated RNA dtr31 for convenience, since this RNA yields a six-nucleotide 5′ product carrying a single adenosine residue. Complete digestion of this 5′ cleavage product with nuclease P yielded two spots of equal intensity; one comigrating with pA (AMP) and the other with pU>p (Fig. 4D), showing that the cleavage results in a 2′,3′-cyclic uridine at the new 3′ end and a free hydroxyl group at the 5′ end. This is the most common end product of RNA cleavage observed in reactions catalyzed by divalent metal ions and ribonucleases (4, 5, 16). These end products are obtained when the cleavage is mediated through an in-line attack by the ribose 2′-OH on the adjacent phosphodiester bond.
In metal ion-catalyzed RNA cleavage, a water-hydrated metal ion abstracts the proton from the ribose 2′-hydroxyl proton, rendering it more nucleophilic for attacking the electrophilic phosphorous center (27). However, metal ions are not necessary for the phosphodiester bond cleavages catalyzed by certain protein enzymes. Ribonuclease A catalyzes RNA cleavage by general acid base catalysis using a specific histidine residue in the active site to abstract the ribose 2′-OH proton and another histidine or lysine residue to stabilize the developing negative charge on the 5′ oxygen on the leaving group by catalytic protonation (33). A similar mechanism may operate in this case. It is evident from Fig. 2C that some functional group is being titrated as the pH is dropped from neutral pH to about 4.0. The protonation of this group could cause a structural change in the RNA molecule to place another group in close proximity to the ribose 2′-hydroxyl group to act as a general base. The developing negative charge on the leaving group 5′ oxygen could be stabilized by the protons in the medium. The few structures of RNA that bind small molecules (reviewed in ref. 34) suggest that intricate pockets and local distortions are commonly found in RNA, enabling it to precisely orient functional groups to facilitate catalysis.
Finally, we and the others have noted the intellectual difficulties in experiments aimed at understanding the RNA world (10, 35). An infinite number of conditions may be chosen to explore the versatility of RNA for catalysis, and to that list one must now add low pH. The results from our study imply that many prebiotic conditions could have impacted the catalytic potential of short oligonucleotides. The prebiotic “RNA world” may have been more robust than is suggested by the limited reactions catalyzed thus far by RNAs under rather more standard laboratory conditions (10).
Acknowledgments
We are grateful to Olke Uhlenbeck, Drew Smith, and Sumedha Jayasena for valuable comments and critical review of the manuscript. We thank Ronald Raines and James Bashkin for helpful discussions. Our thanks are extended to Barry Vant-Hull, Don Burke, Shawn Zinnen, Pat Allen, Timur Shtatland, and Mali Illangasekare for their enthusiasm and many stimulating discussions. We also thank Jack Szostak and David Bartel for editorial comments on the manuscript. This research was supported by Research Grant GM19963 from the National Institutes of Health to L.G. and funds from NeXstar Pharmaceuticals, Inc.
Footnotes
The words “SELEX” and “in vitro evolution” are used interchangeably by us; similarly, “aptamers” and “ligands” are used interchangeably. David Bartel has noted to us that since ligands is the word used to contrive the acronym SELEX, in vitro evolution is a more suitable phrase to use when one seeks a ribozyme.
References
- 1.Kruger K, Grabowski P J, Zaug A, Sands J, Gottschling D E, Cech T R. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Noller H F, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 4.Prody G A, Bakos J T, Buzayan J M, Schneider I R, Bruening G. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 5.Buzayan J M, Gerlach W L, Bruening G. Nature (London) 1986;323:349–353. [Google Scholar]
- 6.Sharmeen L, Kuo M Y P, Dinter-Gottlieb G, Taylor J. J Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein L M, Gall J G. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- 8.Saville B J, Collins R A. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 9.Gold L, Polisky B, Uhlenbeck O C, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 10.Hager A J, Pollard J D, Jr, Szostak J W. Chem Biol. 1996;3:717–725. doi: 10.1016/s1074-5521(96)90246-x. [DOI] [PubMed] [Google Scholar]
- 11.Lorsch J R, Szostak J W. Acc Chem Res. 1996;29:103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 12.Illangasekare M, Sanchez G, Nickles T, Yarus M. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- 13.Pan T, Gutell R R, Uhlenbeck O C. Science. 1991;254:1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- 14.Jayasena V K, Brown D, Shtatland T, Gold L. Biochemistry. 1996;35:2349–2356. doi: 10.1021/bi951879b. [DOI] [PubMed] [Google Scholar]
- 15.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 16.Uhlenbeck O C. Nature (London) 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 17.Watson N, Gurevitz M, Ford J, Apirion D. J Mol Biol. 1984;172:301–323. doi: 10.1016/s0022-2836(84)80028-5. [DOI] [PubMed] [Google Scholar]
- 18.Kierzek R. Nucleic Acids Res. 1992;20:5079–5084. doi: 10.1093/nar/20.19.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosaka H, Ogawa T, Sakamoto K, Yokoyama S, Takaku H. FEBS Lett. 1991;293:204–206. doi: 10.1016/0014-5793(91)81187-d. [DOI] [PubMed] [Google Scholar]
- 20.Surratt C K, Milan S C, Chamberlin M J. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan T, Uhlenbeck O C. Nature (London) 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- 22.Moroney S E, Piccirilly J A. Biochemistry. 1991;30:10343–10349. doi: 10.1021/bi00106a036. [DOI] [PubMed] [Google Scholar]
- 23.Sober H A, Harte R A, Sober E K. Handbook of Biochemistry: Selected Data for Molecular Biology. Cleveland: Chem. Rubber Co.; 1970. pp. G-3–G-98. [Google Scholar]
- 24.Collins R A, Olive J E. Biochemistry. 1993;32:2795–2799. doi: 10.1021/bi00062a009. [DOI] [PubMed] [Google Scholar]
- 25.Brown D M, Todd A R. In: The Nucleic Acids. Chargaff E, Davidson J N, editors. Vol. 1. New York: Academic; 1955. pp. 409–430. [Google Scholar]
- 26.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 27.Pan T, Long D M, Uhlenbeck O C. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 271–302. [Google Scholar]
- 28.Pyle A M. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 29.Dahm S C, Uhlenbeck O C. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 30.Uhlenbeck O C. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 31.Ruffner D E, Dahm S C, Uhlenbeck O C. Gene. 1989;82:31–41. doi: 10.1016/0378-1119(89)90027-9. [DOI] [PubMed] [Google Scholar]
- 32.Perrotta A, Been M D. Biochemistry. 1992;31:16–21. doi: 10.1021/bi00116a004. [DOI] [PubMed] [Google Scholar]
- 33.Richards F M, Wyckoff H W. In: The Enzymes. Boyer P, editor. Vol. 4. New York: Academic; 1971. pp. 647–672. [Google Scholar]
- 34.Feigon J, Dieckmann T, Smith F W. Chem Biol. 1996;3:611–617. doi: 10.1016/s1074-5521(96)90127-1. [DOI] [PubMed] [Google Scholar]
- 35.Gold L, Brown D, He Y, Shtatland T, Singer B, Wu Y. Proc Natl Acad Sci USA. 1997;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]