Abstract
Protein kinase C (PKC) isoforms, α, βI, and γ of cPKC subgroup, δ and ɛ of nPKC subgroup, and ζ of aPKC subgroup, were tyrosine phosphorylated in COS-7 cells in response to H2O2. These isoforms isolated from the H2O2-treated cells showed enhanced enzyme activity to various extents. The enzymes, PKC α and δ, recovered from the cells were independent of lipid cofactors for their catalytic activity. Analysis of mutated molecules of PKC δ showed that tyrosine residues, which are conserved in the catalytic domain of the PKC family, are critical for PKC activation induced by H2O2. These results suggest that PKC isoforms can be activated through tyrosine phosphorylation in a manner unrelated to receptor-coupled hydrolysis of inositol phospholipids.
Protein kinase C (PKC) is activated by 1,2-diacylglycerol produced from receptor-mediated hydrolysis of inositol phospholipids (1). PKC comprises a large family of multiple isoforms with regulatory and catalytic domains in the amino- and carboxyl-terminal halves, respectively. The isoforms are divided into three subgroups, cPKC, nPKC, and aPKC, due to the structural differences in their regulatory domains. Among PKC isoforms, PKC δ has been shown to be phosphorylated on tyrosine in v-ras-transformed keratinocytes (2) and in various cells stimulated with mitogens such as phorbol ester and growth factors (3–6). PKC δ has also reported to be phosphorylated in vitro by various tyrosine kinases such as Fyn (3, 6, 7), c-Src (6–8), and growth factor receptors (3, 6). Thus far, Tyr52 (9) and Tyr187 (10) in its amino-terminal half have been identified as the phosphorylation sites. The functional consequence of this reaction, however, remains controversial; the tyrosine phosphorylation reduces its phorbol ester-dependent catalytic activity in some cells (2, 6), whereas the reaction enhances the enzyme activity in some other cells (3). On the other hand, oxidative stress has been reported to induce prolonged activation of PKC within the cells (11–14). It was postulated that oxidative modification, which occurs probably in the regulatory domain, may generate active enzyme (14), but no evidence is available for such modification. Here, we report that, upon treatment of cells with H2O2, all PKC isoforms examined, including α, βI, and γ of cPKC, δ and ɛ of nPKC, and ζ of aPKC, are tyrosine phosphorylated and catalytically activated, and that tyrosine residues in the catalytic domain are crucial for activation of these enzymes.
MATERIALS AND METHODS
Expression Plasmids.
Hemagglutinin (HA)-epitope-tagged expression plasmids of PKC isoforms were constructed as described (15). Plasmid pTB801 was employed as an expression plasmid of PKC δ without epitope tag (16). Lys376 of PKC δ was replaced with methionine by site-directed mutagenesis to make HA-epitope-tagged kinase-negative PKC δ (17). FLAG-epitope-tagged expression plasmids of PKC δ were constructed by using pECE vector (17). Tyr451, Tyr469, Tyr512, and Tyr523 of PKC δ were replaced with phenylalanine by site-directed mutagenesis. The DNA sequences of these constructs were confirmed by the dideoxynucleotide chain-termination method using a DNA Sequencing System model 373A (Applied Biosystems).
Cells and Transfection.
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum at 37°C in a 5% CO2/95% air incubator. Cells were transfected with the expression plasmids by electroporation, cultured for 48 h (17), and treated as described in each experiment.
Immunoprecipitation.
All procedures were carried out at 0–4°C as described (17). Cells were washed with phosphate-buffered saline and lysed in 20 mM Tris⋅HCl at pH 7.5 containing 1 mM EDTA, 1 mM EGTA, 10 mM 2-mercaptoethanol, 1% Triton X-100, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, and 50 μg/ml phenylmethylsulfonyl fluoride. The lysate was centrifuged for 10 min at 18,000 × g, and the supernatant was incubated for 1 h with an anti-HA monoclonal antibody (Boehringer Mannheim), a polyclonal antibody against PKC δ (Transduction Laboratories, Lexington, KY), or an anti-FLAG monoclonal antibody (Kodak Scientific Imaging Systems). Then, protein A-Sepharose beads (Pharmacia) were added to the mixture and incubated for 30 min. The immunoprecipitates were collected by centrifugation and washed four times with 20 mM Tris⋅HCl at pH 7.5 containing 150 mM NaCl and 1% Triton X-100.
