Abstract
Recently, TAP42 was isolated as a high copy suppressor of sit4−, a yeast phosphatase related to protein phosphatase 2A (PP2A). TAP42 is related to the murine α4 protein, which was discovered independently by its association with Ig-α in the B cell receptor complex. Herein we show that a glutathione S-transferase (GST)–α4 fusion protein bound the catalytic subunit (C) of human PP2A from monomeric or multimeric preparations of PP2A in a “pull-down” assay. In an overlay assay, the GST–α4 protein bound to the phosphorylated and unphosphorylated forms of C that were separated in two-dimensional gels and immobilized on filters. The results show direct and exclusive binding of α4 to C. This is unusual because all known regulatory B subunits, or tumor virus antigens, bind stably only to the AC dimer of PP2A. The α4–C form of PP2A had an increased activity ratio compared with the AC form of PP2A when myelin basic protein phosphorylated by mitogen-activated protein kinase and phosphorylase a were used as substrates. Recombinant α4 cleaved from GST was phosphorylated by p56lck tyrosine kinase and protein kinase C. A FLAG-tagged α4 expressed in COS7 cells was recovered as a protein containing phosphoserine and coimmunoprecipitated with the C but not the A subunit of PP2A. Treatment of cells with rapamycin prevented the association of PP2A with FLAG-α4. The results reveal a novel heterodimer α4–C form of PP2A that may be involved in rapamycin-sensitive signaling pathways in mammalian cells.
Protein phosphatase 2A (PP2A) is involved in signal transduction pathways that regulate multiple functions, including those of yeast and mammalian cells. PP2A exists as a set of ABC heterotrimers, composed of a core dimer of a 60-kDa (A) subunit and a 36-kDa catalytic (C) subunit, plus a variety of distinct regulatory (B) subunits (see refs. 1 and 2). Studies have shown that the B and C subunits do not interact independently. The A subunit, which is made of repeated “armadillo” sequence motifs, is required for assembly of the trimers (3–6).
The different regulatory B subunits of PP2A modify its activity, substrate specificity, and subcellular localization (3, 7). The viral protein simian virus 40 small tumor antigen suppresses the activity of host cell PP2A by binding to the AC core dimer, replacing endogenous B subunits. This results in activation of the mitogen-activated protein (MAP) kinase pathway and serum-independent growth (8). By using the yeast two-hybrid system, other proteins that interact with PP2A have been discovered. The HIV-encoded protein vpr associates with PP2A via the A subunit (9), activating the phosphatase (10) as a means of producing cell cycle arrest. HOX11 is a homeobox protein that binds to the C subunits of both PP1 and PP2A to modify activity and G2/M progression (11). The translation termination factor eRF1 binds to the AC dimer via the C subunit and is thought to recruit PP2A to polysomes (12).
Rapamycin is an immunosuppressant macrolide compound that has antifungal activity that induces G1 phase growth arrest in Saccharomyces cerevisiae. Mutations of sit4, a gene encoding a protein phosphatase (13) related to PP2A, also produce G1 phase arrest (14). A screen for high copy suppressors of ts-sit4− yielded an essential gene, TAP42 (15). The Tap42 protein binds to both the Sit4 phosphatase and the yeast C subunits of PP2A, encoded by the Pph21 and Pph22 genes. Binding to the Pph21 and Pph22 proteins did not require expression of the yeast homologues of the A and B subunits of PP2A, suggesting that Tap42 could bind the phosphatase C subunits independently. Mutation of Tap42 conferred rapamycin resistance, suggesting that this protein is involved in the target of rapamycin signaling pathway (15).
The murine α4 protein is the closest match to Tap42 (24% identity) and was originally discovered as a 52-kDa phosphoprotein associated with Ig-α in the B cell receptor complex (16). The α4 protein was phosphorylated in response to phorbol ester treatment of WEHI-231 cells (16), and it coprecipitated with a 70-kDa phosphotyrosine protein and with Tyr kinase activity, leading to the proposal that α4 may be involved in B cell signaling (17). However, if and how α4 participates in signaling downstream of the IgM receptor is still unknown. With the discovery of Tap42, its association with yeast phosphatases, and its putative involvement downstream of the target of rapamycin protein, it became important to determine whether α4 would bind mammalian PP2A. In this report, we show the direct binding of glutathione S-transferase (GST)–α4 protein to the C subunit of PP2A in binding assays and coimmunoprecipitation of FLAG-α4 and PP2A from cells. Association of α4 and PP2A in cells is blocked by rapamycin. The α4 behaves as a new type of phosphatase regulatory subunit that alters specificity of C subunit and promotes release of the A subunit.
