Abstract
The Tec family of tyrosine kinases are involved in signals emanating from cytokine receptors, antigen receptors, and other lymphoid cell surface receptors. One family member, ITK (inducible T cell kinase), is involved in T cell activation and can be activated by the T cell receptor and the CD28 cell surface receptor. This stimulation of tyrosine phosphorylation and activation of ITK can be mimicked by the Src family kinase Lck. We have explored the mechanism of this requirement for Src family kinases in the activation of ITK. We found that coexpression of ITK and Src results in increased membrane association, tyrosine phosphorylation and activation of ITK, which could be blocked by inhibitors of the lipid kinase phosphatidylinositol 3-kinase (PI 3-kinase) as well as overexpression of the p85 subunit of PI 3-kinase. Removal of the Pleckstrin homology domain (PH) of ITK resulted in a kinase that could no longer be induced to localize to the membrane or be activated by Src. The PH of ITK was also able to bind inositol phosphates phosphorylated at the D3 position. Membrane targeting of ITK without the PH recovered its ability to be activated by Src. These results suggest that ITK can be activated by a combination of Src and PI 3-kinase.
Inducible T cell kinase (ITK) (1–5) belongs to the Tec family of nonreceptor protein tyrosine kinases, which includes Tec I and II (6, 7), Bmx (8), Txk/Rlk (9, 10), and Bruton’s tyrosine kinase (BTK) (11, 12). ITK is expressed primarily in T cells (1–3, 5), with some expression detected in natural killer cells (4). Mutations in BTK, found in B cells, have been shown to be responsible for human X-linked agammaglobulinemia and the murine X-linked immunodeficiency (11–14). Similarly, mice lacking ITK have reduced numbers of mature thymocytes and reduced proliferative responses following T cell receptor (TcR) crosslinking (15). These reduced numbers in the absence of ITK suggest an important role for ITK in T cell development and function.
ITK, while similar to that of the Src family of protein tyrosine kinases, differ from Src kinases in a number of respects. Like Src family kinases, it has Src homology 3 (SH3) and Src homology 2 (SH2) domains. However ITK lacks a negative regulatory tyrosine at the carboxy termini. ITK also has a proline-rich region, included in a Tec homology domain (TH), reported to interact with Src family kinase SH3 domains in vitro and in the yeast 2 hybrid (16). ITK, like the other Tec family members (except for Txk), also has a Pleckstrin homology domain (PH), which is not found in the Src family kinases. PHs have been shown to bind to inositol lipids in vitro (17) and are proposed to anchor their carriers to membrane lipids (18, 19).
Phosphatidylinositol 3-kinase (PI 3-kinase) is a lipid kinase that phosphorylates inositol lipids at the D3 position of the inositol ring (20). It is known that the serine/threonine kinase ribosomal S-6 kinase (RSK) lies downstream of PI 3-kinase based on inhibition studies with PI 3-kinase inhibitors (21). Recently, the serine/threonine kinase AKT has been shown to be activated by inositol lipids generated by PI 3-kinase (22, 23). AKT has a PH in common with ITK, and the activation of AKT by PI 3-kinase may be mediated by binding of the PH of AKT to phosphorylated inositol lipids (23).
Ligation of the T cell receptor, CD28, a cell surface receptor found on T cells, and the FcɛR found on mast cells all result in tyrosine phosphorylation and activation of ITK (24–26). T cell receptor and CD28 induced tyrosine phosphorylation and activation is dependent on the presence of the Src family kinase Lck (25, 27). The association of ITK with CD28 also requires the presence of Lck (27, 28). This behavior is reminiscent of the Syk family kinases, which can be activated by Src family kinases (29). In view of these similarities, and the reported interaction between the TH domain and Src family kinases in vitro, we investigated the relationship between Src and ITK. Cellular Src (C-Src) is expressed in mast cells (30), thymocytes, and the Jurkat T cell line and is induced upon T cell receptor activation in mature T cells (31). It is also activated upon FcɛR signaling in mast cells (30) (which also activates ITK; ref. 26). We therefore used c-Src as a prototypical member of the Src family of tyrosine kinases in our analysis.
We found that coexpression of c-Src and ITK results in the tyrosine phosphorylation and activation of ITK. This tyrosine phosphorylation and activation of ITK by c-Src required the PH of ITK and was sensitive to inhibitors of PI 3-kinase, as well as overexpression of the p85 subunit of PI 3-kinase. In addition, coexpression of c-Src with ITK resulted in increased membrane localization of ITK, which was also dependent on the presence of the PH of ITK. However, an ITK mutant lacking this domain but membrane localized was able to be activated by Src, and no longer inhibitable by PI 3-kinase inhibitor. These data suggest that Src family kinases can activate ITK by a pathway that involves PI 3-kinase via a mechanism involving the PH of ITK.
MATERIALS AND METHODS
Expression Plasmids.