Immunoblot Analysis.
The immunoprecipitates were boiled in SDS sample buffer, and proteins were separated by SDS/PAGE and transferred onto an Immobilon P membrane (Millipore). Immunoblot analysis was carried out using the anti-HA antibody, the anti-FLAG antibody, a monoclonal anti-phosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY), or an antibody against the carboxyl-terminal region of PKC δ (GIBCO/BRL) as the first antibody, and alkaline phosphatase-conjugated second antibodies (Promega) as described (17).
Metabolic Labeling.
Transfected cells were cultured for 1 h in phosphate-free DMEM supplemented with dialyzed fetal calf serum, and then metabolically labeled for 3 h with [32P]orthophosphate (18 MBq/6-cm dish). PKC was then immunoprecipitated and was separated by SDS/PAGE, and phosphoamino acid analysis was carried out as described (18). The radioactive phosphoproteins and phosphoamino acids were visualized by a Bio-Imaging Analyzer (Fuji).
Protein Kinase Assay.
PKC activity was assayed by measuring the incorporation of 32Pi into calf thymus H1 histone from [γ-32P]ATP in the reaction mixture containing 20 mM Tris⋅HCl at pH 7.5, 10 mM MgCl2, 20 μM ATP, 15–50 kBq of [γ-32P]ATP, and 200 μg/ml H1 histone (16, 17). The incubation was carried out for 5 min at 30°C, and the phosphorylated proteins were separated by SDS/PAGE and visualized and quantitated by measuring the intensity of photostimulated luminescence (PSL) using a Bio-Imaging Analyzer. Where indicated, PKC activity was measured in the presence of 8 μg/ml phosphatidylserine, 0.8 μg/ml diacylglycerol, or 0.5 mM CaCl2.
One-Dimensional Peptide Mapping.
FLAG-epitope-tagged PKC δ (approximately 0.5 μg) was purified from the H2O2-treated COS-7 cells by FLAG M2 affinity gel column chromatography (19). The enzyme was digested partially with tosylphenylalanine chloromethyl ketone (TPCK)-treated trypsin (0.4 pmol) at 30°C for the indicated time in 20 mM Tris⋅HCl at pH 7.5 containing 0.5 mM EDTA, 0.5 mM EGTA, and 10 mM 2-mercaptoethanol. The digested samples were subjected to immunoblot analysis.
RESULTS
Tyrosine Phosphorylation of PKC in Cells Exposed to H2O2.
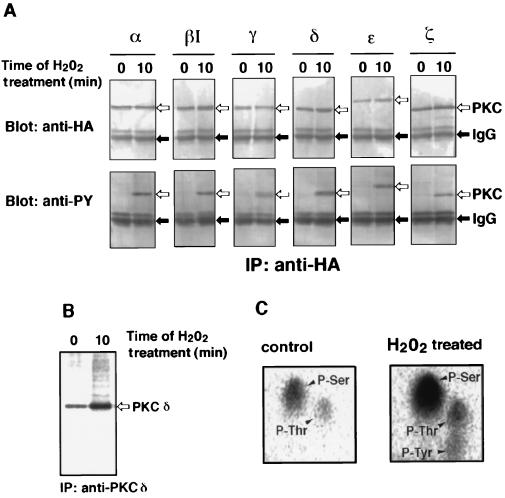
When COS-7 cells that were transfected with epitope-tagged PKC isoform of α, βI, γ, δ, ɛ, or ζ were stimulated with H2O2, all PKC isoforms were tyrosine phosphorylated as judged by immunoblot analysis (Fig. 1A). 32P-Tracer experiment with PKC δ, for example, showed that H2O2 enhances phosphorylation of this isoform (Fig. 1B). Phosphoamino acid analysis revealed that tyrosine residue as well as serine and threonine residues are indeed phosphorylated in response to H2O2 (Fig. 1C).