MATERIALS AND METHODS
Materials.
The AC heterodimer of PP2A was purified from human red blood cells by a modification of the method of Usui et al. (18). The monomeric C subunit of PP2A was prepared by precipitating the AC dimer with 80% ethanol at room temperature, collecting the protein by centrifugation, extracting the pellet with 50 mM Tris⋅HCl (pH 7.4), 5 mM MgCl2, 30 mM 2-mercaptoethanol, and 10% glycerol, followed by centrifugation to remove insoluble protein. This C subunit preparation was free of any A subunit, based on immunoblotting, and all activity was eluted at 36 kDa from Toyopearl 50S gel filtration column (TosoHaas, Montgomeryville, PA). Rabbit antibodies against residues 168–188 of the C subunit of PP2A were prepared and affinity-purified on immobilized peptide. Polyclonal antibodies against residues 48–61 of the C subunit of PP1α were provided by Gregg Gunderson (Columbia University). Recombinant PP1Cα was provided by Ernest Y. Lee (Miami University, Miami, FL). The anti-PP2A and anti-PP1α antibodies did not cross-react with the other phosphatase C subunit. Anti-A subunit antibodies (against residues 7–19) were purchased from Calbiochem. Rabbit polyclonal antibody against GST–α4 fusion protein was raised by immunizing two rabbits with the GST–α4 fusion protein. Ig fraction of antiserum was prepared by 50% ammonium sulfate precipitation and dialysis and used for Western blotting. PKC, phosphatidylserine, and diacylglycerol were purchased from Upstate Biotechnology. COS-7 and 10T1/2 cells were grown in 100-mm dishes in DMEM plus 10% fetal calf serum, at 37°C in a humidified atmosphere of 5% CO2/95% air.
Preparation of GST–α4 and Recombinant α4.
Total RNA was isolated from frozen mouse spleen by a guanidine thiocyanate extraction method (19). Oligonucleotides used for PCR were 5′-CGCGGATCCGCAGCGTCTGAAGACGAGTTACTG-3′ for the forward primer and 5′-CGGAATTCCGCCCATGTTCTGTCGGTTGCCGTAG-3′ for the backward primer. First-strand cDNA synthesis was done with 2.6 μg of RNA, by following the manufacturer’s protocol (SuperScript preamplification system, GIBCO/BRL). Double-stranded cDNA was synthesized by using the forward primer and cDNA was amplified by 25 cycles of PCR, with annealing at 55°C. DNA was digested and subcloned into the BamHI and EcoRI sites of pGEX-4T-2 vector (Pharmacia Biotech). Protein expression was induced by adding 0.5 mM isopropyl β-d-thiogalactoside to transformed Escherichia coli BL-21. The GST–α4 fusion protein was purified with glutathione-agarose (Sigma), as described (20).
The GST–α4 bound to glutathione beads (0.5 ml) was cleaved with tev protease (10 μl) at 4°C for 60 min and the α4 was recovered as two proteins of 42 and 35 kDa, consistent with release of α4 from GST in the fusion proteins (see Fig. 1).
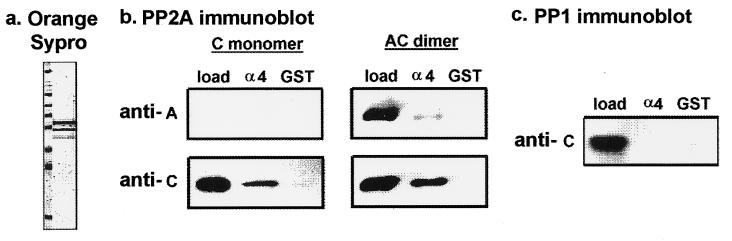
Figure 1.