Full-length wild-type ITK cDNA was cloned in pCMVneo (1). C-Src, the p85 subunit of PI 3-kinase, and kinase-inactive (K295M) c-Src were all cloned into the pMEXneo expression vector. Hemagglutinin (HA)-tagged versions of both the wild-type and PH-deleted ITK were generated by PCR and cloned into pMEXHAneo. The kinase-inactive mutant of ITK (K391R) was made by overlap PCR and sequenced to confirm the mutation. The cellular Kit (c-Kit) fusions were generated by fusing the PH-deleted HA-tagged ITK to the cytoplasmic domain of the murine c-Kit at amino acid 549.
Cell Culture and Transfections.
COS-7 cells (American Type Culture Collection) and were grown in Dulbecco’s essential medium (DEM) containing 10% fetal calf serum and antibiotics at 37°C with 5% CO2. Cells were transiently transfected with calcium phosphate/DNA complexes as described (32). For inhibitor analysis, cells were split 1:2 two days following transfection. On the third day, cells were washed once with serum-free media and incubated in serum-free media for 30 min with the inhibitor or carrier. Cells were then harvested. Ly294002 was from Biomol (Plymouth Meeting, PA) and wortmannin was from Sigma.
Cell Lysis and Immunoprecipitation.
Cells were lysed in Triton X-100 lysis buffer containing phosphatase inhibitors as described (24). ITK was detected with either anti-sera to ITK or mAb to HA (Boehringer Ingleheim), and p85 with a mAb to the p85 subunit of PI 3-kinase. ITK immunoprecipitates were probed with an anti-phosphotyrosine antibody (RC20, Transduction Laboratories, Lexington, KY), then stripped as described (33) and probed with either anti-ITK antiserum or anti-HA antibody. Receptor chimeras were immunoprecipitated using an anti-c-Kit antibody and processed exactly as described above. Membrane and cytosolic fractions were obtained as described (34). Briefly, cells were harvested in hypotonic lysis buffer (20 mM Hepes⋅NaOH, pH 7.6/5 mM NaPPi/5 mM EGTA/1 mM MgCl2/1 mM phenylmethylsulfonyl fluoride/1 mM NaVO3) and Dounce homogenized. The homogenate was then centrifuged at 100,000 × g for 1 hr. The supernatant was taken as the cytosol, and the pellet was dissolved in Triton X-100 lysis buffer and the insoluble material spun out. This was taken as the solubilized membrane fraction.
Kinase Assays.
ITK was immunoprecipitated either with the anti-sera to ITK or antibody to HA and washed. Immune complex kinase assays were then performed. Kinase assays were performed using the src peptide as a substrate exactly as described (24). Briefly, the kinase reaction was performed for 15 min at room temperature in 50 μl kinase buffer (25 mM Hepes, pH 7.5/5 mM MnCl2) containing 10 μCi γ32P-ATP (3,000 Ci/mmol; 1 Ci = 37 GBq) and 5 μg RRsrc peptide (Sigma). Aliquots were spotted on phosphocellulose paper, washed with 1% phosphoric acid, and counted. Results are expressed as fold increase over ITK expressed alone, whose enzymatic activity was equated to 1, with activities corrected for expression levels. Contamination of the immunoprecipitates by Src was ruled by the failure of the PH-deleted mutants of ITK to be activated in the presence of Src using the same assay, as well as failure to detect Src in the immunoprecipitates of ITK by immunoblotting.
Inositol Lipids Binding Analysis.
The binding of inositol phosphates were performed exactly as described (35). Briefly, inositol phosphates (H3 inositol 1-; 1,3,4-; 1,4,5-; 1,3,4,5-; and 1,2,3,4,5- phosphates) from DuPont/NEN were incubated with glutathione S-transferase fusions of the PH of ITK, or the SH3 domain of c-Yes in 50 μl of Hepes⋅KOH (pH 7.2) for 10 min at 4°C. One microliter of Ig (50 mg/ml) was added, then 51 μl of 30% PEG6000 in 50 mM Hepes⋅KOH was then mixed in on ice for 5 min. Samples were centrifuged for 5 min at 10,000 × g, the pellet solubilized in scintillant, and counted. Background binding to the glutathione S-transferase/Yes SH3 was subtracted from all values.
RESULTS
Induction of Tyrosine Phosphorylation and Activation of ITK by c-Src.
