Abstract
The use of low molecular weight organic compounds to induce dimerization or oligomerization of engineered proteins has wide-ranging utility in biological research as well as in gene and cell therapies. Chemically induced dimerization can be used to activate intracellular signal transduction pathways or to control the activity of a bipartite transcription factor. Dimerizer systems based on the natural products cyclosporin, FK506, rapamycin, and coumermycin have been described. However, owing to the complexity of these compounds, adjusting their binding or pharmacological properties by chemical modification is difficult. We have investigated several families of readily prepared, totally synthetic, cell-permeable dimerizers composed of ligands for human FKBP12. These molecules have significantly reduced complexity and greater adaptability than natural product dimers. We report here the efficacies of several of these new synthetic compounds in regulating two types of protein dimerization events inside engineered cells—–induction of apoptosis through dimerization of engineered Fas proteins and regulation of transcription through dimerization of transcription factor fusion proteins. One dimerizer in particular, AP1510, proved to be exceptionally potent and versatile in all experimental contexts tested.
Many cellular processes are mediated by inducible protein–protein interactions (1). Chemical inducers of dimerization, or “dimerizers,” are a promising new class of reagents that enable the controlled association of proteins or protein domains and so provide a means of triggering specific signal transduction, protein recruitment, or gene transcription events independently of other cellular activities (2). Dimerizers can be used to trigger signal transduction pathways that are naturally regulated by inducible protein dimerizations when those proteins are engineered to contain drug-binding domains (3–8). Certain pathways can also be activated through dimerizer-dependent recruitment of proteins to the inner surface of the plasma membrane (4, 9, 10). Dimerizer-controlled transcription of a target gene is achieved through the drug-dependent reconstitution of a transcription factor comprising separately expressed DNA-binding and activation domains, each fused to drug-binding domains (10–12). In all cases, the use of dimerizers allows these activities to be promoted at will and in a dose-dependent and reversible fashion, suggesting a variety of experimental and, potentially, therapeutic applications.
To date, dimerizers have been based on natural-product compounds, such as FK506 (3, 10), rapamycin (11, 12), cyclosporin (6, 10), and coumermycin (13). FK1012, the most widely reported reagent, is a semisynthetic dimer of FK506 that can simultaneously bind two molecules of the FK506-binding protein FKBP12 (FKBP), thereby promoting the dimerization of FKBP-containing fusion proteins (3). Although FK1012 is an effective dimerizer in a number of applications, it is a large and complex molecule that is difficult to modify. Here, we describe a series of wholly synthetic FKBP dimerizers that are much simpler molecules than FK1012 and thereby can serve as a platform for tailoring dimerizers for specific applications and for optimizing pharmacological properties. We tested the activities of these synthetic dimerizers in two systems with potentially different geometric and kinetic constraints: induction of Fas-mediated apoptosis and activation of gene transcription.
MATERIALS AND METHODS
Chemical Synthesis.
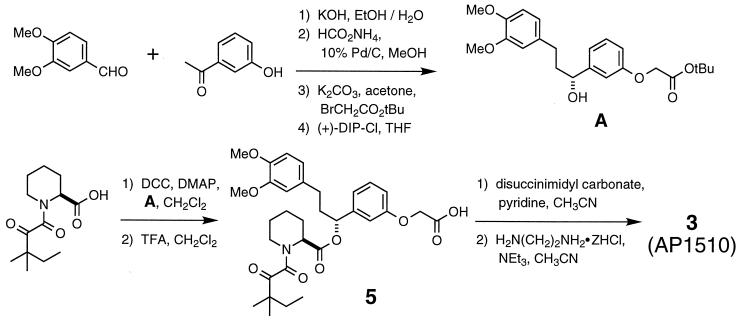
Details of this procedure will be published elsewhere. AP1510 was obtained by standard synthetic methods, as shown in Fig. 2.
Figure 2.
Synthesis of compound 3 (AP1510). Abbreviations: (+)-DIP-Cl, (+)-B-chlorodiisopinocampheylborane; THF, tetrahydrofuran; DCC, 1,3-dicyclohexylcarbodiimide; DMAP, 4-dimethylaminopyridine; TFA, trifluoroacetic acid; Me, methyl; Et, ethyl; tBu, tert-butyl.
Fas Construct.
