Abstract
Endurance exercise training induces mitochondrial biogenesis in skeletal muscle. The peroxisome proliferator activated receptor co-activator 1α (PGC-1α) has recently been identified as a nuclear factor critical for coordinating the activation of genes required for mitochondrial biogenesis in cell culture and rodent skeletal muscle. To determine whether PGC-1α transcription is regulated by acute exercise and exercise training in human skeletal muscle, seven male subjects performed 4 weeks of one-legged knee extensor exercise training. At the end of training, subjects completed 3 h of two-legged knee extensor exercise. Biopsies were obtained from the vastus lateralis muscle of both the untrained and trained legs before exercise and after 0, 2, 6 and 24 h of recovery. Time to exhaustion (2 min maximum resistance), as well as hexokinase II (HKII), citrate synthase and 3-hydroxyacyl-CoA dehydrogenase mRNA, were higher in the trained than the untrained leg prior to exercise. Exercise induced a marked transient increase (P < 0.05) in PGC-1α transcription (10- to > 40-fold) and mRNA content (7- to 10-fold), peaking within 2 h after exercise. Activation of PGC-1α was greater in the trained leg despite the lower relative workload. Interestingly, exercise did not affect nuclear respiratory factor 1 (NRF-1) mRNA, a gene induced by PGC-1α in cell culture. HKII, mitochondrial transcription factor A, peroxisome proliferator activated receptor α, and calcineurin Aα and Aβ mRNA were elevated (≈2- to 6-fold; P < 0.05) at 6 h of recovery in the untrained leg but did not change in the trained leg. The present data demonstrate that exercise induces a dramatic transient increase in PGC-1α transcription and mRNA content in human skeletal muscle. Consistent with its role as a transcriptional coactivator, these findings suggest that PGC-1α may coordinate the activation of metabolic genes in human muscle in response to exercise.
Endurance exercise training enhances the oxidative capacity and metabolic efficiency of skeletal muscle (Saltin & Gollnick, 1983). This adaptive response is in part attributed to the training-induced increase in expression of proteins involved in the transport and oxidation of metabolic substrates and an overall increase in mitochondrial content (Booth & Baldwin, 1996; Williams & Neufer, 1996). The underlying process responsible for these cellular adaptations appears to result at least in part from the cumulative effects of transient changes in gene expression that occur in response to each exercise bout (Williams & Neufer, 1996; Hood, 2001).
A number of factors, including calcium, energy charge (AMP/ATP), redox state (NAD/NADH), oxygen tension, mechanical stretch, neuroregulatory peptides, free radicals, growth factors, and cytokines (IL-6), are all known to be altered in skeletal muscle in response to exercise/contractile activity (reviewed in Booth & Baldwin, 1996; Williams & Neufer, 1996; Olson & Williams, 2000; Winder, 2001; Scarpulla, 2002). Although a number of these initiating stimuli activate specific signalling pathways, the molecular mechanism(s) linking exercise to the transcriptional regulation of exercise-responsive genes remains to be fully elucidated. Among the most promising of nuclear proteins thought to be involved in providing this link are nuclear respiratory factor-1 (NRF-1) and nuclear respiratory factor-2α (NRF-2α). NRF-1 and NRF-2α bind to and activate the promoters of various nuclear genes that encode for components of the respiratory chain in the mitochondria as well as factors required for mitochondrial DNA transcription and replication (Scarpulla, 2002). Knockout of the NRF-1 gene severely limits mitochondrial DNA formation and is lethal during early embryonic development (Huo & Scarpulla, 2001), suggesting that NRF-1 plays a vital role during mitochondrial biogenesis. Although NRF-1 mRNA has been reported to increase by ≈35 % in soleus muscle of rats in response to an acute bout of exercise (Murakami et al. 1998), the potential role(s) that NRF-1 or NRF-2α may play in mediating gene activation and mitochondrial biogenesis in response to endurance exercise training in humans has not been investigated.
In addition to the NRFs, a transcriptional coactivator of peroxisome proliferator activated receptor gamma (PPARγ), known as PGC-1α, has recently emerged as a regulatory factor thought to be involved in coordination of the activation of nuclear genes that encode for mitochondrial proteins in several tissues including skeletal muscle (Scarpulla, 2002). PGC-1α does not bind to DNA itself but rather interacts with selected transcription factors already bound to the promoter region of target genes. The positive effects of PGC-1α on transcription are thought to be mediated by its ability to bind to specific transcription factors (e.g. NRF-1 and myocyte enhancer factor 2c (MEF2c)), recruit additional coactivators that possess intrinsic histone acetyl transferase activity, and bind RNA polymerase II and various splicing factors, thereby enhancing the overall efficiency of the transcriptional machinery (reviewed in Puigserver et al. 1999; Knutti & Kralli, 2001). Over-expression of PGC-1α induces mitochondrial biogenesis in both skeletal myocytes grown in culture and in cardiac muscle of transgenic mice (Wu et al. 1999; Lehman et al. 2000), a response that in myotubes is associated with a dramatic induction of NRF-1 and NRF-2α (Wu et al. 1999). In mice, forced expression of PGC-1α specifically in skeletal muscle causes the muscle to appear much redder, increases expression of the slow troponin I and myoglobin genes (two genes normally expressed only at low levels in type II fibres), and increases the expression of several genes involved in mitochondrial oxidative metabolism (Lin et al. 2002). Collectively these findings provide evidence that increased expression of the endogenous PGC-1α gene is critical for the activation of genes involved in oxidative metabolism in skeletal muscle.