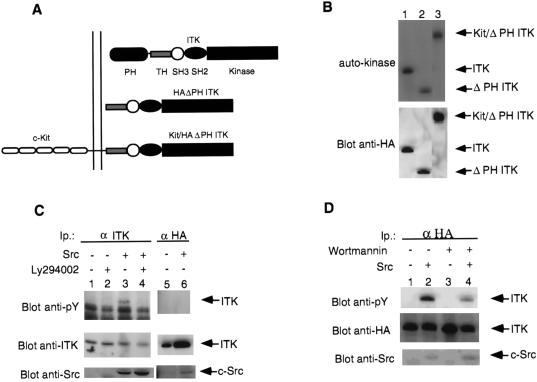
The similarities between Syk family kinases and ITK in their requirement for Src family kinases for activation prompted us to cotransfected expression plasmids carrying c-Src and wild-type ITK (see Fig. 1A for constructs, Fig. 1B for expression of the different proteins) into COS-7 cells to determine whether c-Src could activate ITK. Upon coexpression of c-Src and ITK, ITK becomes tyrosine phosphorylated (Fig. 1C), accompanied by up to 6-fold increase in its enzymatic activity against the exogenous substrate RRsrc (Fig. 2A). That the activity of endogenous Src is insufficient to activate ITK may reflect the fact that endogenous Src and family members are not activated, and that upon overexpression, the increased level of introduced Src results in its activation. Indeed, there is no difference in the ability of overexpressed wild-type c-Src and a constitutively active c-Src mutant Y527F in their ability to activate ITK (A.A. and H.H., unpublished work). A kinase-inactive form of c-Src was unable to induce tyrosine phosphorylatqion or activate ITK (Fig. 1C; note that we could not overexpress the kinase-inactive c-Src to the same level as the wild type, probably due to potential negative effects of such a mutant on cells). C-Src did not induce detectable tyrosine phosphorylation of a kinase-inactive mutant of ITK (A.A. and H.H., unpublished work). We have also observed that other Src family kinases, Lck and Fyn, have similar effects on ITK (ref. 27; A.A. and H.H., unpublished work). Together, these data indicate that c-Src can induce the tyrosine phosphorylation and activation of ITK.
Figure 1.
Structure of the different ITKs used in this study. (A) HAΔPH ITK: HA-tagged PH-deleted ITK; Kit/ΔPH ITK: murine c-Kit/HAΔPH ITK fusion. (B) Expression and kinase activity of ITK and mutants. Immunoprecipitates of ITK or mutants from transfected COS-7 cells were analyzed for autokinase activity (Top). Lanes: 1, ITK (wild-type); 2, ΔPH ITK; 3, Kit/ΔPH ITK. (Bottom) Probed with anti-HA. Arrows point to wild-type ITK or the mutants. (C) Tyrosine phosphorylation of ITK in the presence of Src and inhibition by Ly294002. COS-7 cells transfected with untagged wild-type ITK (except for lanes 5 and 6, where HA-tagged wild-type ITK was used) in the presence or absence of wild-type or kinase-inactive (K295M) c-Src as indicated. ITK immunoprecipitates were analyzed for enzymatic activity (see Fig. 2A) or phosphotyrosine (Top). Lanes 1, 2, and 5 contain ITK without c-Src; lanes 3, 4, and 6 contain ITK with c-Src. Lane 6 contains ITK cotransfected with the kinase-inactive c-Src. Lanes 2 and 4 were from cells incubated with Ly294002. (D) Inhibition of Src-induced tyrosine phosphorylation of ITK by wortmannin. COS-7 cells transfected with untagged wild-type ITK in the presence or absence of wild-type c-Src as indicated. ITK was immunoprecipitated and analyzed for phosphotyrosine (Top). Lanes: 1 and 3, ITK without c-Src; 2 and 4, ITK with c-Src. Lanes 3 and 6 were from cells incubated with wortmannin. Probes: (C and D Top) anti-phosphotyrosine antibodies; (Middle) anti-ITK antibodies; and (Bottom) anti-Src antibodies on whole cell lysates. Arrow indicates ITK or c-Src.
Figure 2.
Enzymatic activity of ITK and mutants in the presence or absence of Src and inhibitor. (A) Immunoprecipitates of ITK or mutants expressed in the presence of absence or Src and/or Ly294002 were assayed for kinase activity. ▪, ITK alone; ░⃞, ITK plus Src; □, ITK plus Ly294002. Assays were corrected for expression and expressed as n-fold increase over ITK expressed in the absence of Src. (B) Enzymatic activity of ITK and mutants in the presence or absence of Ly294002. COS-7 cells transfected with the indicated mutants and either left untreated or treated with Ly294002. ITK immunoprecipitates were assayed for in vitro kinase activity, then probed with anti-HA antibody (to detect ITK) (Lower). The filter was treated with KOH and exposed to x-ray film (Upper). Arrow indicates ITK or mutants.
Inhibition of c-Src-induced Activation of ITK By the PI 3-Kinase inhibitor Ly294002.
ITK has a PH and these domains have been demonstrated to bind to phosphorylated inositol lipids (17). The SH3 domain of Src can bind the p85 subunit of PI 3-kinase that can result in the activation of PI 3-kinase (35–39). Viral Src (v-Src) can be found in a complex with PI 3-kinase and D3 phosphorylated inositol lipids are elevated in cells transformed by v-Src (20). Src family kinases can also phosphorylate PI 3-kinase that may also result in the activation of PI 3-kinase (40, 41). Considering these data, we tested the inhibitors of PI 3-kinase, Ly294002 (21, 42), and wortmannin for their effect on the Src-induced activation of ITK. Incubation of COS-7 cells transfected either with ITK alone or ITK plus c-Src with Ly294002 (100 μM) resulted in marked inhibition of both the Src-induced tyrosine phosphorylation (Fig. 1C) and the enzymatic activation of ITK (Fig. 2A). Ly294002 added at a 5-fold-lower concentration (20 μM) inhibited tyrosine phosphorylation 50% without affecting enzymatic activity. At a 10-fold-lower (10 μM) concentration these effects were less apparent (A.A. and H.H., unpublished work). Similar results were obtained with wortmannin (10−7 M) (Fig. 1D). We continued with Ly294002 as it has been reported to specifically inhibit PI 3-kinase without having any effect on other serine kinases, in contrast to wortmannin (21, 42). Ly294002 did not affect the in vivo tyrosine kinase activity of Src (ref. 42; A.A. and H.H., unpublished work) or the enzymatic activity of ITK in the absence of Src (Fig. 2B).