Figure 1.
Tyrosine phosphorylation of PKC isoforms in transfected COS-7 cells exposed to H2O2. (A) Tyrosine phosphorylation of PKC isoforms. Cells transfected with each HA-epitope-tagged PKC expression plasmid were incubated with 5 mM H2O2 for 10 min, and PKC isoforms were immunoprecipitated (IP) by the anti-HA antibody. Immunoblot analysis was carried out using the anti-HA antibody (Upper) or the anti-phosphotyrosine antibody (anti-PY) (Lower). Positions of PKC isoforms and IgG are indicated by open and closed arrows, respectively. (B) Phosphorylation of PKC δ. Cells transfected with the expression plasmid of PKC δ without epitope tag were labeled metabolically with [32P]orthophosphate and incubated with 5 mM H2O2 for 10 min. PKC δ was immunoprecipitated by the antibody against PKC δ. The phosphorylated PKC δ was visualized by a Bio-Imaging Analyzer. The position of PKC δ is indicated by an open arrow. (C) Phosphoamino acid analysis of PKC δ. The phosphorylated PKC δ recovered form the gel in B was subjected to phosphoamino acid analysis. Positions of phosphoserine (P-Ser), phosphothreonine (P-Thr), and phosphotyrosine (P-Tyr) are indicated by arrowheads.
Activation of PKC in Cells Exposed to H2O2.
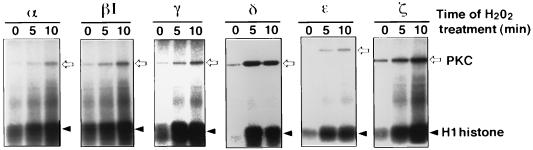
PKC isoforms obtained from the H2O2-treated cells were assayed for their enzyme activity with H1 histone as substrate (Fig. 2). The results showed that PKC δ, ɛ, and ζ are activated severalfold and that PKC α, βI, and γ are stimulated to lesser extent (Table 1). Autophosphorylation of these PKC isoforms was also enhanced (Fig. 2). Menadione, which generates superoxide anion, also induced tyrosine phosphorylation as well as activation of PKC isoforms (data not shown).
Figure 2.
Activation of PKC isoforms by H2O2 in transfected COS-7 cells. Cells transfected with each HA-epitope-tagged PKC expression plasmid were incubated with 5 mM H2O2 for the indicated time. PKC isoforms were immunoprecipitated by the anti-HA antibody, and PKC activity was assayed with H1 histone as substrate. Phosphorylated proteins were separated by SDS/PAGE and visualized by using a Bio-Imaging Analyzer.
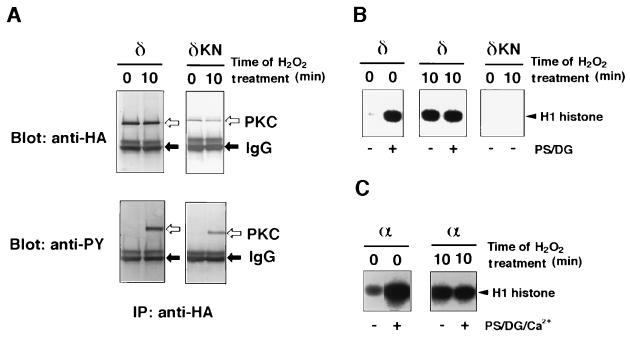
Next, catalytic properties of tyrosine-phosphorylated PKC isoforms were studied (Fig. 3). In response to H2O2, a kinase-negative mutant of PKC δ was tyrosine phosphorylated as well (Fig. 3A), although it did not show any enzyme activity either before or after H2O2 stimulation (Fig. 3B). Thus, the protein kinase activity measured in this experiment represents PKC δ itself, but is not due to any other protein kinases that might contaminate the immunoprecipitates. It is worth noting that PKC δ obtained from the H2O2-treated cells was not activated by phosphatidylserine and diacylglycerol, whereas the enzyme from the untreated cells required these lipids for enzyme activity (Fig. 3B). Similarly, PKC α prepared from the H2O2-treated cells did not require Ca2+, phosphatidylserine, or diacylglycerol (Fig. 3C). The results are given quantitatively in Table 2. It was concluded that PKC isoforms phosphorylated on tyrosine are almost fully active without Ca2+ and lipid activators. The stoichiometry of tyrosine phosphorylation is unknown, but PKC isoforms recovered form the H2O2-treated cells seem to be phosphorylated on at least one tyrosine residue, because the enzyme was almost quantitatively trapped by an anti-phosphotyrosine agarose column (data not shown). H2O2-induced tyrosine phosphorylation and activation of the PKC isoforms were observed also for NIH 3T3 cells, which were employed as host cells instead of COS-7 cells (data not shown).