The retroviral expression vector pSRα–myristoylation–2FKBP–Fas-E was constructed in the retroviral expression vector pSRα–MSV–tkneo (ref. 14; gift of D. Afar and O. Witte), modified by the addition of a BamHI site downstream of the original EcoRI cloning site and the elimination of a BamHI site elsewhere. The insert was assembled by standard subcloning procedures from the following DNA components: (i) the myristoylation sequence from v-src (residues 1–14) flanked by EcoRI and XbaI sites, derived by PCR of a plasmid encoding MF2FE (5), provided by David Spencer; (ii) an XbaI–2FKBP–SpeI fragment; (iii) XbaI–human Fas amino acids 175–304 followed by a SpeI site, a termination codon, and a BamHI site, derived by PCR from the plasmid encoding MF2FE; and (iv) a 16-amino acid Haemophilus influenzae hemaglutinin epitope flanked by XbaI and SpeI sites, derived from pCGNN (15).
Transcription Factor Expression Constructs.
Transcription factor fusions were expressed from the tricistonic vector pCGNN–F3p65/Z1F3/Neo (FKBPx3–p65–IRES–ZFHD1–FKBPx3–IRES–Neo). Details of the expression vector, pCGNN, and the cloning of individual components of the transcription factors have been described (11). To insert the ZFHD1–FKBPx3 and neo coding sequences 3′ to the encephalomyocarditis virus (EMCV) internal ribosome entry sequence (IRES), 3 nucleotides, ACC, were added immediately 5′ to the 11th ATG of the EMCV IRES to create a Kozak consensus sequence and a NcoI site that encompasses the ATG. The ZFHD1–FKBPx3 and neo genes were engineered to contain NcoI or compatible sites encompassing their ATGs, which were then used to fuse the genes to the 11th ATG of the IRES. Fragments encoding chimeric proteins and internal ribosome entry sequences were all generated with 5′ flanking XbaI ends and 3′ flanking SpeI and BamHI ends and assembled by stepwise insertion of XbaI–BamHI fragments into SpeI–BamHI opened vectors, as described (11).
Cell Line Expressing Fas Fusion Protein.
293T cells (16) were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum, penicillin, and streptomycin. HT1080 cells (ATCC CCL-121), derived from a human fibrosarcoma, were grown in minimal essential medium supplemented with nonessential amino acids, 10% fetal bovine serum, penicillin, and streptomycin.
Helper-free retroviruses were produced by transient cotransfection of 293T cells with a Psi(−) amphotropic packaging vector (a gift of D. Afar and O. Witte) and the Fas construct described above. HT1080 cells infected with retroviral stock in the presence of 8 μg/ml Polybrene were selected in the presence of 500 μg/ml G418. M45-11, an individual clone with a high sensitivity to dimerizer treatment, was used for the experiments shown.
Assay for Inducible Fas Activation in Cell Lines.
Cells were plated at 104 cells/well in 96-well plates. The next day, medium was removed and fresh medium containing 50 ng/ml Actinomycin D and dimerizer was placed on triplicate wells. The ethanol concentration was at or below 0.1%, which alone has no effect on the cells (data not shown). The next day, medium was replaced with medium containing 10% Alamar Blue (Accumed, Westlake, OH), a viability indicator that measures reducing equivalents released during cell metabolism. At intervals after addition, the relevant optical densities of the wells (OD570–OD600) were measured and normalized to the OD of untreated cells (=100%).
Transient Transfection and Measurement of Transcriptional Activation.
A HT1080 clone (HT1080L) containing a retrovirally integrated secreted alkaline phosphatase (SEAP) reporter gene under control of a minimal interleukin 2 gene promoter and 12 ZFDH1 binding sites was generated as described (11). HT1080L cells plated in 96-well dishes (1.5 × 104 cells/well) were transfected with 50 ng pCGNN–F3p65/Z1F3/Neo. Following lipofection, 200 μl medium containing the indicated amounts of test compound was added to each well. After 18–24 h, medium was removed and assayed for SEAP activity as described (11). Background SEAP activity, measured from mock-transfected HT1080 cells, was subtracted from each value.
Cell Line Expressing Transcription Factor Fusion Proteins.