The purpose of the present study was to test in humans the hypothesis that prolonged exercise activates transcription and/or mRNA content of the NRF and PGC-1α genes in skeletal muscle, and to determine whether such a response is dependent on the training state of the muscle. In addition, the effects of exercise and exercise training on the mRNA expression of two other candidate transcription factors, PPARα and mitochondrial transcription factor A (mtTFA, also known as TFAM), were also determined. Finally, in the light of recent evidence suggesting that activation of the calcium-calmodulin-dependent protein kinase (CaMK) signalling pathway induces mitochondrial biogenesis via PGC-1α (Lin et al. 2002; Wu et al. 2002), we also examined whether the mRNA expression of components of the calcium signalling system (CaMKI and calcineurin) are influenced by exercise and exercise training. Subjects performed 4 weeks of one-legged knee extensor exercise after which the molecular responses to 3 h of two-legged knee extensor exercise were compared in the untrained and trained leg.
Methods
Subjects
Seven healthy, normally physically active subjects not involved in any type of endurance activities participated in the present study. The subjects ranged in age from 19 to 24 years and had an average height of 184 ± 3 cm (mean ± s.e.m.) and a mean weight of 84 ± 7 kg. After being given both written and oral information on the experimental protocol and procedures, the subjects gave their informed, written consent to participate. The study conformed with the guidelines laid down in the Declaration of Helsinki and was approved by the Copenhagen and Frederiksberg Ethics Committee, Denmark and the Human Investigations Committee, Yale University, USA.
Experimental design
A modified bicycle ergometer (Andersen et al. 1985) was applied for both one-legged and two-legged knee extensor exercise. Before initiating the training period, each of the subjects completed a one-legged knee extensor exercise performance test with each leg separately to determine the maximal load that could be maintained for 2 min (2 min max) (Pilegaard et al. 2002). The leg to be trained was randomly chosen. The subjects trained 5 days per week for 4 weeks by one-legged knee extensor exercise. The first training session was performed at 70 % of 2 min max until exhaustion (≈1 h). As performance improved, the resistance was increased keeping the duration approximately the same.
The subjects were provided with a standardized dinner and evening snack meal the day before the experiment. On the day of the experiment, the subjects arrived at the laboratory after consuming a light breakfast. A resting muscle biopsy was taken (≈2.5 h after breakfast) from the middle portion of the vastus lateralis muscle of each leg using the percutaneous needle biopsy technique with suction (Bergstrom, 1962). The subjects performed two-legged knee extensor exercise for 3 h at the one-legged 2 min max of the untrained leg (i.e. per leg, 50 % of the 2 min max of the untrained leg). Additional muscle biopsies were obtained from both legs at the end of exercise and after 2, 6 and 24 h of recovery. Food intake was restricted throughout the experiment until after the 6 h biopsy after which the subjects were provided with standardized meals including dinner and breakfast the next morning (2.5 h before the 24 h biopsy). Force development during the 3 h exercise bout was recorded for each leg using a force transducer connected to a strain-gauge amplifier showing that the two legs performed similar amounts of work.
Muscle analyses
The major part of the muscle biopsies (≈110–130 mg) was used for determining transcriptional activity. The remaining portion of the biopsy was frozen in liquid N2 for RNA analysis. Samples were stored at - 80 °C.
Isolation of nuclei
Nuclei were isolated from fresh muscle biopsies as previously described (Pilegaard et al. 2000). In brief, the muscle tissue was rapidly placed in a pre-cooled aluminum block, weighed and placed in ice-cold buffer. The tissue was thoroughly minced, gently homogenized and the nuclei spun down. The crude nuclear pellet was gently resuspended in buffer containing Triton X-100 and filtered through pre-wetted cheesecloth. Nuclei were repelleted and gently resuspended in rinsing buffer. The final nuclear pellet was resuspended in 200 µl of storage buffer, quick-frozen in liquid N2, and stored at - 80 °C.
Isolation of gDNA
Genomic DNA (gDNA) was isolated from 20 µl of each nuclei preparation as previously described (Hildebrandt & Neufer, 2000; Pilegaard et al. 2000). Final gDNA pellets were resuspended overnight (4 °C) in 50 µl nuclease-free water containing 0.1 mm EDTA, 10 mm Tris (pH 8.0). The relative gDNA content of the nuclei samples was determined by PCR amplification of the 18S gene (described below: Reverse transcription and PCR), and used to adjust the volumes of the nascent RNA transcript samples for differences in nuclei content prior to the nuclear run-on reaction.
Nuclear run-on
Relative transcriptional activity of the selected genes was determined by a reverse transcription (RT)-PCR-based nuclear run-on technique as previously described (Hildebrandt & Neufer, 2000; Pilegaard et al. 2000). Briefly, incomplete transcripts were allowed to proceed to completion in the presence of non-radioactive nucleotides ATP, CTP, GTP and UTP. After lysing the nuclei, nuclear proteins and genomic DNA were enzymatically digested by proteinase K and DNase I, respectively, and nascent RNA transcripts were isolated by extraction (TriZol, Invitrogen) and precipitation. The RNA product was then subjected to a second DNase digestion and precipitation followed by thorough rinsing and resuspension overnight (4 °C) in 22 µl of diethyl pyrocarbonate (DEPC)-treated H2O containing 0.1 mm EDTA and 10 mm Tris (pH 8.0).