Figure 3.
Properties of tyrosine-phosphorylated PKC recovered from transfected COS-7 cells. (A) Expression of PKC δ and kinase-negative PKC δ. Cells transfected with HA-epitope-tagged plasmid of either PKC δ or kinase-negative PKC δ (δKN) were incubated with 5 mM H2O2 for 10 min. The expressed proteins were immunoprecipitated by the anti-HA antibody. Immunoblot analysis was carried out using the anti-HA antibody (Upper) or the anti-phosphotyrosine antibody (Lower). Positions of PKC δ and IgG are indicated by open and closed arrows, respectively. (B) Protein kinase assay of PKC δ. Protein kinase activity of the immunoprecipitates recovered in A was assayed with H1 histone as substrate in the presence or absence of 8 μg/ml phosphatidylserine (PS) and 0.8 μg/ml diacylglycerol (DG). (C) Protein kinase assay of PKC α. HA-epitope-tagged PKC α was expressed, immunoprecipitated as in A, and assayed as in B in the presence of 0.5 mM Ca2+, phosphatidylserine, and diacylglycerol.
Tyrosine Phosphorylation Sites in PKC Induced by H2O2 Treatment.
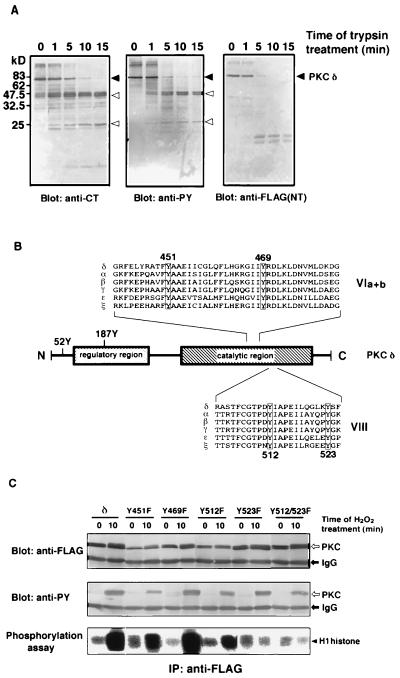
Tyr52 (9) and Tyr187 (10) in the amino-terminal half of PKC δ were previously shown to be phosphorylated. Mutated molecules of PKC δ deleting the amino-terminal region, including these two tyrosine residues, were still phosphorylated on tyrosine in response to H2O2 (data not shown). The molecule of PKC δ recovered from the H2O2-treated cells was digested partially with trypsin and subjected to immunoblot analysis to explore the region of tyrosine phosphorylation (Fig. 4A). Tryptic fragments with approximate molecular mass of 25 and 45 kDa were detected by the antibody against phosphotyrosine as well as the antibody recognizing the carboxyl terminus of PKC δ, but not by the antibody against the epitope tag linked to the amino terminus of the enzyme. The results indicated that PKC δ is phosphorylated on tyrosine in its carboxyl-terminal half region including the catalytic domain.