A cell line that also contains the transcription factor fusion genes stably integrated was generated by transfecting HT1080L cells (Lipofectamine; GIBCO/BRL) with 2 μg pCGNN–F3p65/Z1F3/Neo and 0.2 μg pBabeNeo (17). Clones were selected in medium containing 500 μg/ml G418. Twenty clones were screened by Western blot for the presence of the activation domain fusion. Of the 13 clones that clearly expressed an appropriately sized protein, 5 were further tested for response to AP1510. All 5 exhibited at least 20-fold inducibility, and clone HT28-18, which produced the most SEAP in response to AP1510, was selected for further analysis.
RESULTS
Molecular Design.
Previous work has identified a series of pipecolyl α-ketoamides that bind to FKBP12 at low nanomolar concentrations, as measured by inhibition of the rotamase enzymatic activity of the protein (18). X-ray crystallographic analysis showed that these molecules bind in a fashion analogous to FK506. However, because they recapitulate only the FKBP-binding moiety of FK506, they are considerably lower in molecular weight and easier to synthesize. These studies provided a basis for the design of simple synthetic FKBP ligands and also suggested positions to which linker moieties could be connected for the construction of dimeric molecules.
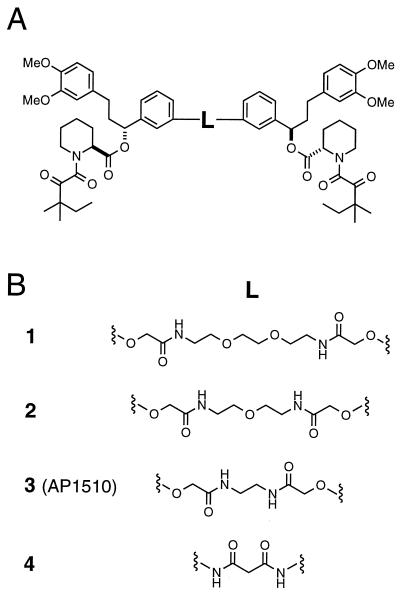
We synthesized and characterized a large set of compounds based on the pipecolyl α-ketoamides and other monomers (Fig. 1). Model building indicated that connecting two monomeric ligands with a linker of length ≈6–9 Å or greater, installed in the meta configuration on their phenyl rings (Fig. 1A), should sterically permit the simultaneous binding of two molecules of FKBP (data not shown). Homodimeric compounds of this general design were therefore prepared, incorporating a variety of monomeric subunits and linkers. We focus here on a set of four dimerizers based on a single pipecolyl α-ketoamide monomer joined by linkers of various lengths and compositions (Fig. 1B). The synthesis of one of the compounds, AP1510, is outlined in Fig. 2. More complete synthetic descriptions and structure–activity studies will be reported elsewhere.
Figure 1.
Chemical structures of synthetic dimerizers.
Inducible Fas Activation.
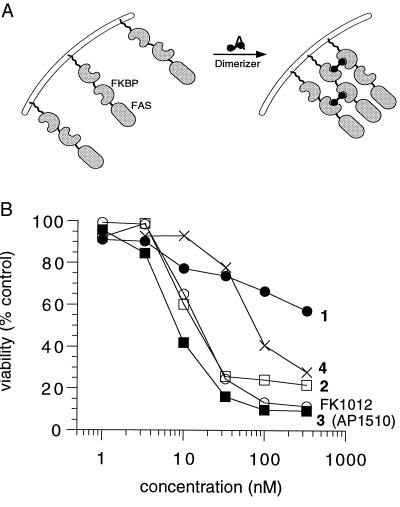
Previous studies have shown that apoptosis can be induced by dimerizers through the regulated oligomerization of fusion proteins carrying the intracellular signaling domain of the Fas receptor (5, 6, 19). The fusion protein consists of a myristoyl anchor to localize the protein to the plasma membrane, followed by two FKBPs and the intracellular domain of the human Fas protein (Fig. 3A). Jurkat cells or primary mouse thymocytes that express this fusion protein undergo efficient apoptosis in response to FK1012 (ref. 5; our unpublished observations). FK1012-induced apoptosis presumably proceeds through dimerizer-induced clustering of the signaling domain of Fas and subsequent activation of the endogenous Fas-induced signaling pathway.
Figure 3.
(A) Scheme for obtaining dimerizer-dependent Fas activation. The fusion construct used consists of a myristoylation sequence for membrane anchoring, followed by two FKBP12 domains and amino acids 175–304 of human Fas, which includes the cell-killing domain (20). Addition of dimerizer is presumed to induce clustering of multiple Fas death domains, thereby activating the Fas-mediated apoptotic pathway. The presence of two FKBP domains on each protein may enable clustering of more than two death domains. (B) Dimerizer-induced killing of cells expressing the dimerizer-dependent Fas construct. M45-11, a clone of HT1080 cells that is stably transduced with the retrovirus pSRα–myristoylation–2FKBP-Fas-E, was treated overnight with the concentration of dimerizer shown, and viability was then measured. Values shown are the means of triplicate wells.