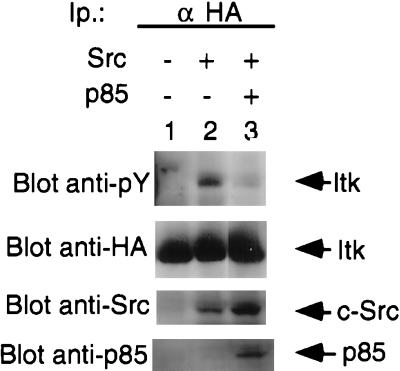
PI 3-kinase is composed of a regulatory subunit (p85) and a catalytic subunit (p110) (43). It has been shown that overexpression of the regulatory p85 subunit can act as a dominant negative for PI 3-kinase. We therefore overexpressed the p85 subunit and determined if it affected the ability of Src to induce tyrosine phosphorylation of ITK. We found that overexpression of the p85 subunit of PI 3-kinase significantly inhibited the ability of Src to induce tyrosine phosphorylation of ITK (Fig. 3).
Figure 3.
Overexpression of the p85 subunit of PI 3-kinase inhibits Src-induced tyrosine phosphorylation of ITK. ITK immunoprecipitates from COS-7 cells transfected with HA-tagged wild-type ITK in the presence or absence of c-Src and the p85 subunit of PI 3-kinase as indicated, were analyzed for phosphotyrosine (Top). Lanes: 1, ITK without c-Src; 2 and 3, ITK with c-Src; 3, ITK with c-Src and p85. (Upper Middle) Anti-HA immunoblot of the first panel demonstrating expression of ITK. (Lower Middle) Whole cell lysates probed for Src. (Bottom) Whole cell lysates probed for p85. Arrows indicate ITK, Src, and p85, respectively.
These results demonstrate that c-Src can induce the tyrosine phosphorylation of ITK that involves a PI 3-kinase pathway. As lipids generated by PI 3-kinase can activate protein kinase C-ζ (44, 45), we examined the effect of the protein kinase C inhibitor calphostin C on the activation of ITK. No inhibition of the c-Src-induced tyrosine phosphorylation of ITK was observed, suggesting that this activation was not through protein kinase C (A.A. and H.H., unpublished work).
Requirement for the PH of ITK for Membrane Localization and Activation in the Presence of Src.
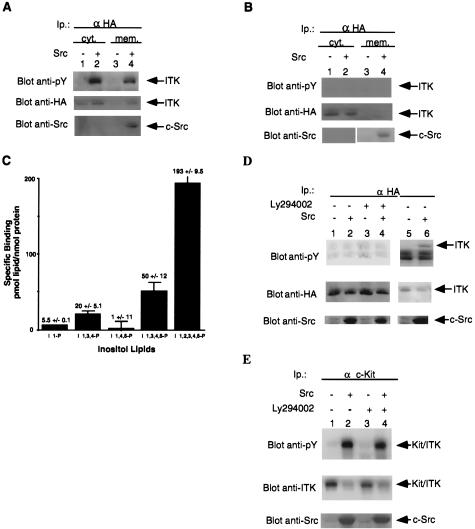
As PHs have been demonstrated to bind phospholipids (17) and to anchor proteins carrying them to membranes (18, 19), we tested ITK for membrane localization and activation in the presence or absence of Src. ITK immunoprecipitates from membrane and cytosolic fractions of COS-7 cells transfected with ITK with or without Src were tested for increased tyrosine phosphorylation and activity. In the presence of Src, there was increased association of ITK with the membrane, in addition to the previously noted increase in tyrosine phosphorylation (Fig. 4A) and enzymatic activity in both the membrane and cytosolic fractions. A mutant of ITK lacking the PH (ΔPH ITK) was not membrane associated in the presence or absence of Src, and was not tyrosine phosphorylated (Fig. 4D) or activated (Fig. 2A). Src protein could, however, be detected only in the membrane fractions (Fig. 4 A and B, Lower). This PH-deleted mutant possessed kinase activity equivalent to wild-type in vitro in the absence of Src (Fig. 2B). Our results therefore show that Src can induce the membrane association and activation of ITK through a PI 3-kinase pathway, which requires the PH of ITK. As activation of PI 3-kinase by Src results in the production of inositol D3 phosphorylated lipids at the membrane, we determined if ITK’s PH could bind to inositol D3 phosphates, thus explaining its Src-induced membrane association. We found that the PH of ITK could indeed bind to inositol phosphates only when they were phosphorylated at the D3 position (Fig. 4C).
Figure 4.