Binding of GST–α4 with purified PP2A. (a) Bacterial GST–α4 fusion protein was purified, analyzed by SDS/PAGE, and visualized by fluorescent protein staining with Orange Sypro (Molecular Probes). Molecular mass markers shown in the left lane are 205, 116, 97, 84, 66, 55, 45, and 30 kDa. (b) Immunoblotting with anti-PP2A subunit antibodies was used to assay binding of PP2A to GST–α4. Human PP2A (C monomer or AC dimer) was incubated with equimolar amounts of either GST–α4 protein (α4) bound to glutathione-agarose or GST alone (GST) bound to glutathione-agarose. After washing and elution, 50% of the eluted proteins were analyzed by immunoblotting with antibodies against A subunit (anti-A) or C subunit (anti-C) of PP2A. For visual comparison, 5% of the PP2A added to the assay (load) was stained. (c) The C subunit of PP1 was incubated with GST–α4 (α4) or GST alone (GST), as described above. Immunoblotting used anti-peptide antibody against the PP1 C subunit (anti-C).
PP2A Binding Assays.
In vitro binding assay (“pull down”) was performed with purified PP2A and GST–α4 protein bound to glutathione-agarose beads, in a buffer containing 25 mM Tris⋅HCl (pH 7.5), 75 mM NaCl, 0.25% Nonidet P-40, and 15 mM 2-mercaptoethanol for 3 h at 4°C. The beads were washed once briefly with ice-cold buffer containing 25 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl (TBS) and 5× Laemmli sample buffer was added and boiled for 1 min. Proteins were resolved by SDS/PAGE and quantitated by FluorImaging Western blot (20) detection system (Amersham).
The α4 cDNA was subcloned into pGEX-2TK vector to provide a site for phosphorylation by cAMP-dependent protein kinase (PKA). The GST–RRASV–α4 fusion protein and GST–RRASV protein as a control were purified and phosphorylated by PKA according to the manufacturer’s protocol (Pharmacia Biotech). Lysates of mouse fibroblasts (10T1/2 cells) were subjected to two-dimensional electrophoresis and transferred onto filters as described (21). The membrane was blotted against anti-PP2A (residues 168–188) antibody and then incubated with 32P-labeled GST–RRASV–α4 protein or GST–RRASV protein. Binding of the radiolabeled fusion protein was detected with PhosphorImager (Molecular Dynamics).
Phosphatase Activity Measurement and Phosphorylation of α4.
Substrates for PP2A included 32P-labeled phosphorylase a (phos a), prepared as described (22), and 32P-labeled myelin basic protein (MBP) prepared by using activated MAP kinase. Assay of PP2A used an established protocol (22), with either purified PP2A heterodimer or the heterodimer bound to GST–α4, with correction for the amount of PP2A activity associated with GST alone. Phos a was used at 15 μM and MBP was used at 1 μM. The assays were linear with respect to time and concentration under the conditions used.
Recombinant α4 was cleaved from the GST fusion protein and recovered as α4 proteins of 42 and 35 kDa. They were phosphorylated in a reaction containing 20 mM Tris⋅HCl (pH 7.4), 20 mM β-glycerophosphate, 10 mM MgCl2, 0.5 mM MnCl2, 0.1% Nonidet P-40, and 1 mM ATP with [γ-32P]ATP (1,000 dpm/pmol), plus 1 mM orthovanadate and 5 nM calyculin A. The p56lck protein Tyr kinase, isolated from LSTRA cell membranes, or PKC, premixed with CaCl2, phosphatidylserine, and diacylglycerol, was added as the kinase to start the reaction and samples were analyzed at various times by SDS/PAGE and autoradiography.
Expression of FLAG-α4 Fusion Protein in COS7 Cells.
Full-length α4 cDNA was subcloned into BamHI and EcoRI sites of FLAG2 vector. COS7 cells were grown to 50% confluency and transfected by lipofection according to the manufacturer’s instructions (GIBCO/BRL). Plasmid DNA (10 μg per 100-mm dish) was transiently transfected overnight with serum-free medium and cells were then incubated with 10% fetal bovine serum for 36–48 h. The cells were metabolically labeled for 3 h with [32P]orthophosphate at 0.2 mCi/ml in phosphate-free DMEM (1 Ci = 37 GBq). The labeled cells were washed with TBS and lysed on the plate with 0.4 ml of ice-cold lysis buffer containing 0.5% Nonidet P-40, 25 mM Tris⋅HCl (pH 7.4), 75 mM NaCl, 30 mM 2-mercaptoethanol, aprotinin (20 μg/ml), leupeptin (20 μg/ml), 2 mM Pefablock (Boehringer-Mannheim), 1 mM Na3VO4, and 1 μM okadaic acid. Lysed cells were scraped off, transferred into microtubes, and centrifuged at 10,000 × g for 5 min. The supernatants (detergent-soluble fractions) were immunoprecipitated with anti-FLAG M2 affinity gel (Kodak) and immunoblotted with polyclonal antibody against α4 and anti-peptide (residues 168–188) antibody against C subunit of PP2A.