Figure 4.
Tyrosine phosphorylation sites in PKC. (A) One-dimensional peptide mapping analysis by partial proteolysis with trypsin. FLAG-epitope-tagged PKC δ purified from the H2O2-treated cells was digested with trypsin for the indicated times. Immunoblot analysis was carried out using either the antibody against the carboxyl-terminal region of PKC δ (anti-CT) (Left), the anti-phosphotyrosine antibody (anti-PY) (Center), or the anti-FLAG antibody [anti-FLAG(NT)] (Right). Positions of FLAG-epitope-tagged PKC δ and of tyrosine-phosphorylated fragments are indicated by closed and open arrows, respectively. Molecular mass markers are shown in kDa. (B) Structure and phosphorylation sites of PKC δ. The structure of PKC δ is schematically shown, and amino acid sequences of subdomain VIa, VIb, and VIII of rat PKC isoforms are aligned. Two phosphorylation sites identified in the regulatory domain of PKC δ, Tyr52 and Tyr187, are indicated as 52Y and 187Y. The tyrosine residues conserved in the carboxyl-terminal half of the PKC family are highlighted. (C) Site-directed mutagenesis of PKC δ. COS-7 cells transfected with the expression plasmid of FLAG-epitope-tagged PKC δ or with mutated molecules replacing tyrosine by phenylalanine were incubated with 5 mM H2O2 for 10 min, and FLAG-epitope-tagged molecules were immunoprecipitated. The mutants replacing Tyr451, Tyr469, Tyr512, Tyr523, and Tyr512/Tyr523 were designated as Y451F, Y469F, Y512F, Y523F, and Y512/523F, respectively. Immunoblot analysis was carried out using either the anti-FLAG antibody (anti-FLAG) (Top) or the anti-phosphotyrosine antibody (anti-PY) (Middle). PKC activity in the immunoprecipitates was assayed with H1 histone as substrate (Bottom).
Because all PKC isoforms examined were phosphorylated on tyrosine, we focused our attention on the tyrosine residues commonly present in the carboxyl-terminal half of the PKC family. Four tyrosine residues, Tyr451, Tyr469, Tyr512, and Tyr523 of PKC δ, are conserved in the carboxyl-terminal half of the PKC family (Fig. 4B) (1). Thus, each of these tyrosine residues was replaced by phenylalanine, and the mutated molecules were expressed in COS-7 cells (Fig. 4C). The replacement of either Tyr512 or Tyr523 in subdomain VIII by phenylalanine attenuated H2O2-induced activation of the enzyme. The mutation of both Tyr512 and Tyr523 almost abolished enzyme activation, but the mutated enzyme was still tyrosine phosphorylated to some extent in response to H2O2. On the other hand, PKC δ mutated on Tyr451 or Tyr469 in subdomains VIa and VIb was fairly activated and tyrosine phosphorylated. We conclude, therefore, that PKC δ is phosphorylated on more than one tyrosine residue upon treatment with H2O2 and that the tyrosine residues, especially Tyr512 and Tyr523 in subdomain VIII, are critical for PKC activation.
DISCUSSION
It has been described that H2O2 induces inositol phospholipid hydrolysis in some cultured cells (20, 21). In general, however, oxidative stress does not appear to produce diacylglycerol. It has also been postulated that H2O2 induces oxidative modification of PKC, thereby activating the enzyme (14), but no substantial evidence is available for such modification. The present study indicates that all PKC isoforms examined are tyrosine phosphorylated and exhibit enhanced enzyme activity in response to H2O2. Tyrosine kinases such as Lck and Syk are known to be activated by oxidative stress (22–24). Some PKC isoforms were tyrosine phosphorylated in vitro by Lck and Syk, and in fact, the phosphorylated isoforms showed enzyme activity higher than the nonphosphorylated isoforms (data not shown). In addition to PKC δ, PKC α has been reported to be tyrosine phosphorylated in insulin-stimulated cells (25). However, the tyrosine kinases that phosphorylate PKC isoforms in vivo remain to be identified. Both PKB (26) and PKN (27) have a catalytic domain highly homologous to the PKC family, but were not tyrosine phosphorylated in vitro or in the H2O2-treated cells (data not shown). Thus, phosphorylation on tyrosine does not appear to be a mechanism of activation commonly observed for protein kinases related to the PKC family.