Requirement for the PH of ITK for Src-induced membrane localization and activation. (A) Src induces membrane localization of ITK. COS-7 cells were transfected with HA-tagged ITK in the presence or absence of Src. ITK was then immunoprecipitated from cytosolic and soluble membrane fraction and analyzed for phosphotyrosine content. Lanes: 1, cytosolic fraction/ITK alone; 2, cytosolic fraction/ITK plus Src; 3, membrane fraction/ITK alone; 4, membrane fraction/ITK plus Src. (B) The PH is required for Src-induced membrane association of ITK. COS-7 cells were transfected with HA-tagged ΔPH ITK in the presence or absence of Src. ΔPH ITK was immunoprecipitated from each fraction described above and analyzed for phosphotyrosine content. Lanes: 1, cytosolic fraction/ΔPH ITK alone; 2, cytosolic fraction/ΔPH ITK plus Src; 3, membrane fraction/ΔPH ITK alone; 4, membrane fraction/ΔPH ITK plus Src. Top was probed with anti-phosphotyrosine; Middle with anti-HA. Bottom shows equal amounts of protein from each fraction probed with anti-Src. Arrows point to ITK and Src, respectively. (C) Binding of the PH of ITK to inositol phosphates. Purified glutathione S-transferase-PH proteins were incubated with the indicated inositol phosphates as described in Materials and Methods. I 1-P, inositol 1-monophosphate; I 1,3,4-P; inositol 1,3,4-triphosphate; I 1,4,5-P, inositol 1,4,5-triphosphate; I 1,3,4,5-P, inositol 1,3,4,5-tetraphosphate; I 1,2,3,4,5-P, inositol 1,2,3,4,5-hexaphosphate. (D) Requirement for the PH of ITK for Src-induced activation. COS-7 cells were transfected with the HA-tagged ΔPH mutant of ITK in the presence or absence of c-Src. ITK immunoprecipitates were then analyzed for enzymatic activity (Fig. 2A) and phosphotyrosine content. Lanes: 1 and 3, ΔPH ITK without c-Src; 2 and 4, ΔPH ITK with c-Src. Lanes 3 and 4 were from cells treated with Ly294002. Probes: anti-phosphotyrosine (Top); anti-HA antibody (Middle); and (Bottom) anti-Src antibodies on whole cell lysates. Arrows point to the different mutants of ITK or c-Src. (E) Membrane bound ΔPH ITK recovers ability to be activated by Src. COS-7 cells were transfected with either Kit/ΔPH ITK alone or in the presence of c-Src. ITK was immunoprecipitated and analyzed for phosphotyrosine content. Lanes: 1 and 3, Kit/ΔPH ITK alone; 2 and 4, Kit/ΔPH ITK plus c-Src. Lanes 3 and 4 were from cells treated with Ly294002. Probes: (Top) anti-phosphotyrosine, (Middle) anti-ITK; and (Bottom) anti-src antibodies on whole cell lysates. Arrows indicate ITK or c-Src.
Membrane Targeting of the PH-Deleted ITK Recovers Its Ability To Be Activated by Src.
Removing the PH of ITK resulted in a kinase that was refractory to activation by Src. If the role of the PH is indeed to localize ITK to the membrane upon Src activation of PI 3-kinase, then membrane localization of the PH-deleted ITK mutant should recover its ability to be activated by Src. We therefore generated a membrane localized form of the PH-deleted ITK by fusing the murine c-Kit extracellular and transmembrane domains to the PH-deleted ITK (Kit/ΔPH ITK) (Fig. 1 A and B). While expression of the membrane targeted protein, Kit/ΔPH ITK without c-Src resulted in some tyrosine phosphorylation (Fig. 4E), coexpression with c-Src resulted in a large increase in tyrosine phosphorylation of this protein and increased enzymatic activity that was not inhibitable by the PI 3-kinase inhibitor (Figs. 4E and 2A). This suggests that membrane targeting bypassed the membrane recruitment step that required the PH. In the absence of Src, this protein had enzymatic activity equivalent to that of the wild-type (Fig. 1B).
DISCUSSION
Nonreceptor tyrosine kinases have been demonstrated to play important roles in signaling through receptors in lymphoid cells, particularly the antigen receptors. In this report, we have taken advantage of the fact that transient coexpression of Src family kinases and downstream effectors in COS-7 cells results in the activation of these downstream enzymes (46). We should, however, note that this system may not fully reflect all the signaling events during lymphocyte activation. We demonstrated that coexpression of ITK with c-Src resulted in membrane association, tyrosine phosphorylation, and activation of ITK, that required the activity of PI 3-kinase and the PH of ITK. We have also shown that the PH of ITK can bind inositol phosphates only when they are phosphorylated at the D3 position. Src and PI 3-kinase can thus cooperate in activating ITK.