RNA isolation
Total RNA was isolated from ≈20 mg of tissue by a modified guanidinium thiocyanate-phenol-chloroform extraction method adapted from Chomczynski & Sacchi (1987) and described previously (Pilegaard et al. 2000). RNA was resuspended overnight (4 °C) in 2 µl (mg original tissue weight)−1 of DEPC-treated H2O containing 0.1 mm EDTA.
Reverse transcription and PCR
Reverse transcription of both nascent RNA (from the nuclear run-on reactions) and of total RNA was performed using the Superscript II RNase H-system (Invitrogen) as previously described (Hildebrandt & Neufer, 2000; Pilegaard et al. 2000). RT products of nascent nuclear run-on RNA were diluted with nuclease-free H2O based on the relative gDNA content of each nuclei preparation, with the average volume set to 150 µl. For the RT products from total RNA samples, each sample originating from 11 µl of total RNA was diluted to a total of 150 µl.
Nascent RNA transcripts, gDNA of nuclei samples, and mRNA were determined by real-time PCR (ABI PRISM 7700 Sequence Detection System, Applied Biosystems). Forward and reverse primers and the TaqMan probe were designed from human specific sequence databases (Entrez-NIH and Ensembl, Sanger Institute), using computer software (Primer Express, Applied Biosystems) and are given in Table 1. For each of the genes, a Blast Search revealed that sequence homology was obtained only for the target gene. One oligonucleotide from each set was designed to span an exon-exon junction, eliminating the possibility of amplifying genomic DNA. All self-designed TaqMan probes were labelled with 5′-6-carboxyfluorescein (FAM) and 3′-6-carboxy-N,N,N’,N’-tetramethylrhodamine (TAMRA). Prior optimization was conducted for each set of self-designed oligonucleotides, which consisted of determining optimal primer concentrations, the probe concentration and verifying the efficiency of the amplification. GAPDH transcription and GAPDH mRNA were amplified using a 5′-VIC- and 3′-TAMRA-labelled pre-developed assay reagent (Applied Biosystems). Validating tests were performed to ensure that the amplification efficiency was similar for each of the target genes and the endogenous control (GAPDH). Finally, the relative nuclei content of the gDNA samples was determined by amplifying the 18S gene using a 5′-VIC- and 3′-TAMRA-labelled pre-developed assay reagent designed to amplify genomic DNA (Applied Biosystems). PCR amplification was performed (in triplicate) as singleplex reactions in a total reaction volume of 25 µl. The reaction mixture consisted of 2.5 µl diluted template, the TaqMan probe and forward and reverse primers as determined from the prior optimization procedure, nuclease-free water and 2 × TaqMan Universal MasterMix (Applied Biosystems) containing AmpliTaq Gold DNA polymerase, AmpErase uracil N-glycosylase, dNTPs with dUTP, ROX as passive reference and buffer components. An identical PCR cycle profile was used for all genes: 50 °C for 2 min + 95 °C for 10 min + [95 °C for 15 s + 60 °C for 1 min] × 40 cycles. Data were analysed using a comparative critical threshold (Ct) method where the amount of target normalized to the amount of endogenous control and relative to the control sample is given by 2−ΔΔCt (Applied Biosystems). For each subject, all samples were run together allowing relative comparisons of the samples from a given subject.
Table 1.
Primer and probe sequences used for real-time PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| PGC-1α | 5′-CAAGCCAAACCAACAACTTTATCTCT-3′ | 5′-CACACTTAAGGTGCGTTCAATAGTC-3′ |
| NRF-1 | 5′-GGCACTGTCTCACTTATCCAGGTT-3′ | 5′-CAGCCACGGCAGAATAATTCA-3′ |
| NRF-2α | 5′-AAATTGAGATTGATGGAACAGAGAA-3′ | 5′-TATGGCCTGGCTTACACATTCA-3′ |
| mtTFA | 5′-AGATTCCAAGAAGCTAAGGGTGATT-3′ | 5′-TTTCAGAGTCAGACAGATTTTTCCA-3, |
| PPARα | 5′-AGCTTTGGCTTTACGGAATACCA-3′ | 5′-CCACAGGATAAGTCACCGAGGA-3′ |
| Cal B1 | 5′-CACAGATGGGAATGGAGAAGTAGA-3′ | 5′-GCAAACCTCAATTTCTGCTCCTT-3′ |
| Cal Aα | 5′-GAGGGTGCATCAATTCTTCGA-3′ | 5′-CAAATCAAAGAATTGTCCATGAATGT-3′ |
| Cal Aβ | 5′-AGTGCCACAGTTGAGGCTATTGA-3′ | 5′-CCAAACCCTTTGCCTCTTCA-3′ |
| CaMKI | 5′-TTGCTGGTCCATAGGTGTCATC-3′ | 5′-AGAGTTTGGCATCATTCTCGTCAT-3′ |
| HKII | 5′-TTGTCCGTAACATTCTCATCGATT-3′ | 5′-TGTCTTGAGCCGCTCTGAGAT-3′ |
| CS | 5′-GACTACATCTGGAACACACTCAACTCA-3′ | 5′-CGCGGATCAGTCTTCCTTAGTAC-3′ |