For PKC δ, tyrosine phosphorylation in subdomain VIII is critical for activation in response to H2O2. The significance of phosphorylation on serine and threonine residues concurrently observed in response to H2O2 (Fig. 1C) was not investigated in the present study. It is possible that the PKC enzyme activated in the H2O2-treated cells autophosphorylates its serine and threonine residues. However, it cannot be ruled out that some other serine and threonine protein kinases take part in PKC activation. Thr500 in subdomain VIII of PKC β has been shown to be phosphorylated nearly stoichiometrically in vivo (28, 29). If Thr505 in PKC δ, which is equivalent to Thr500 in PKC β, is constitutively phosphorylated, PKC δ could be phosphorylated on both threonine and tyrosine residues in subdomain VIII upon H2O2 stimulation. Note that mitogen-activated protein kinases are phosphorylated for their activation dually on both threonine and tyrosine residues in subdomain VIII on the activation loop (30). Recently, however, it was reported that phosphorylation of Thr505 in PKC δ is not a prerequisite for its enzyme activity (31). More detailed studies are needed to understand fully the activation mechanisms of the PKC family involving protein phosphorylation.
Platelet-derived growth factor may generate H2O2, and H2O2 is required for signaling of growth factors in some cultured cells (32). An obvious possibility arises that PKC can be activated in two independent signaling pathways in response to growth factors. One is the classical diacylglycerol-dependent pathway through activation of phospholipases C and D (1). The other could be a pathway through direct phosphorylation on tyrosine in subdomain VIII. Transcription factors NF-κB and AP-1 have been reported to be activated in two different ways: one is through PKC, and the other is by H2O2 in a manner independent of PKC (33, 34). The results described herein suggest that H2O2 could activate these transcription factors through activation of PKC involving tyrosine phosphorylation. The physiological picture of PKC activation by tyrosine phosphorylation will be substantiated by further investigations.
Table 1.
PKC activity in transfected COS-7 cells treated with H2O2
| PKC isoform | H2O2 treatment, min | PKC activity
|
|
|---|---|---|---|
| PSL | % | ||
| α | 0 | 20,300 | 100 |
| α | 5 | 33,500 | 165 |
| α | 10 | 54,600 | 269 |
| βI | 0 | 24,200 | 100 |
| βI | 5 | 35,600 | 147 |
| βI | 10 | 56,900 | 235 |
| γ | 0 | 22,700 | 100 |
| γ | 5 | 46,100 | 203 |
| γ | 10 | 65,500 | 289 |
| δ | 0 | 9,240 | 100 |
| δ | 5 | 49,200 | 532 |
| δ | 10 | 42,000 | 455 |
| ɛ | 0 | 10,400 | 100 |
| ɛ | 5 | 34,000 | 327 |
| ɛ | 10 | 46,800 | 450 |
| ζ | 0 | 14,100 | 100 |
| ζ | 5 | 53,000 | 376 |
| ζ | 10 | 67,100 | 476 |
Phosphorylation of H1 histone by PKC isoforms described in Fig. 2 was quantitated by measuring PSL using a Bio-Imaging Analyzer. The activity of each PKC isoform recovered from the cells without H2O2 treatment was used as 100%.
Table 2.