RESULTS
Binding of PP2A with GST–α4.
The cDNA for α4 protein was prepared from mouse spleen total RNA by reverse transcription-coupled PCR and used to produce a GST–α4 fusion protein in bacteria. About half the fusion protein was recovered full length (70 kDa) and there were two additional forms truncated from the C terminus by less than 10 kDa (Fig. 1a). This was presumably due to partial degradation of the fusion protein during isolation. This preparation of GST–α4 bound to glutathione beads was used in a binding assay with different purified forms of human PP2A. Both the AC heterodimer and the monomeric C subunit forms of PP2A bound to GST–α4 in a pull-down assay, indicative of stable association (Fig. 1b, α4 lanes). The same amount of PP2A, based on content of C by immunoblotting, was added to each assay, and the same fraction of the total C was recovered bound to GST–α4. Therefore, association of C monomer and AC heterodimer with GST–α4 was about the same. Under these conditions the AC heterodimer is stable in solution, with no evidence for spontaneous dissociation. However, with the AC heterodimer a barely detectable amount of the A subunit was recovered bound to GST–α4 (Fig. 1b). This indicated that the A subunit was displaced from C bound to GST–α4.
There were two different controls to demonstrate specificity of the binding interaction. (i) GST alone was incubated with both forms of PP2A and showed no stable association (Fig. 1b, GST lanes). (ii) The purified C subunit of PP1 did not show binding to either GST or GST–α4 (Fig. 1c). The results show specific binding of GST–α4 directly to the C subunit of PP2A.
The GST–α4 also was incubated with the ABC heterotrimer of PP2A from red blood cells that had been purified by DEAE chromatography. The fraction of the total C subunit of PP2A that bound to GST–α4 was about the same as for monomeric C or heterodimer AC in the assay (data not shown). Again, a much smaller fraction of the A or B subunits, relative to C subunit, was detected by immunoblotting proteins bound to GST–α4. The results lead us to speculate that there was transient formation of a GST–α4–ABC tetramer, and release of the A and B subunits to yield GST–α4–C.
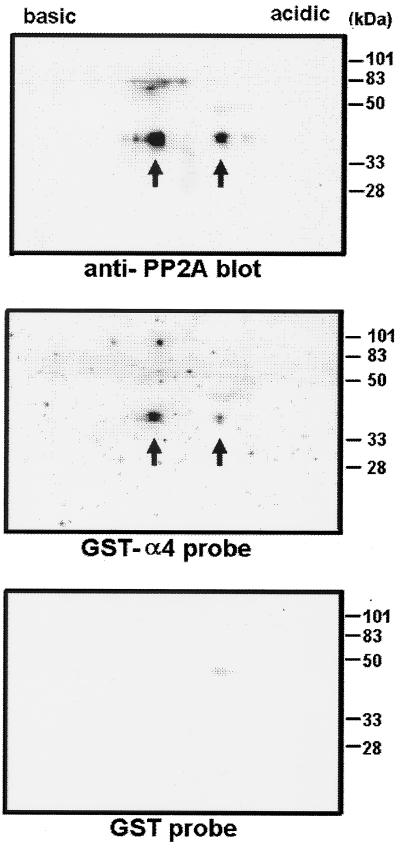
Another assay used radiolabeled GST–α4 fusion protein as a probe for immobilized C subunit of PP2A. Only the GST–RRASV–α4 protein was phosphorylated by PKA, not the GST–4T2–α4 protein, showing that the only site of PKA phosphorylation was in the RRASV insert in the spacer region, not in α4 itself. Both GST–RRASV–α4 and GST–RRASV were 32P-labeled to the same specific radioactivity and used to probe PP2A separated by two-dimensional gel electrophoresis. Only the GST–RRASV–α4 bound to the PP2A C subunit, not the GST–RRASV (Fig. 2). The location of PP2A was detected with a specific anti-peptide antibody. The immunoblotting revealed the phosphorylated and unphosphorylated forms of the C subunit at 36 kDa (Fig. 2 Top). The GST–RRASV–α4 appeared to bind to both these forms of PP2A (Fig. 2 Middle) that were isolated from mouse 10T1/2 fibroblasts, showing that phosphorylation of the C subunit of PP2A did not influence binding of GST–α4. As a control the GST–RRASV did not show any significant binding (Fig. 2 Lower) to PP2A C subunit.