| HAD | 5′-CATAGCGACCAGCACGGAT-3′ | 5′-GATGGCTTCCACCACCAAGT-3′ |
| Gene | Probe | |
| PGC-1α | 5′-AGTCACCAAATGACCCCAAGGGTTCC-3′ | |
| NRF-1 | 5′-ACCACGGTCACCGTTGCCCAA-3′ | |
| NRF-2α | 5′-AAGCATTGTAGAACAAACCTACGCGCCAG-3′ | |
| mtTFA | 5′-CCGCAGGAAAAGCTGAAGACTGTAAAGGA-3′ | |
| PPARα | 5′-TCACGGACACGCTTTCACCAGCTTC-3′ | |
| Cal B1 | 5′-TTTAAAGAATTCATTGAGGGCGTCTCTCAGTTC-3′ | |
| Cal Aα | 5′-ACAAACAGTGACTGGCGCATCAATATCCA-3′ | |
| Cal Aβ | 5′-AAGCAATACGAGGATTCTCTCCACCACATAGAA-3′ | |
| CaMKI | 5′-CCTACATCTTGCTCTGCGGTTACCCTCC-3′ | |
| HKII | 5′-ACCAAGCGTGGACTGCTCTTCCGA-3′ | |
| CS | 5′-ACGGGTTGTTCCAGGCTATGGCCA-3′ | |
| HAD | 5′-CAGCCTCCGTTGTCCACAGCACA-3′ | |
PGC-1 α, peroxisome proliferator activated receptor gamma co-activator 1α; NRF-1, nuclear respiratory factor 1; NRF- 2α a, nuclear respiratory factor 2 α; mtTFA, mitochondrial transcription factor A; PPAR α, peroxisome proliferator activated receptor a; Cal B1, calcineurin subunit B1; Cal A α, calcineurin subunit A α; Cal A β, calcineurin subunit A β; CaMKI, calcium-calmodulin-dependent protein kinase I; HKII, hexokinase II; CS, citrate synthase; HAD, 3-hydroxyacyl-CoA dehydrogenase.
Statistics
Transcriptional activity and mRNA content were normalized to GAPDH transcription and mRNA levels, respectively, and samples were expressed relative to the corresponding untrained pre-exercise sample, which was set to 1. Values are means ± s.e.m. Two-way ANOVA for repeated measures was used to evaluate the effect of time and training state on the response of transcription and mRNA content and one-way ANOVA for repeated measures was used to evaluate the effect of time for each leg separately. Student-Newman-Keuls post hoc test was used to locate differences. A paired t test was used to compare pre-exercise transcription and pre-exercise mRNA content in the untrained and the trained legs. Differences were considered significant at P < 0.05.
Results
Performance
After training, knee extensor exercise at the load used in the first training session (70 % of 2 min max load) could easily be sustained for more than 100 min with the trained leg (before training, exhaustion occurred after ≈60 min). Average time to exhaustion in an all-out bout at ≈110 % of the 2 min max resistance of the untrained leg was significantly (P < 0.05) improved by training (3.6 ± 0.7 min for untrained and 7.2 ± 1.7 min for trained).
Transcription
Prior to exercise, transcription of the PGC-1α gene was ≈10-fold higher (P < 0.05) in the trained leg than in the untrained leg (Fig. 1). In the untrained leg, PGC-1α increased in response to exercise in all subjects, although the timing and magnitude of increase varied considerably among subjects such that mean changes in PGC-1α transcription were not significant. In the trained leg, exercise also induced a marked transient increase (P < 0.05) in PGC-1α transcription, peaking immediately after exercise (> 40-fold over the pre-exercise value in the untrained leg). Although relative workload was much lower in the trained leg, PGC-1α transcription was significantly (P < 0.05) higher in the trained compared with the untrained leg immediately after exercise.
Figure 1. Transcription and mRNA content of peroxisome proliferator activated receptor co-activator 1α (PGC-1α) in untrained and trained legs in response to 3 h of two-legged knee extensor exercise.
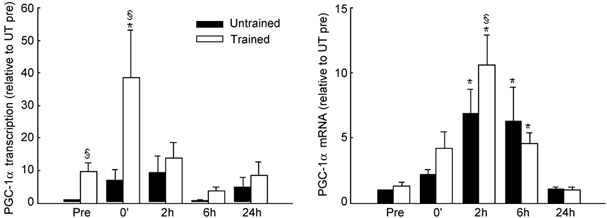
Muscle biopsies were obtained from both legs before (Pre), immediately at the end of exercise (0′) and after 2, 6 and 24 h of recovery. The PGC-1α response was normalized to GAPDH transcription and mRNA, respectively, and expressed relative to the pre-exercise value in the untrained leg (Untrained Pre), which was set to 1. Values are means ± s.e.m., n = 5–7. * Significantly different from Pre in the same leg (P < 0.05). §Different from Untrained (P < 0.05).
Content of mRNA
Muscle PGC-1α mRNA content was similar in the untrained and trained leg before exercise (Fig. 1). In response to exercise, PGC-1α mRNA increased (P < 0.05) during recovery, peaking in both the untrained (7-fold) and the trained (10-fold) leg 2 h after exercise. This increase in PGC-1α mRNA was transient in both legs, returning to resting levels within 24 h after exercise. PGC-1α mRNA was significantly higher in the trained than in the untrained leg after 2 h of recovery.