To test the ability of the new synthetic dimerizers to trigger apoptosis, the fibrosarcoma cell line HT1080 was engineered to stably express the myristoylated FKBP–Fas construct. Test compounds were added to cells, and viability was measured ≈18 h later using an Alamar blue assay (Fig. 3B). FK1012 elicited the expected dose-dependent reduction in viability of the engineered cells, with half-maximal activity at roughly 20 nM. The new synthetic dimerizers also exhibited dose-dependent killing, but with a range of potencies. We noted an apparent inverse correlation of linker length and activity among compounds 1, 2, and 3 (Fig. 1). Compound 1, with a long 16-atom linker, elicited limited cell killing with a very shallow dose-response curve. By contrast compound 3 (AP1510), with a 10-atom linker, was more potent than FK1012 and showed half-maximal activity at about 6 nM. However, a dimerizer with a linker further shortened to only 5 atoms (compound 4) suffered a 10-fold drop in potency compared with 3, consistent with model-building suggestions that a linker of at least 6–9 Å should be optimal. None of the compounds had any effect on control cells that did not express the Fas–FKBP fusion protein, at concentrations up to 1 μM (data not shown).
Inducible Transcriptional Activation.
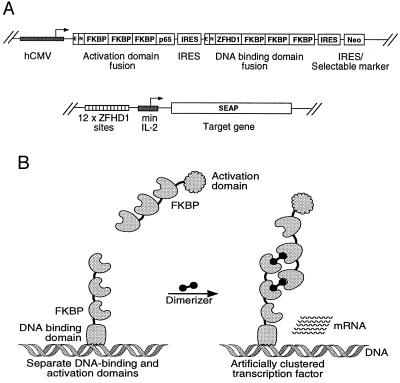
We next examined the utility of the synthetic dimerizers for regulating transcription. Here, compounds are required to mediate the formation of nuclear hetero-oligomers between DNA-binding and activation domains, in contrast to the membrane-bound homo-oligomers that elicit Fas activation. We assembled two genes encoding the human composite DNA-binding domain ZFHD1 (21) and the transcriptional activation domain of the human NF-κB p65 subunit (22), each fused to three FKBPs (Fig. 4A). For ease of use, we combined these genes for expression on a single polycistronic mRNA. The resulting plasmid was transiently transfected into an HT1080-derived cell line containing a retrovirally integrated SEAP reporter gene under the control of 12 binding sites for ZFHD1 (Fig. 4A). Thus, addition of dimerizer to these cells should lead to recruitment of p65 activation domains to the reporter gene, activating SEAP production (Fig. 4B). Transfected cells were treated with compounds for 18–24 h, and SEAP activity in the medium was assayed.
Figure 4.
Regulation of gene expression by synthetic dimerizers. (A) Schematic diagrams of plasmids used in the experiments. Transcription factor fusion proteins were produced from a tricistronic transcript under the control of the human cytomegalovirus (hCMV) immediate early promoter and enhancer. E represents an epitope tag and N represents the simian virus 40 (SV40) T-antigen nuclear localization sequence, both fused to the N-termini of the transcription factors. The SEAP reporter gene was driven by a minimal interleukin 2 (IL-2) promoter flanked by 12 tandemly reiterated binding sites for ZFHD1. (B) Scheme for obtaining dimerizer-dependent gene expression. Eukaryotic transcription factors are generally organized into two discrete functional domains, a DNA-binding domain and an activation domain. By fusion of a DNA-binding domain and an activation domain to the human protein FKBP, the interaction of the two fusion proteins can be made dependent on the presence of an FKBP-dimerizing drug. This interaction will result in dimerizer-dependent expression of a target gene carrying binding sites for the DNA-binding domain. Note that the fusion of three FKBPs to both the DNA-binding and activation domains gives each DNA-binding domain the potential to recruit a large number of activation domains: first, by recruiting multiple activation domain fusions directly, and then by having unoccupied FKBP domains on the activation domain fusion recruit additional activation domain fusions.