Figure 2.
Binding of 32P-labeled GST–α4 probe to PP2A from fibroblasts. Cell lysates of mouse 10T1/2 fibroblasts were subjected to two-dimensional gel electrophoresis and transferred onto nitrocellulose filters. The filters were immunoblotted with antibody against C subunit of PP2A (Top). Unphosphorylated and phosphorylated PP2A was detected as two forms (arrows) at 36 kDa, separated by isoelectric focusing. By using duplicate filters, one was incubated for 30 min with 32P-labeled GST–α4 probe (Middle) and the other was incubated with 32P-labeled GST probe (Bottom). Molecular mass standards are shown on the right.
Effects of GST–α4 on PP2A Activity.
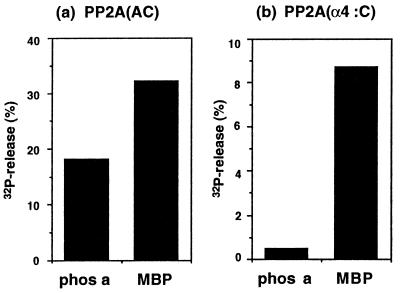
The phosphatase activity of PP2A was measured by release of 32Pi, using as substrates 32P-labeled phos a and MBP 32P-phosphorylated at Thr-97 by MAP kinase (23). Under the conditions of the assay, purified PP2A heterodimer hydrolyzed MBP in preference to phos a (Fig. 3). The preference is evident even with phos a at a 15-fold higher concentration than MBP in the assays. The activity ratio of MBP/phos a of 1.5 to 1.8 was observed in multiple assays at a range of reactions times and with various dilutions of PP2A. By comparison, the PP2A bound to GST–α4 on glutathione beads hydrolyzed MBP with a much higher preference over phos a, giving a ratio of MBP/phos a of 19. Addition of soluble GST–α4 to assays gave a dose-dependent increase in activity of PP2A toward MBP, relative to addition of the same amount of GST alone (data not shown). These assays measured the increase in activity of the small fraction of the PP2A in the assay that was bound to GST–α4. When all the PP2A in the assay was bound to immobilized GST–α4, the MBP/phos a ratio gauged effects of GST–α4 on activity of PP2A, regardless of the total amount of activity in the assay. The 10-fold increase in the activity ratio shows that the interaction of α4 with PP2A was regulatory, insofar as it affected activity of the enzyme with different substrates.
Figure 3.
Effects of α4 binding on PP2A phosphatase activity. Phosphatase activity was measured by using 32P-labeled phos a and 32P-labeled MBP as substrates. (a) The AC dimer of PP2A was incubated with 15 μM 32P-labeled phos a or 1 μM 32P-labeled MBP for 1 h at 30°C. (b) The AC dimer of PP2A was bound to GST–α4 on agarose beads or beads with GST alone, as described in text. The PP2A bound to GST–α4 was assayed as the difference in activity between the GST–α4 and GST alone. Results shown are the average of two experiments.
Phosphorylation of α4 in Vitro and in Vivo.
The GST–α4 protein purified from bacteria was not radiolabeled in vitro by [γ-32P]ATP plus either PKC or p56lck, though both kinases had robust activity with MBP in parallel control reactions (data not shown). Perhaps the N-terminal GST interfered with phosphorylation by blocking access to or altering the conformation of α4. When the immobilized fusion protein was cleaved, α4 proteins of 42 and 35 kDa were recovered, corresponding to full-length and C-terminal truncated forms. Reaction of these proteins with either PKC or p56lck gave time-dependent radiolabeling of both proteins (data not shown). This indicated that recombinant α4 could be phosphorylated by either kinases. The sites of Tyr and Ser/Thr phosphorylation apparently were not in the C-terminal 7-kDa that was missing from the smaller α4 protein that was labeled efficiently. Integration of the total radiolabeling of the α4 proteins showed that the same plateau was attained within 2 h with either kinase, but reaction with both kinases was not additive. We conclude that α4 can be phosphorylated by one or the other kinase but not both.