To determine the effect of exercise and exercise training on the mRNA content of transcription factors associated with mitochondrial biogenesis, we also measured the mRNA content of NRF-1, NRF-2α, mtTFA and PPARα (Table 2). NRF-1 and NRF-2α mRNA were not different in the untrained and trained legs before exercise and failed to show any significant increase in response to exercise (Table 2). Exercise did induce a significant (P < 0.05) increase in mtTFA (> 3-fold) and PPARα (1.8-fold) mRNA in the untrained leg at 6 h of recovery (Table 2).
Table 2.
The mRNA content of selected mitochondrial proteins transcription factors and calcium signaling proteins in response to exercise
| Pre | 0′ | 2 h | 6 h | 24 h | ||
|---|---|---|---|---|---|---|
| Transcription factors | ||||||
| NRF−1 | UT | 1 | 1.2 ± 0.1 | 1.0 ± 0.1 | 1.7 ± 0.5 | 0.8 ± 0.1 |
| T | 1.2 ± 0.2 | 1.0 ± 0.1 | 1.2 ± 0.2 | 1.1 ± 0.1 | 1.4 ± 0.4 | |
| NRF−2α | UT | 1 | 1.5 ± 0.4 | 2.0 ± 0.7 | 3.0 ± 0.8 | 2.0 ± 0.6 |
| T | 1.7 ± 0.5 | 2.0 ± 0.6 | 1.9 ± 0.7 | 1.9 ± 0.5 | 1.8 ± 0.4 | |
| mtTFA | UT | 1 | 1.8 ± 0.4 | 1.8 ± 0.5 | 3.4 ± 0.8* | 1.5 ± 0.3 |
| T | 1.6 ± 0.4 | 1.7 ± 0.3 | 2.2 ± 0.4 | 1.9 ± 0.4 | 2.1 ± 0.7 | |
| PPARα | UT | 1 | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.5 ± 0.3 | 1.1 ± 0.2 |
| T | 1.1 ± 0.2 | 0.8 ± 0.1 | 1.8 ± 0.5 | 1.8 ± 0.2* | 0.8 ± 0.2 | |
| Ca2+ signaling proteins | ||||||
| Calcinerium Aα | UT | 1 | 1.3 ± 0.3 | 1.1 ± 0.2 | 2.5 ± 0.6* | 1.3 ± 0.2 |
| T | 1.5 ± 0.3 | 1.0 ± 0.2 | 1.5 ± 0.3 | 1.7 ± 0.3 | 1.4 ± 0.5 | |
| Calcineurin Aβ | UT | 1 | 1.2 ± 0.2 | 0.9 ± 0.1 | 1.6 ± 0.3* | 0.9 ± 0.1 |
| T | 1.0 ± 0.2 | 1.4 ± 0.6 | 1.2 ± 0.4 | 0.9 ± 0.2 | 0.8 ± 0.1 | |
| Calcineurin B1 | UT | 1 | 1.2 ± 0.2 | 1.1 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.1 |
| T | 1.4 ± 0.3 | 0.9 ± 0.1 | 1.2 ± 0.2 | 1.0 ± 0.1 | 0.9 ± 0.1 | |
| Calcineurin B1 | UT | 1 | 1.2 ± 0.2 | 1.1 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.1 |
| T | 1.4 ± 0.3 | 0.9 ± 0.1 | 1.2 ± 0.2 | 1.0 ± 0.1 | 0.9 ± 0.1 | |
| CaMKI | UT | 1 | 1.5 ± 0.3 | 1.5 ± 0.2 | 2.2 ± 0.7 | 1.3 ± 0.2 |
| T | 2.3 ± 0.3† | 1.8 ± 0.3 | 2.6 ± 0.8 | 2.1 ± 0.5 | 2.5 ± 0.8 | |
| Metabolic enzymes | ||||||
| HKII | UT | 1 | 2.2 ± 0.4 | 4.0 ± 1.7 | 6.2 ± 2.3* | 1.1 ± 0.1 |
| T | 2.2 ± 0.4† | 4.5 ± 1.3† | 4.5 ± 1.3 | 4.6 ± 1.0 | 2.7 ± 0.7 | |
| CS | UT | 1 | 1.1 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.2 | 1.2 ± 0.1 |
| T | 1.5 ± 0.1† | 1.4 ± 0.2 | 1.7 ± 0.2† | 1.3 ± 0.1 | 1.4 ± 0.2 | |
| HAD | UT | 1 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.5 ± 0.4 | 0.8 ± 0.1 |
| T | 1.7 ± 0.2† | 2.0 ± 0.7 | 1.9 ± 0.4 | 1.3 ± 0.3 | 1.4 ± 0.5 | |
The mRNA content of the transcription factors nuclear respiratory factor 1 (NRF−1), Nuclear respiratory factor 2α (NRF−2α), Mitochondrial transcription factor A (mtTFA) and peroxisome proliferators activated receptor α (PPAR α), Selected calcium-signalling proteins calcineurin submit Aα, calcineriun subnit Aβ, Calcineurin suunit B1 and CaMKI (calcium-calmodulin-dependent protein Kinase I), and the matabolioc enzymes hexokinase II (HKII), Citrate synthase (CS) and 3-hydroxyacl-CoA dehydrogenase (HAD) in the hexokinase II (HKII), citrate synthase (CS) and 3-hydroxyacl-CoA dehydrogenase (HAD) in the untrained (UT) and trained (T) leg before (Pre) and immediately after (0′) exercise and at 2,6 and 24 h of recovery from 3 h of two-legged Knee extensor exercise. Values are presented as fold changes relative to Pre, which was set to 1. Values are means ± s.e.m.