PHs have been demonstrated to bind phosphorylated inositol lipids and as such are postulated to localized proteins carrying such domains to these regions of the membrane (19). Indeed, a mutation on the PH of BTK (E41K) results in its enrichment in membrane fractions when expressed in fibroblasts, and it was suggested that this may have resulted in its constitutive activation (47). The wild-type BTK has also been detected to undergo changes in membrane localization upon activation (48). We have also detected increases in membrane bound ITK coincident with activation in the presence of Src. Both events were dependent on the PH of ITK, suggesting an important role for the PH in membrane localization and the activation of ITK. However, our data suggest that membrane localization of ITK per se is not enough for full activation of ITK, and most likely that localization to activated Src kinases, as well as PI 3-kinase, may be important in the regulation of ITK. We postulated that membrane localization of ITK would also result in tyrosine phosphorylation of ITK similar to the case with the E41K mutant of BTK. However, significant tyrosine phosphorylation of membrane localized ITK continued to be dependent on the increased expression (and activity) of Src. This suggests that activation of ITK requires not only membrane localization, but we speculate, also proximity to activated Src. Activated PI 3-kinase could fulfill the requirement for this localization, as this enzyme would be active at the sites of activated Src kinase, and would then generate D3 phosphorylated lipids. Indeed, an active role for Src in the activation of ITK is suggested by the experiments where incubation of ITK with phosphatidyl inositol phosphates (PIP) lipids in vitro or coexpression of activated PI 3-kinase alone with ITK does not result in tyrosine phosphorylation or activation of ITK in the absence of coexpressed Src (A.A. and H.H., unpublished work). Interestingly, a membrane-targeted PH-deleted kinase-inactive mutant of ITK was tyrosine phosphorylated when coexpressed with Src (A.A. and H.H., unpublished work). This result suggests that ITK may be able to serve as a substrate of Src.
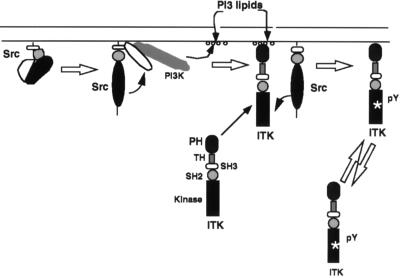
The data suggest a stepwise process of activation of ITK (see Fig. 5). Activation of Src results in the activation of PI 3-kinase, which then generates phosphorylated inositol lipids in this microenvironment. These lipids recruit ITK to those areas of activated Src and PI 3-kinase that require the PH of ITK. Once localized to Src, ITK is activated by Src, probably by direct phosphorylation, although we cannot rule out other mechanisms of activation. Our data demonstrate that the PH is required for the Src-induced membrane association and activation gives strong support for this proposal. The model is also supported by the finding that the PH of ITK can bind to inositol phosphates only when they are phosphorylated at the D3 position. Additional support comes from the finding that the PH of BTK can bind to inositol phosphates phosphorylated at the D3 position (35, 49). It has been reported that Src kinases can induce the tyrosine phosphorylation and activation of BTK (50, 51). However, these groups did not examine the role of PI 3-kinase in this activation and it remains to be seen if this enzyme also plays a role in the Src-induced activation of BTK.
Figure 5.
Proposed mechanism of activation of ITK by Src. See Materials and Methods for details. The arrow from Src to ITK is meant to indicate that Src acts on ITK in some fashion to induce its tyrosine phosphorylation and activation. Curved arrows are meant to indicate one enzyme acting on the other and straight arrows indicate the next step in the activation of ITK by Src.
Recently (52), the structure of the isolated TH and SH3 domains of ITK was solved by NMR. The structure suggested that the SH3 domain of ITK binds to the TH domain. It was proposed (52) that this would result in the inhibition of the enzyme. While we have not examined the role of the TH domain in the Src-induced activation of ITK, our results do not preclude a separate level of regulation of ITK, i.e., through its SH3/TH domain interaction.
Src family kinases have been reported to bind and activate PI 3-kinase (33, 34, 36–39). The consequence of this activation have hitherto been unknown. Our data suggest one function for this property of Src family kinases, namely, activation of Tec family kinases such as ITK (and probably BTK, and possibly the other family members). Interestingly, all the cell surface receptors reported to date that activate ITK also activate PI 3-kinase (33, 53–55), and we speculate a role for PI 3-kinase in their signaling pathways for activating ITK. If our data can be applied to BTK, they suggest a possible explanation for the both activating (BTKE41K) (47), as well as inactivating, mutations in the PH of BTK such as BTKR28C (13, 14). Increased association at the membrane, such as that seen in the BTK E41K mutant, would lead to an enzyme that is easier to activate by Src family kinases. By contrast, a mutant kinase with reduced ability to localize to the membrane may be compromised in its ability to be activated by Src family kinases.
Acknowledgments
We thank Drs. Spencer Gibson and Gordon Mills (M. D. Anderson Cancer Center, Houston) for the kind gift of anti-ITK antiserum and cDNA; Greg Stella and Dr. Peter Besmer (Memorial–Sloan Kettering Cancer Center, New York) for the murine c-Kit cDNA; Dr. Alvaro Monteiro for the c-Src plasmid; and other members of the Hanafusa laboratory for various reagents. Drs. Shinya Tanaka, Alvaro Monteiro, and Chris Marshall are thanked for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (CA-08748 and AI-37294 to B.D. and CA-44356 to H.H.). A.A. was supported in part by National Institutes of Health Training Grant CA-09673. A.A. is the recipient of a postdoctoral fellowship from the National Science Foundation.