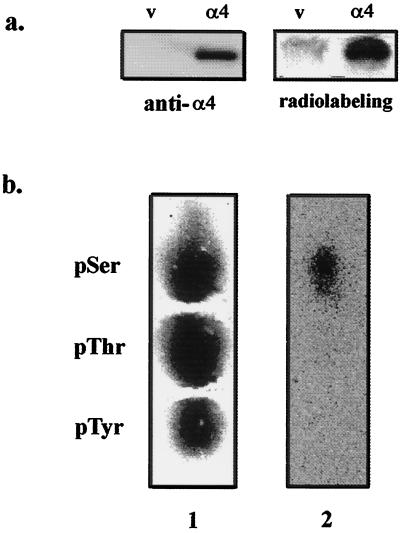
To investigate phosphorylation of α4 in intact cells an epitope-tagged version of α4 was transiently expressed in COS7 cells. As a control, cells were transfected with empty vector lacking the FLAG-α4 coding sequence. Metabolic 32P-labeling of the cells and immunoprecipitation with anti-FLAG M2 antibody showed that an α4 protein was only recovered from cells transfected with vector encoding FLAG-α4 (Fig. 4a Left). This FLAG-α4 was a 32P-phosphoprotein (Fig. 4a Right) that was excised from the gel, hydrolyzed, and found to contain only phosphoserine, based on phosphoamino acid analysis by high-voltage electrophoresis (Fig. 4b).
Figure 4.
FLAG-α4 in COS7 cells is phosphorylated exclusively on serine. (a) Cells transfected with empty vector (lanes v) or FLAG-tagged α4 (lanes α4) were metabolically labeled with 32P. After immunoprecipitation with anti-FLAG M2 affinity gel, proteins were analyzed by immunoblotting using anti-α4 antiserum (Left) and for 32P by PhosphorImaging (Right). FLAG-α4 migrated as a protein of 47 kDa. Results were replicated in three independent experiments. (b) The FLAG-α4 protein was excised from the gel and digested with trypsin, and the tryptic peptides were acid-hydrolyzed to yield phosphoamino acids that were analyzed with internal standards by high-voltage electrophoresis (24, 25). The standards, phosphoserine (pSer), phosphothreonine (pThr), and phosphotyrosine (pTyr) were stained with ninhydrin (lane 1) and radioactivity in the same lane was detected by phosphorimaging (lane 2), showing that the FLAG-α4 only contained radiolabeled phosphoserine.
In Vivo Binding of α4 with PP2A and Effect of Rapamycin.
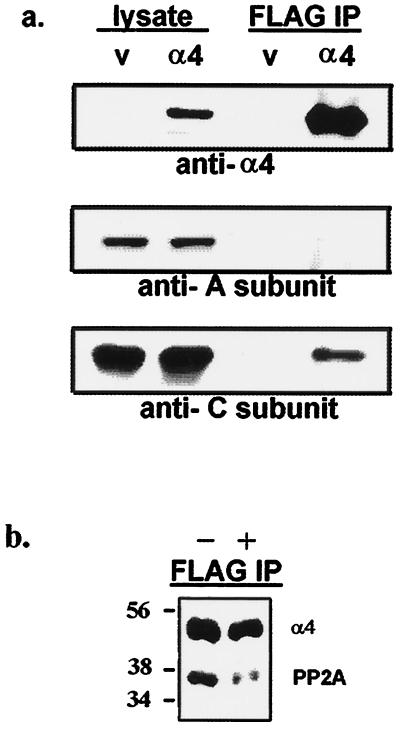
The FLAG-tagged version of α4 was expressed in COS7 cells to test for its interaction with endogenous PP2A. Cells were transfected with either empty vector or with a vector encoding FLAG-α4. Overexpression of the tagged α4 did not noticeably affect cell growth over a period of 2 days, and the 47-kDa protein was detected in cell lysates by immunoblotting with a rabbit anti-α4 antibody (Fig. 5a Top). The 36-kDa C subunit of PP2A was specifically coprecipitated with FLAG-α4, using anti-FLAG M2 affinity gel. (Fig. 5a Bottom). Only a relatively small fraction of the total PP2A in the cells was immunoprecipitated with the FLAG-α4 (estimated as 2–5%). Immunoblotting with an anti-peptide antibody revealed the 60-kDa A subunit in lysates, but there was no A subunit detected in the FLAG immunoprecipitates (Fig. 5a Middle). This result is consistent with the in vitro binding assays showing selective recovery of C subunit, with GST–α4 (Fig. 1). If the cells expressing FLAG-α4 were treated with 100 nM rapamycin in serum-free medium for 30 min before lysis and anti-FLAG precipitation, then the FLAG-α4 was still recovered but not the PP2A (Fig. 5b). Rapamycin interfered with the association of PP2A and FLAG-α4 in living cells.