Significantly different from Pre in the same leg, P < 0.05
significantly different from untrained leg, P < 0.05.
The Ca2+-activated proteins calcineurin and CaMK have recently been implicated in the control of mitochondrial gene expression in skeletal muscle (Lin et al. 2002; Wu et al. 2002). In the present study, exercise induced small but significant (P < 0.05) increases in the mRNA content of the catalytic calcineurin Aα (2.5-fold) and calcineurin Aβ (1.6-fold) subunits at 6 h of recovery in the untrained leg, but no changes were observed in the trained leg (Table 2). Although CaMKI mRNA was elevated in the trained leg before exercise (≈2-fold), exercise did not affect CaMKI or calcineurin B1 regulatory subunit mRNA.
Before exercise, the mRNA content of hexokinase II (HKII), citrate synthase (CS) and 3-hydroxyacyl-CoA dehydrogenase (HAD) were all signficantly (P < 0.05) increased in the trained leg (1.5- to 2.2-fold; Table 2). In response to exercise, HKII mRNA increased dramatically in the untrained leg, peaking 6 h after exercise (≈6-fold) while in the trained leg, HKII mRNA was ≈2- to 5-fold higher than pre-exercise untrained levels at all time points (Table 2). Training induced a significant overall increase in CS mRNA (main effect, P = 0.025), although no specific increase in CS or HAD mRNA was detected in response to exercise in either the untrained or trained leg.
Enzyme activity
Basal CS activity averaged 17.9 ± 1.2 µmol min−1 g−1 in the untrained leg and 21.7 ± 0.9 µmol min−1 g−1 in the trained leg, a difference of 22 % (P < 0.1). Analyses of resting samples (pre-exercise) from the trained and untrained legs revealed that the difference in CS activity between the two legs was highly correlated with the difference in CS mRNA (R = 0.85, P < 0.05).
Discussion
The main finding of the present study is that prolonged exercise induces a marked increase in transcription and mRNA content of the transcription factor coactivator PGC-1α in skeletal muscle of humans. The induction of PGC-1α was transient, peaking within the first 2 h after exercise. Interestingly, PGC-1α transcription and mRNA reached significantly higher levels in the trained leg despite the fact that the relative work intensity during the experiment was much lower for the trained than the untrained leg. These findings suggest that the mechanisms regulating the exercise-induced activation of the PGC-1α gene in muscle may become more ‘sensitive’ to exercise with endurance training.
PGC-1α was originally cloned using a yeast two-hybrid system to identify proteins in brown adipose cells that interact with the PPARγ transcription factor (Puigserver et al. 1998). Interaction of PPARγ with PGC-1α was found to dramatically enhance the activation of transcription induced by PPARγ. Subsequent studies revealed that PGC-1α is also expressed in liver and skeletal muscle tissue where it co-activates the transcription of genes required for gluconeogenesis and mitochondrial biogenesis, respectively (Wu et al. 1999; Yoon et al. 2001). In C2C12 myotubes, forced expression of PGC-1α causes a dramatic increase in the expression of NRF-1, NRF-2α and mtTFA, as well as GLUT4 and several nuclear genes that encode for components of the respiratory chain (Wu et al. 1999; Michael et al. 2001). The net result in these cells is an increased glucose transport, an increased mitochondrial DNA content, and an overall proliferation of mitochondria (Wu et al. 1999). Promoter deletion and protein-protein binding studies have confirmed that PGC-1α mediates these effects, at least in part, via direct interation with the NRF-1 and MEF2 transcription factors (Wu et al. 1999; Michael et al. 2001). More recently, PGC-1α has also been shown to enhance the activation of genes encoding for mitochondrial enzymes of the fatty acid β-oxidation pathway via coactivation of the PPARα transcription factor (Vega et al. 2000). Activation of AMP-activated protein kinase (AMPK), an energy-sensing kinase, increases PGC-1α expression (Terada et al. 2002) and is associated with increased NRF-1 binding activity and mitochondrial biogenesis in rat skeletal muscle (Bergeron et al. 2001). Thus, induction of PGC-1α has been proposed to play an important role in coordinating the activation of various genes required for mitochondrial biogenesis in skeletal muscle.