Properties of PKC isoforms recovered from the H2O2-treated cells
| PKC isoform | H2O2 treatment, min | Activators | PKC activity
|
|
|---|---|---|---|---|
| PSL | % | |||
| δ | 0 | — | 9,100 | 100 |
| δ | 0 | PS, DG | 37,400 | 441 |
| δ | 10 | — | 41,900 | 460 |
| δ | 10 | PS, DG | 40,600 | 446 |
| α | 0 | — | 20,500 | 100 |
| α | 0 | PS, DG, Ca2+ | 133,000 | 650 |
| α | 10 | — | 86,100 | 420 |
| α | 10 | PS, DG, Ca2+ | 80,000 | 390 |
Phosphorylation of H1 histone by the PKC isoforms described in Fig. 3 in the presence or absence of phosphatidylserine (PS), diacylglycerol (DG), and Ca2+ was quantitated by measuring PSL using a Bio-Imaging Analyzer. The activity of each PKC isoform recovered from the cells without H2O2 treatment in the absence of these activators was used as 100%.
Acknowledgments
We thank Drs. R. M. Perlmutter and Y. Minami for providing the Lck cDNA, Dr. H. Yamamura for providing the Syk cDNA, and Ms M. Inagaki for secretarial assistance. This study was supported in part by research grants from the Scientific Research Funds of the Ministry of Education, Science, Sports and Culture of Japan, the Sankyo Research Institute for Neurosciences, and the Suntory Institute for Bioorganic Research.
ABBREVIATIONS
- PKC
protein kinase C
- HA
hemagglutinin
- PSL
photostimulated luminescence
References
- 1.Nishizuka Y. FASEB J. 1995;9:486–496. [PubMed] [Google Scholar]
- 2.Denning M F, Dlugosz A A, Howett M K, Yuspa S H. J Biol Chem. 1993;268:26079–26081. [PubMed] [Google Scholar]
- 3.Li W, Mischak H, Yu J-C, Wang L-M, Mushinski J F, Heidaran M A, Pierce J H. J Biol Chem. 1994;269:2349–2352. [PubMed] [Google Scholar]
- 4.Haleen-Smith H, Chang E-Y, Szallasi Z, Blumberg P M, Rivera J. Proc Natl Acad Sci USA. 1995;92:9112–9116. doi: 10.1073/pnas.92.20.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soltoff S P, Toker A. J Biol Chem. 1995;270:13490–13495. doi: 10.1074/jbc.270.22.13490. [DOI] [PubMed] [Google Scholar]
- 6.Denning M F, Dlugosz A A, Threadgill D W, Magnuson T, Yuspa S H. J Biol Chem. 1996;271:5325–5331. doi: 10.1074/jbc.271.10.5325. [DOI] [PubMed] [Google Scholar]
- 7.Gschwendt M, Kielbassa K, Kittstein W, Marks F. FEBS Lett. 1994;347:85–89. doi: 10.1016/0014-5793(94)00514-1. [DOI] [PubMed] [Google Scholar]
- 8.Zang Q, Lu Z M, Curto M, Barile N, Shalloway D, Foster D A. J Biol Chem. 1997;272:13275–13280. doi: 10.1074/jbc.272.20.13275. [DOI] [PubMed] [Google Scholar]
- 9.Szallasi Z, Denning M F, Chang E-Y, Rivera J, Yuspa S H, Lehel C, Olah Z, Anderson W B, Blumberg P M. Biochem Biophys Res Commun. 1995;214:888–894. doi: 10.1006/bbrc.1995.2370. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Chen X-H, Kelley C A, Alimandi M, Zhang J, Chen Q, Bottaro D P, Pierce J H. J Biol Chem. 1996;271:26404–26409. doi: 10.1074/jbc.271.42.26404. [DOI] [PubMed] [Google Scholar]
- 11.Palumbo E J, Sweatt D, Chen S-J, Klann E. Biochem Biophys Res Commun. 1992;187:1439–1445. doi: 10.1016/0006-291x(92)90463-u. [DOI] [PubMed] [Google Scholar]
- 12.Brawn M K, Chiou W J, Leach K L. Free Radical Res. 1995;22:23–37. doi: 10.3109/10715769509147525. [DOI] [PubMed] [Google Scholar]