Figure 5.
Binding of FLAG-α4 to PP2A in COS7 cells is inhibited by rapamycin. (a) Cells were transfected with empty vector (lanes v) or FLAG-tagged α4 (lanes α4). Lysates (5% of total, left lanes) and immunoprecipitates with anti-FLAG M2 affinity gel (50% of total, right lanes) were analyzed by immunoblotting using anti-α4 antiserum (Top), anti-A subunit of PP2A antibody (Middle), and anti-C subunit of PP2A antibody (Bottom). Expression of FLAG-α4 in these cells did not alter the level of endogenous A (60 kDa) and C (36 kDa) subunits of PP2A. Results were replicated in several experiments. (b) Cells were transfected and processed as in above, except 100 nM rapamycin (lane +) or vehicle alone (lane −) was added in serum-free medium for 30 min before harvesting. The anti-FLAG precipitates were resolved by SDS/PAGE and immunoblotted with a mixture of anti-α4 and anti-C subunit antibodies to detect both proteins in a single step. Immunoblotting with each antibody separately confirmed the specificity of staining. Cells transfected with empty vector gave no signals as shown above. The results were replicated in independent experiments.
DISCUSSION
The results of this study show, to our knowledge, for the first time that murine α4 protein binds directly to the C subunit of PP2A and this occurs with the exclusion of other subunits. Binding to GST–α4 was about the same for either the monomeric C subunit or the AC heterodimer of PP2A, and an insignificant fraction of the A subunit was recovered bound to GST–α4. To explain this observation, one can imagine that α4 and A subunit compete for the same site on the C subunit or that they bind at separate sites but binding of GST–α4 promotes dissociation of A. Either circumstance would distinguish α4 from other known PP2A regulatory subunits, such as B subunits or tumor virus antigens, because they require both A and C subunits to form stable heterotrimers.
Binding of α4 altered the substrate specificity of PP2A, with an increase in activity toward MBP labeled by MAP kinase, relative to phosphorylase. The increased activity ratio may reflect suppression of activity with the phosphorylase substrate and/or enhancement of activity with the MBP substrate. In this regard, α4 behaves like other B regulatory subunits of PP2A. Binding of different B subunits to the AC form of PP2A is known to produce suppression and enhancement of reactivity with different substrates (see ref. 1). At the least the results exclude the possibility that α4 simply acts as an inhibitor of PP2A, because the GST–α4–C heterodimer was an active phosphatase.
Examination of the interaction between α4 and PP2A was prompted by the isolation of Tap42 in yeast. Sit4 is a yeast phosphatase, and Tap42 was found as a high copy suppressor of ts-sit4 (15). The Tap42 protein stably associated with Sit4, and also with Pph21 and Pph22, the yeast PP2A homologues. Some 10–20% of each of the protein partners was complexed to one another, estimated from coprecipitation. Recently the human version of Sit4, called PP6, was cloned and its sequence was determined, but the recombinant protein was inactive in inclusion bodies (26). This may be the primary partner for α4 in mammalian cells, though PP2A also is involved. Results with the yeast proteins predicted that mammalian α4 would bind to mammalian C subunit of PP2A, and the other PP2A subunits would not be required for the interaction. What could not have been anticipated from the genetics, and would not have been guessed from the known biochemistry of PP2A, is that binding of α4 directly to the C subunit seemed to cause release of the A subunit. There is general agreement that most, if not all, PP2A in cells is present as ABC heterotrimers (1, 6). On the basis of our binding assays, α4 can bind monomer, dimer, and trimer forms of PP2A, so we imagine there even may be ABC–α4 tetramers transiently formed in cells, and these are proposed as intermediates that release subunits to give an α4–C heterodimer form of PP2A.