Two previous studies have examined the acute effects of exercise on PGC-1α expression (Terada et al. 2002; Tunstall et al. 2002). In rats, PGC-1α mRNA was found to increase by ≈7-fold in epitrochlearis muscle immediately after 6 h of swimming exercise (Terada et al. 2002). In humans, however, Tunstall et al. (2002) recently reported that PGC-1α mRNA was unchanged in vastus lateralis muscle immediately after exercise and after 3 h of recovery in subjects tested both before and after 9 days of exercise training. Thus, these latter findings are in direct contrast with the results from the present study. In fact, in the present study, exercise elicited a marked increase in PGC-1α expression in muscle from both the untrained and trained legs in every subject. Increases in PGC-1α transcription ranged from 3- to > 100-fold and increases in PGC-1α mRNA ranged from 3- to > 20-fold. The reasons for the disparate results in the study by Tunstall et al. (2002) and the present study are not clear but may be related to differences in the mode (cycling vs. knee extension), duration (1 h vs. 3 h) and/or intensity (63 % of peak oxygen uptake vs. 50 % of one-legged 2 min max of untrained leg) of exercise in the two studies. Another possibility is that the subjects in the former study (Tunstall et al. 2002) consumed a high carbohydrate (71 %) diet for the 24 h prior to each trial which may have elevated muscle glycogen levels prior to exercise. We have previously found that the transcriptional activation of metabolic genes in response to exercise is blunted when pre-exercise muscle glycogen content is high whereas transcriptional activation is enhanced when pre-exercise muscle glycogen content is low, raising the possibility that signalling mechanisms regulating exercise-responsive genes may be sensitive to muscle gycogen content (Pilegaard et al. 2002).
In contrast to PGC-1α, exercise failed to elicit any increase in NRF-1 or NRF-2α mRNA, nor was there any difference in basal expression between the untrained and trained legs. These findings were somewhat surprising given that NRF-1 and NRF-2α mRNA increase dramatically in response to forced expression of PGC-1α in C2C12 myotubes grown in culture (Wu et al. 1999) and NRF-1 mRNA has been reported to increase (≈35 %) in rat soleus muscle during recovery from acute treadmill exercise (Murakami et al. 1998). The NRF transcription factors are thought to play a critical role in the activation of mitochondrial biogenesis (Scarpulla, 2002). In the present study, basal (pre-exercise) CS and HAD mRNAs were signficantly elevated in muscle from the trained leg, providing evidence that mitochondrial biogenesis was induced by the 4 week endurance training programme. To further examine this issue, we analysed muscle RNA samples from two previously published studies (Pilegaard et al. 2000) in which subjects performed 5 consecutive days of one-legged knee extensor exercise (≈70 % of 2 min maximum workload to exhaustion, ≈80 min on the 5th day) or a single bout of cycling (≈45 % of peak oxygen uptake, 4 h), and again found no change in muscle NRF-1 mRNA up to 4 h after the end of exercise or in response to exercise training (Table 3). Thus, despite a dramatic increase in PGC-1α, together these findings provide evidence that exercise does not induce a rapid or dramatic increase in the expression of the NRF genes in skeletal muscle of humans, implying that the existing concentration of these transcription factors, if involved in the adaptive response to exercise, is sufficient to meet the needs of the cell.
Table 3.
NRF-1 mRNA content in response to two different exercise protocols
| Pre | 0′ | 15′ | 1h | 2h | 4h | |
|---|---|---|---|---|---|---|
| One-legged knee extensor exercise | 1 | 1.2 ± 0.2 | 1.0 ± 0.1 | 1.2 ± 0.1 | 1.2 ± 0.2 | 1.4 ± 0.3 |
| Cycling exercise | 1 | 0.9 ± 02 | 08 ± 02 | 0.9 ± 0.3 | 1.2 ± 0.4 | 1.3 ± 0.5 |
The mRNA content of the nuclear respiratory factor 1 in human skeletal muscle in response to exhaustive one-legged knee extensor exercise (70% of 2 min maximum workload, ˜80 min) and to 4 h of cycling at approximately 50% of peak oxygen uptake. Muscle biopsies were obtained from the vastus lateralis muscle before (Pre) and immediately after (0′) exercise and after 15 min (15′), 1h, 2h and 4h of recovery. Values are presented as fold changes relative to Pre, which was set to 1. Values are means 1 S.E.M. These data were obtained from analysis of RNA samples from two studies previously described in detail (Pilegaard et al. 2000). The samples were analysed by conventional RT-PCR using a set of primers (FP 5′-TGGGCGGGAAGACCTTTTGTATG-3′; RP 5′-ATGCTTGCGTCGTCTGGATGGT-3′) different from the set used in the present study.
Enzymes regulated by sustained increases in intracellular calcium, including calcineurin, a calcium-calmodulin-dependent protein phosphatase, and CaMK have recently been implicated in the control of the slow-oxidative type I fibre phenotype (Olson & Williams, 2000; Wu et al. 2002). Interestingly, PGC-1α mRNA is significantly upregulated in muscle from transgenic mice expressing a truncated constitutively active form of CaMKIV (Wu et al. 2002), thus raising the possibility that calcium signalling mechanisms may mediate the exercise-induced activation of the PGC-1α gene. Although activity of the calcium signalling enzymes was not determined in the present study, exercise did induce a significant increase in mRNA for both the α and β isoforms of the calcineurin A catalytic subunit in the untrained leg (Table 2), raising the interesting possibility that training may increase the sensitivity of the calcineurin signalling pathway in muscle.