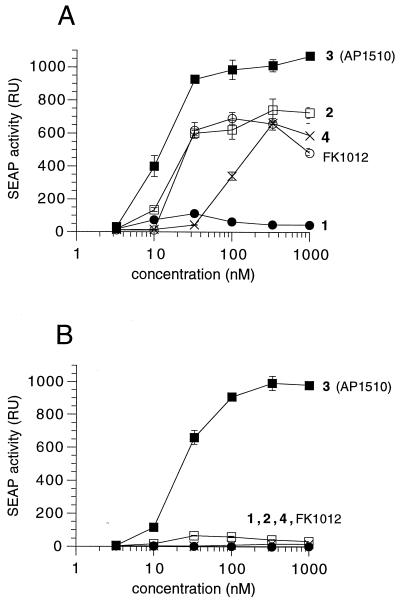
All of the dimerizers elicited dose-dependent transcriptional activation in these cells (Fig. 5A), although their potencies (measured as EC50, the concentration giving half-maximal activation) and the peak levels of SEAP expression varied widely. FK1012 activated with an EC50 of about 20 nM, as previously observed in yeast cells (12). Interestingly, the synthetic dimerizers 1, 2, and 3 again showed a clear inverse correlation between linker length and activity. Compound 1 activated to only a very low plateau level, whereas the peak expression with 2 was equivalent to that of FK1012 and that of compound 3 (AP1510) exceeded FK1012 levels by 50%. AP1510 also showed a slightly greater potency (EC50 10–20 nM). Finally, the very short 5-atom linker of compound 4 was again detrimental and reduced potency 10-fold although maximal expression was still equivalent to FK1012 and 2. Thus, the transcriptional data resemble the results observed for activation of Fas signaling.
Figure 5.
Dose-responsive activation of gene expression by synthetic FKBP dimerizers. HT1080L cells, which contain the pZHWTx12–IL2–SEAP reporter gene stably integrated, were (A) transiently or (B) stably transfected with a plasmid encoding FKBPx3–p65 and ZFHD1–FKBPx3 fusion proteins. SEAP activity secreted into the growth medium was measured following incubation of cells with the indicated concentration of dimerizer. Assays were performed in triplicate, and mean values (in relative units, RU) ± SD were plotted.
We observed substantially lower SEAP production when the transcription factor fusions contained fewer than three FKBP modules (data not shown). Presumably this reflects the role of avidity in assembling a stable complex of the fusion proteins and/or in recruiting a large number of activation domains to the promoter through multivalent clustering (Fig. 4B). As expected, transcription was also extremely low in the absence of dimerizer, so that the activation observed here with AP1510 represents an induction of several hundred-fold (data not shown). Overall, our data indicate that synthetic dimerizers, and in particular AP1510, can efficiently regulate target gene transcription in transiently transfected cells.
Inducible Transcriptional Activation in Stable Cell Lines.
Many regulated gene expression applications require the use of stably transfected cells in which the engineered transcription factors are expressed from genes integrated into chromatin. Because expression levels of the transcription factors are often considerably lower than those obtained transiently, stable cell lines provide a more stringent test for dimerizer performance. We therefore re-investigated the activity of our synthetic compounds using an HT1080 cell line that stably incorporated both the reporter gene and the expression plasmids for the transcription factor fusion proteins. As expected, Western blot and immunofluorescence analyses indicated that the per-cell expression level of the transcription factor fusion proteins was roughly 10-fold lower in this cell line than in transiently transfected cells (data not shown).
Interestingly, we found that several dimerizer molecules, including FK1012, were virtually inactive at inducing reporter gene expression in the stable cell line (Fig. 5B). The notable exception was AP1510 (compound 3), which elicited an equivalent induction to that seen in transiently transfected cells and with almost the same potency (EC50 20–25 nM). Therefore, within the set of compounds we tested, AP1510 was uniquely able to activate transcription in both transiently and stably transfected cells.
Transcriptional Activation by AP1510 Can Be Inhibited by Monomeric FKBP Ligands.