- 13.Whisler R L, Goyette M A, Grants I S, Newhouse Y G. Arch Biochem Biophys. 1995;319:23–35. doi: 10.1006/abbi.1995.1263. [DOI] [PubMed] [Google Scholar]
- 14.Gopalakrishna R, Anderson W B. Proc Natl Acad Sci USA. 1989;86:6758–6762. doi: 10.1073/pnas.86.17.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill G N, Kikkawa U. J Biol Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- 16.Ono Y, Fujii T, Ogita K, Kikkawa U, Igarashi K, Nishizuka Y. J Biol Chem. 1988;263:6927–6932. [PubMed] [Google Scholar]
- 17.Konishi H, Matsuzaki H, Tanaka M, Ono Y, Tokunaga C, Kuroda S, Kikkawa U. Proc Natl Acad Sci USA. 1996;93:7639–7643. doi: 10.1073/pnas.93.15.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle W J, van der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzaki H, Konishi H, Tanaka M, Ono Y, Takenawa T, Watanabe Y, Ozaki S, Kuroda S, Kikkawa U. FEBS Lett. 1996;396:305–308. doi: 10.1016/0014-5793(96)01120-9. [DOI] [PubMed] [Google Scholar]
- 20.Shasby D M, Yorek M, Shasby S S. Blood. 1988;72:491–499. [PubMed] [Google Scholar]
- 21.Zick Y, Sagi-Eisenberg R. Biochemistry. 1990;29:10240–10245. doi: 10.1021/bi00496a013. [DOI] [PubMed] [Google Scholar]
- 22.Schieven G L, Kirihara J M, Burg D L, Geahlen R L, Ledbetter J A. J Biol Chem. 1993;268:16688–16692. [PubMed] [Google Scholar]
- 23.Qin S, Inazu T, Yamamura H. Biochem J. 1995;308:347–352. doi: 10.1042/bj3080347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardwick J S, Sefton B M. Proc Natl Acad Sci USA. 1995;92:4527–4531. doi: 10.1073/pnas.92.10.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Roth R A. Biochem Biophys Res Commun. 1994;200:1570–1577. doi: 10.1006/bbrc.1994.1630. [DOI] [PubMed] [Google Scholar]
- 26.Mukai H, Ono Y. Biochem Biophys Res Commun. 1994;199:897–904. doi: 10.1006/bbrc.1994.1313. [DOI] [PubMed] [Google Scholar]
- 27.Konishi H, Kuroda S, Tanaka M, Matsuzaki H, Ono Y, Kameyama K, Haga T, Kikkawa U. Biochem Biophys Res Commun. 1995;216:526–534. doi: 10.1006/bbrc.1995.2654. [DOI] [PubMed] [Google Scholar]
- 28.Keranen L M, Dutil E M, Newton A C. Curr Biol. 1995;5:1394–1403. doi: 10.1016/s0960-9822(95)00277-6. [DOI] [PubMed] [Google Scholar]
- 29.Tsutakawa S E, Medzihradszky K F, Flint A J, Burlingame A L, Koshland D E., Jr J Biol Chem. 1995;270:26807–26812. doi: 10.1074/jbc.270.45.26807. [DOI] [PubMed] [Google Scholar]
- 30.Seger R, Krebs E G. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 31.Stempka L, Girod A, Muller H J, Rincke G, Marks F, Gschwendt M, Bossemeyer D. J Biol Chem. 1997;272:6805–6811. doi: 10.1074/jbc.272.10.6805. [DOI] [PubMed] [Google Scholar]
- 32.Sundaresan M, Yu Z-X, Ferrans V J, Irani K, Finkel T. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 33.Meyer M, Schreck R, Baeuerle P A. EMBO J. 1993;12:2005–2015. doi: 10.1002/j.1460-2075.1993.tb05850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stauble B, Boscoboinik D, Tasinato A, Azzi A. Eur J Biochem. 1994;226:393–402. doi: 10.1111/j.1432-1033.1994.tb20064.x. [DOI] [PubMed] [Google Scholar]