ABBREVIATIONS
- ITK
inducible T cell kinase
- PH
Pleckstrin homology domain
- SH2 and SH3
Src homology domains 2 and 3
- TH
Tec homology domain
- BTK
Bruton’s tyrosine kinase
- HA
hemagglutinin
- PI 3-kinase
phosphatidylinositol 3-kinase
- c-Src
cellular Src
- c-Kit
cellular Kit
References
- 1.Gibson S, Leung B, Squire J A, Hill M, Arima N, Goss P, Hogg D, Mills G B. Blood. 1993;82:1561–1572. [PubMed] [Google Scholar]
- 2.Heyeck S D, Berg L J. Proc Natl Acad Sci USA. 1993;90:669–673. doi: 10.1073/pnas.90.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siliciano J D, Morrow T A, Desiderio S V. Proc Natl Acad Sci USA. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka N, Asao H, Ohtani K, Nakamura M, Sugamura K. FEBS Lett. 1993;324:1–5. doi: 10.1016/0014-5793(93)81520-a. [DOI] [PubMed] [Google Scholar]
- 5.Yamada N, Kawakami Y, Kimura H, Fukamachi H, Baier G, Altman A, Kato T, Inagi Y, Kawakami T. Biochem Biophys Res Commun. 1993;192:231–240. doi: 10.1006/bbrc.1993.1404. [DOI] [PubMed] [Google Scholar]
- 6.Mano H, Ishikawa F, Nishida J, Hirai H, Takahu F. Oncogene. 1993;5:1781–1786. [PubMed] [Google Scholar]
- 7.Mano H, Mano K, Tang B, Koehler M, Yi T, Gilbert D J, Jenkins N A, Copeland N G, Ihle J N. Oncogene. 1993;8:417–424. [PubMed] [Google Scholar]
- 8.Tamagonone L, Lahtinen I, Nustonen T, Virtaneva K, Francis F, Muscatelli F, Alitalo R, Smith C I E, Larsson C, Alitalo K. Oncogene. 1994;9:3683–3688. [PubMed] [Google Scholar]
- 9.Haire R N, Ohta Y, Lewis J E, Fu S M, Kroisel P, Litman G W. Hum Mol Genet. 1994;3:897–901. doi: 10.1093/hmg/3.6.897. [DOI] [PubMed] [Google Scholar]
- 10.Hu Q, Davidson D, Schwartzberg P L, Macchiarini F, Lenardo M J, Bluestone J A, Matis L A. J Biol Chem. 1995;279:1928–1934. doi: 10.1074/jbc.270.4.1928. [DOI] [PubMed] [Google Scholar]
- 11.Tsukuda S, Saffron D C, Rawlings D J, Parolini O, Allen R C, Klisak I, Sparkes R S, Kubagawa H, Mohandas T, Quan S, Belmont J W, Cooper M D, Conley M E, Witte O N. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 12.Vetrie D, Vorechovsky I, Sideras P, Holland J, Davies A, Flinterf F, Hammarstrom L, Kinnon C, Levinsky R, Bobrow M, Smith C I E, Bentley D R. Nature (London) 1993;361:226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 13.Rawlings D J, Saffran D C, Tsukuda S, Largaespada A, Grimaldi J C, Cohen L, Mohr R N, Bazan J F, Howard M, Copeland N G, Jenkins N A, Witte O N. Science. 1993;261:358–361. doi: 10.1126/science.8332901. [DOI] [PubMed] [Google Scholar]
- 14.Thomas J D, Sideras P, Smith C I E, Vorechovsky I, Chapman V, Paul W E. Science. 1993;261:355–358. doi: 10.1126/science.8332900. [DOI] [PubMed] [Google Scholar]
- 15.Liao X C, Littman D R. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 16.Cheng G, Ye Z-S, Baltimore D. Proc Natl Acad Sci USA. 1994;91:8152–8155. doi: 10.1073/pnas.91.17.8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlan J E, Hajduk P J, Yoon H S, Fesik S W. Nature (London) 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson K M, Lemmon M A, Schlessinger J, Sigler P B. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 19.Lemmon M A, Ferguson K M, Schlessinger J. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 20.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 21.Cheatham B, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgering B M, Coffer P J. Nature (London) 1995;376:553–554. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 23.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 24.August A, Gibson S, Kawakami Y, Kawakami T, Mills G B, Dupont B. Proc Natl Acad Sci USA. 1994;91:9347–9351. doi: 10.1073/pnas.91.20.9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson S, August A, Kawakami T, Kawakami Y, Dupont B, Mills G B. J Immunol. 1996;156:2716–2722. [PubMed] [Google Scholar]
- 26.Kawakami Y, Yao L, Tashiro M, Gibson S, Mills G B, Kawakami T. J Immunol. 1995;155:3556–3562. [PubMed] [Google Scholar]
- 27.Gibson S, August A, Branch D, Dupont B, Mills G B. J Biol Chem. 1996;271:7079–7083. doi: 10.1074/jbc.271.12.7079. [DOI] [PubMed] [Google Scholar]