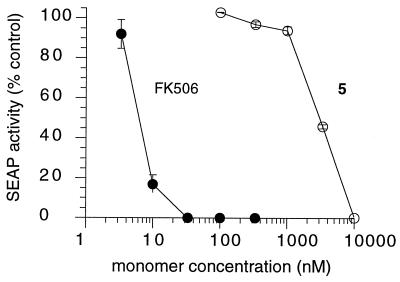
If the stimulation of transcription by synthetic dimerizers such as AP1510 is mediated by cross-linking of FKBP fusion proteins (Fig. 4B), then monomeric FKBP ligands should act as competitive inhibitors of gene activation. We therefore tested the abilities of FK506 and compound 5 (the monomeric precursor of synthetic dimerizers 1-4; Fig. 2) to inhibit the expression of SEAP induced by 100 nM AP1510, a maximally effective concentration (Fig. 5B), in the stable cell line. As expected, both compounds competitively inhibited the induction of SEAP expression, supporting the proposed mechanism of action of AP1510 (Fig. 6). The IC50 value for FK506 was about 6 nM, whereas the IC50 value for compound 5 was more than 300-fold higher. We expected FK506 to compete more potently because it has 25- to 50-fold higher affinity than 5 for FKBP [KD 0.4 nM vs. 10–20 nM (18)]. The ability to actively reverse dimerization using competitive monomers has a number of uses, including kinetic analysis of dimerizer-induced events (12).
Figure 6.
Dose-responsive inhibition of dimerizer-mediated gene expression by monomers. HT28-18 cells, which contain the pZHWTx12–IL2–SEAP reporter gene and a vector expressing the FKBPx3–p65 and ZFHD1–FKBPx3 fusion proteins stably integrated, were incubated with 100 nM AP1510 in the absence or presence of the indicated concentration of monomer. Eighteen hours later, SEAP activity secreted into the growth medium was measured. Assays were performed in triplicate, and mean values (in relative units) ± SD were plotted.
DISCUSSION
AP1510 Is a Potent Synthetic Dimerizer in Multiple Applications.
Our data demonstrate that simple, wholly synthetic dimerizers can be used to regulate protein-protein interactions inside cells. In particular, AP1510 potently activates Fas signaling and transcriptional activation in cells expressing appropriate FKBP fusion proteins. AP1510 has also been successfully used to activate signaling from several receptor tyrosine kinases including c-kit (C. A. Blau, personal communication) and epidermal growth factor receptor (S. Muthuswamy and M.G., unpublished results). Therefore, AP1510 appears to be a broadly applicable dimerizer, able to direct formation of both homo-oligomers and hetero-oligomers, to act in the nucleus and the cytoplasm and to function even when the proteins are expressed at low levels.
Factors Influencing Dimerizer Activity.
The ability to vary the design of dimerizers should allow a comprehensive study of structure–activity relationships. Here, we chose a single monomer subunit and examined the consequences of varying the linker. The substantial differences we observed suggest that the architecture of the dimeric protein complexes enforced by each compound—the proximity of the proteins, their relative orientation, and conformational flexibility—affects the biological potency of the complexes. Remarkably, despite the presumed different stoichiometric and geometric requirements of Fas signaling (Fig. 3A) and transcriptional activation (Fig. 4B), very similar trends were observed with this set of compounds. A dimerizer with a long linker (compound 1) led to very poor activation, suggesting that a minimum proximity or rigidity between the proteins may be required for biological activity. Activity then increased as linkers shortened, culminating in the 10-atom linker of AP1510 which is close to the optimal length suggested by model building. Further shortening (compound 4) still permitted full signaling, but with about ten-fold lower potencies (higher EC50 values), consistent with the notion that formation of active complexes is disfavored but that the complexes, once formed, are active. However, the potencies of signaling complexes assembled by different dimerizers will also be heavily influenced by the ratio of productive to nonproductive dimerization events they promote. Nonproductive interactions, such as dimerization between FKBP domains within the same protein or interactions with endogenous FKBP, may be minimized by the optimal linker of AP1510.
In addition to the intrinsic potencies of the dimerized complexes, other factors may be contributing to the different activities we observed. These include potential variations in the cell penetrability of the compounds and their physicochemical properties in aqueous solution. Also, despite the fact that the monomer subunits of each dimerizer are the same, the functional affinities of each dimerizer could be different because of positive or negative effects of FKBP-FKBP proximity. Nevertheless, our data show that the activity of synthetic dimerizers can be dramatically altered by appropriate engineering of the linker, as well as other portions of the molecules (unpublished results).