The dramatic induction of PGC-1α observed in the present study suggests that a rapid increase in the amount of this transcriptional coactivator may play an important role in initiating, at least in part, the transcriptional activation of exercise-responsive genes in muscle (Pilegaard et al. 2000). For example, exercise in the untrained leg induced a marked transient increase (≈6-fold) in HKII mRNA (Table 2), a gene that is often associated with adaptations to endurance training. In apparent contrast, however, no acute increases in CS or HAD mRNA were detected during recovery from exercise in either the untrained or trained leg despite the higher basal levels of each mRNA in the trained leg (Table 2). Although it is possible that the peak in mRNA for some genes such as CS or HAD may occur much later in recovery (e.g. after 6–24 h), it is also clear that some genes such as HKII are much more responsive to exercise. It is therefore reasonable to speculate that there may be a temporal heirarchy to the transcriptional activation of genes during recovery from exercise initially involving immediate early/ stress response genes, transcription factors/co-activators such as PGC-1α, and select genes potentially required for metabolic recovery such as HKII, pyruvate dehydrogenase kinase 4 and uncoupling protein 3 (Pilegaard et al. 2000).
In summary, the present data show that exercise induces a dramatic transient increase in transcription of the PGC-1α gene in human skeletal muscle during recovery from exercise. Activation of PGC-1α appears to be enhanced with training and occurs in the absence of marked changes in the mRNA expression of putative upstream calcium signalling proteins or downstream transcription factor targets (e.g. NRF-1 or NRF-2α). The ability of PGC-1α to transform a predominately glycolytic muscle to an oxidative phenotype when selectively expressed in muscle of transgenic mice (Lin et al. 2002) raises the exciting possibility that the transient induction of PGC-1α in response to exercise may be involved in coordinating the subsequent changes in gene expression that underly the adaptive responses to endurance exercise in skeletal muscle of humans.
Acknowledgments
We wish to thank the subjects who participated in the study for their extraordinary effort. The technical assistance of Kristina Møller Kristensen, Michael Kristensen, and Bjarki Haraldsson is gratefully acknowledged. We also thank Eva Blomstrand for performing the CS activity analyses. The study was supported by grants from the Danish National Research Foundation (504-14), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR-45372), Bethesda, MD, USA. H.P. was in part supported by the Ministry of Culture Committee on Sports Research, Denmark, and additional support was obtained from Team Denmark, Denmark.
References
- Andersen P, Adams RP, Sjogaard G, Thorboe A, Saltin B. Dynamic knee extension as model for study of isolated exercising muscle in humans. J Appl Physiol. 1985;59:1647–1653. doi: 10.1152/jappl.1985.59.5.1647. [DOI] [PubMed] [Google Scholar]
- Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- Bergstrom J. Muscle electrolytes in man. Scand J Clin Lab Invest. 1962;68:1–110. [Google Scholar]
- Booth FW, Baldwin KM. Muscle plasticity: energy demand and supply processes. In: Rowell LB, Shepherd JT, editors. The Handbook of Physiology. Exercise: Regulation and Integration of Multiple Systems. Bethesda: American Physiological Society; 1996. pp. 1075–1123. [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Hildebrandt AL, Neufer PD. Exercise attenuates the fasting-induced transcriptional activation of metabolic genes in skeletal muscle. Am J Physiol Endocrinol Metab. 2000;278:E1078–1086. doi: 10.1152/ajpendo.2000.278.6.E1078. [DOI] [PubMed] [Google Scholar]
- Hood DA. Contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90:1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Michael LF, Wu Z, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, Kelly DP, Spiegelman BM. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc Natl Acad Sci U S A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Shimomura Y, Yoshimura A, Sokabe M, Fujitsuka N. Induction of nuclear respiratory factor-1 expression by an acute bout of exercise in rat muscle. Biochim Biophys Acta. 1998;1381:113–122. doi: 10.1016/s0304-4165(98)00018-x. [DOI] [PubMed] [Google Scholar]
- Olson EN, Williams RS. Calcineurin signaling and muscle remodeling. Cell. 2000;101:689–692. doi: 10.1016/s0092-8674(00)80880-6. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Keller C, Steensberg A, Helge JW, Pedersen BK, Saltin B, Neufer PD. Influence of pre-exercise muscle glycogen content on exercise-induced transcriptional regulation of metabolic genes. J Physiol. 2002;541:261–271. doi: 10.1113/jphysiol.2002.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Ordway GA, Saltin B, Neufer PD. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. Am J Physiol Endocrinol Metab. 2000;279:E806–814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Saltin B, Gollnick PD. Skeletal muscle adaptability: significance for metabolism and performance. In: Peachey LD, Adrian RH, Geiger SR, editors. The Handbook of Physiology. Skeletal Muscle. Baltimore, USA: Williams & Wilkins; 1983. pp. 555–631. [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun. 2002;296:350. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- Tunstall RJ, Mehan KA, Wadley GD, Collier GR, Bonen A, Hargreaves M, Cameron-Smith D. Exercise training increases lipid metabolism gene expression in human skeletal muscle. Am J Physiol Endocrinol Metab. 2002;283:E66–72. doi: 10.1152/ajpendo.00475.2001. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Neufer PD. Regulation of gene expression in skeletal muscle by contractile activity. In: Rowell LB, Shepherd JT, editors. The Handbook of Physiology. Exercise: Regulation and Integration of Multiple Systems. New York, USA: Oxford University Press; 1996. pp. 1124–1150. [Google Scholar]
- Winder WW. Energy-sensing and signaling by AMP-activated protein kinase in skeletal muscle. J Appl Physiol. 2001;91:1017–1028. doi: 10.1152/jappl.2001.91.3.1017. [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]