- 28.Raab M, Cai Y-C, Bunnell S C, Heyeck S D, Berg L J, Rudd C E. Proc Natl Acad Sci USA. 1995;92:8891–8895. doi: 10.1073/pnas.92.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howe L R, Weiss A. Trends Biochem Sci. 1995;20:59–64. doi: 10.1016/s0968-0004(00)88958-6. [DOI] [PubMed] [Google Scholar]
- 30.Eisenman E, Bolen J B. Nature (London) 1992;355:78–80. doi: 10.1038/355078a0. [DOI] [PubMed] [Google Scholar]
- 31.Branch D R, Mills G B. J Immunol. 1995;154:3678–3685. [PubMed] [Google Scholar]
- 32.Sabe H, Okada M, Nakagawa H, Hanafusa H. Mol Cell Biol. 1992;12:4706–4713. doi: 10.1128/mcb.12.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.August A, Dupont B. Int Immunol. 1994;6:769–774. doi: 10.1093/intimm/6.5.769. [DOI] [PubMed] [Google Scholar]
- 34.Fujioka Y, Matazaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada T, Kasuga M. Mol Cell Biol. 1996;16:6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukuda M, Kojima T, Kabayama H, Mikoshiba K. J Biol Chem. 1996;271:30303–30306. doi: 10.1074/jbc.271.48.30303. [DOI] [PubMed] [Google Scholar]
- 36.Fukui Y, Hanafusa H. Mol Cell Biol. 1989;9:1651–1658. doi: 10.1128/mcb.9.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad K V S, Kapeller R, Janssen O, Repke H, Duke-Cohan J S, Cantley L C, Rudd C E. Mol Cell Biol. 1993;13:7708–7717. doi: 10.1128/mcb.13.12.7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee L B, Fujita D J. Mol Cell Biol. 1993;13:7408–7417. doi: 10.1128/mcb.13.12.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pleiman C M, Hertz W M, Cambier J C. Science. 1994;263:1609–1612. doi: 10.1126/science.8128248. [DOI] [PubMed] [Google Scholar]
- 40.Karnitz L M, Sutor S L, Abraham R T. J Exp Med. 1994;179:1799–1808. doi: 10.1084/jem.179.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Willebrand M, Baier G, Couture C, Burn P, Mustelin T. Eur J Immunol. 1994;24:234–238. doi: 10.1002/eji.1830240137. [DOI] [PubMed] [Google Scholar]
- 42.Vlahos C J, Matter W F, Hui K Y, Brown R. J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 43.Hiles I D, Otsu M, Volinia S, Fry M J, Gout I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty N F, Hsuan J J, Courtneidge S A, Parker P J, Waterfield M D. Cell. 1992;70:419–429. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi H, Brewer K A, Exton J H. J Biol Chem. 1993;268:13–17. [PubMed] [Google Scholar]
- 45.Toker A, Meyer M, Reddy K K, Falck J R, Aneja R, Aneja S, Parra A, Burns D J, Ballas L M, Cantley L C. J Biol Chem. 1994;269:32358–32367. [PubMed] [Google Scholar]
- 46.Chan A C, Iwashima M, Turck C W, Weiss A. Cell. 1992;71:649–662. doi: 10.1016/0092-8674(92)90598-7. [DOI] [PubMed] [Google Scholar]
- 47.Li T, Tsukuda S, Satterthwaite A, Havlik M H, Park H, Takatsu K, Witte O N. Immunity. 1995;2:451–460. doi: 10.1016/1074-7613(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 48.Kawakami Y, Yao L, Miura T, Tsukuda S, Witte O N, Kawakami T. Mol Cell Biol. 1994;14:5108–5113. doi: 10.1128/mcb.14.8.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salim K, Bottomley M J, Querfurth E, Zvelebil M J, Gout I, Scaife R, Margolis R L, Gigg R, Smith C I E, Driscoll P C, Waterfield M D, Panayotou G. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 50.Mahanjan S, Fargnoli J, Burkhardt A L, Kut S A, Saouaf S J, Bolen J B. Mol Cell Biol. 1995;15:5304–5311. doi: 10.1128/mcb.15.10.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rawlings D J, Schrenberg A M, Park H, Wahl M I, Lin S, Kato R M, Fluckinger A-C, Witte O N, Kinet J-P. Science. 1996;271:822–825. doi: 10.1126/science.271.5250.822. [DOI] [PubMed] [Google Scholar]
- 52.Andreotti A H, Bunnell S C, Feng S, Berg L J, Schreiber S L. Nature (London) 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 53.Exley M, Varticovski L, Peter M, Sancho J, Terhorst C. J Biol Chem. 1994;269:15140–15146. [PubMed] [Google Scholar]
- 54.Pages F, Ragueneau M, Rottapel R, Truneh A, Nunes J, Imbert J, Olive D. Nature (London) 1994;369:327–329. doi: 10.1038/369327a0. [DOI] [PubMed] [Google Scholar]
- 55.Prasad K V, Cai Y C, Raab M, Duckworth B, Cantley L, Shoelson S E, Rudd C E. Proc Natl Acad Sci USA. 1994;91:2834–2838. doi: 10.1073/pnas.91.7.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]