One important observation from this work is that synthetic dimerizers such as AP1510 are more active than FK1012 in a variety of settings, despite their presumed affinity for FKBP being 25- to 50-fold lower. Indeed, in stable cell lines only AP1510 is able to activate transcription appreciably (Fig. 5B). This result illustrates dramatically that dimerizer activity does not correlate simply with binding affinity. Further study of matched sets of dimerizers with different binding affinities should clarify the features responsible. One possibility is that the faster exchange kinetics associated with lower affinity interactions may permit rapid dissolution of nonproductive interactions and formation of productive ones, whereas slower dissociation may create a kinetic barrier to the assembly of large, productive oligomers.
Future Directions.
The development of wholly synthetic dimerizers should allow their optimization for individual applications. Although we found here that AP1510 is broadly active, some future applications may require molecules with different geometries, such as a longer linker. In addition, the ability to readily modify the compounds should facilitate efforts to improve their pharmacological properties for use in animals and eventually in the clinic. Finally, these compounds provide a starting point for designing second-generation molecules; for example, those which have been engineered (“bumped”) to bind only to a modified FKBP fusion protein and not to the endogenous wild-type protein, potentially diminishing nonproductive interactions and further improving performance.
Acknowledgments
We thank David M. Spencer for sharing plasmids and ideas and Owen Witte for sharing retroviral expression plasmids. We also thank Carl T. Rollins, Roberta Crowton, Lauren Stevenson for technical assistance, and David Berstein for helpful comments on the manuscript. AP1510 and plasmids that can be used for dimerizer-induced gene activation and protein oligomerization are being distributed to the research community. They can be requested through our web site at http://www.ariad.com.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: FKBP, human FK506 binding protein 12; SEAP, secreted alkaline phosphatase.
References
- 1.Austin D J, Crabtree G R, Schreiber S L. Chem Biol. 1994;1:131–136. doi: 10.1016/1074-5521(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 2.Spencer D M. Trends Genet. 1996;12:181–187. doi: 10.1016/0168-9525(96)10013-5. [DOI] [PubMed] [Google Scholar]
- 3.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 4.Holsinger L J, Spencer D M, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9810–9814. doi: 10.1073/pnas.92.21.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer D M, Belshaw P J, Chen L, Ho S N, Randazzo F, Crabtree G R, Schreiber S L. Curr Biol. 1996;6:839–847. doi: 10.1016/s0960-9822(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 6.Belshaw P J, Spencer D M, Crabtree G R, Schreiber S L. Chem Biol. 1996;3:731–738. doi: 10.1016/s1074-5521(96)90249-5. [DOI] [PubMed] [Google Scholar]
- 7.Luo Z, Tzivion G, Belshaw P J, Vavvas D, Marshall M, Avruch J. Nature (London) 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 8.Blau C A, Peterson K R, Drachman J G, Spencer D M. Proc Natl Acad Sci USA. 1997;94:3076–3081. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spencer D M, Graef I, Austin D J, Schreiber S L, Crabtree G R. Proc Natl Acad Sci USA. 1995;92:9805–9809. doi: 10.1073/pnas.92.21.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera V M, Clackson T, Natesan S, Pollock R, Amara J F, Keenan T, Magari S R, Phillips T, Courage N L, Cerasoli F, Jr, Holt D A, Gilman M. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 12.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 13.Farrar M A, Alberol-Ila J, Perlmutter R M. Nature (London) 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 14.Muller A J, Young J C, Pendergast A-M, Pondel M, Landau N R, Littman D R, Witte O. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attar R M, Gilman M Z. Mol Cell Biol. 1992;12:2432–2443. doi: 10.1128/mcb.12.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt D A, Luengo J I, Yamashita D S, Oh H-J, Konialian A L, Yen H-K, Rozamus L W, Brandt M, Bossard M L, Levy M A, Eggleston D S, Liang J, Schultz L W, Stout T J, Clardy J. J Am Chem Soc. 1993;115:9925–38. [Google Scholar]
- 19.Freiberg R A, Spencer D M, Choate K A, Peng P D, Schreiber S L, Crabtree G R, Khavari P A. J Biol Chem. 1996;271:31666–31669. doi: 10.1074/jbc.271.49.31666. [DOI] [PubMed] [Google Scholar]
- 20.Itoh N, Nagata S. J Biol Chem. 1993;268:10932–10937. [PubMed] [Google Scholar]
- 21.Pomerantz J L, Sharp P A, Pabo C O. Science. 1995;267:93–96. doi: 10.1126/science.7809612. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz M L, Baeuerle P A. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]