Interestingly, Tap42 conferred rapamycin sensitivity to yeast, and binding of Tap42 to Sit4 and Pph21/22 was inhibited by rapamycin treatment or nutrient withdrawal (14). Herein we show that rapamycin treatment of mammalian cells prevented association of FLAG-α4 with PP2A. This implies that a rapamycin-sensitive pathway is conserved for formation of Tap42–Sit4 and Tap42–Pph21/22 complexes in yeast and of α4–C heterodimers (where the C could be either PP2A or PP6) in mammalian cells. Because mutations of Tap42 in yeast afforded a rapamycin-resistant phenotype, one might speculate knock out of the mouse α4 would give a similar phenotype. A remaining challenge is to discover the basis for the effects of rapamycin on Tap42 and α4, which may involve blocking phosphorylation by target of rapamycin or another kinase.
Studies have revealed interesting differences between proteins that function as phosphatase regulatory subunits in yeast. The Sit4 phosphatase binds Tap42 and Sap155/Sap190 proteins (14, 27), whereas the Pph21/Pph22 (PP2A) phosphatase binds Tap42 and Tpd3 (A subunit) and Cdc55 (B subunit). The Sit4 does not bind Tpd3 and the Pph21/Pph22 do not bind Sap155/Sap190 (27). There must be divergent regions of Sit4 and Pph21/Pph22 dictating the specificity for regulatory subunits. Tap42 is unusual in binding to both these “cousins” of the phosphoserine/phosphothreonine phosphatase family, presumably to sites composed of common sequences. It will be important to investigate whether the same regulatory patterns are conserved with the mammalian proteins.
When first described, α4 was a 52-kDa phosphoprotein (pp52) that associated with the Ig-α protein of the B cell receptor complex. The phosphorylation of p52 was induced by phorbol 12-myristate 13-acetate in WEHI-231 B cells, and in vitro kinase reactions with anti-α4 immunoprecipitates showed phosphotyrosine, raising the possibility that α4 may be phosphorylated on both Tyr and Ser/Thr residues in cells (17). We found that FLAG-α4 expressed in COS cells was a phosphoprotein that only contained phoshphoserine. Finding only phosphoserine was consistent with our in vitro phosphorylation results. Recombinant α4 was phosphorylated by either a Tyr kinase (p56lck) or a Ser/Thr kinase (PKC), but the reactions were not additive, implying that the two modifications were mutually exclusive. The GST–α4 fusion protein was not phosphorylated under the same conditions, presumably due to steric interference by the GST. Though PP2A bound to GST–α4, the kinases did not react with the fusion protein, so we speculate that kinases and phosphatases interact at different sites to the N and C termini of α4, respectively. Physiological conditions that lead to Tyr phosphorylation of α4 have yet to be found, but perhaps this occurs transiently in response to activation of the B cell receptor. Phosphorylation of α4 was not essential for binding to PP2A but deserves more attention because it may modulate either binding affinity for PP2A, the regulatory effects on PP2A activity, or association with other proteins. Conversely, phosphorylation of the C subunit of PP2A, which occurs at Tyr-307 near the C terminus (21), did not seem to affect binding of α4.
One property of α4 remains curious. The 340 residue protein has an calculated mass of 42 kDa, and cleavage of the GST–α4 fusion protein yielded a 42-kDa protein. In addition, the FLAG-α4 protein migrated near the predicted size 47 kDa. The yeast homologue Tap42 as the number in its name implies, exhibited the predicted migration in gel electrophoresis. However, the α4 protein was originally purified from B cells as pp52. How α4 becomes pp52, or if these are identical, is not yet established. An anti-pp52 mAb was used to isolate the α4 clone. Our polyclonal serum against GST–α4 detected a protein of 42 kDa expressed in highest levels in mouse skeletal muscle, but it was present as well in mouse heart, kidney, liver, and spleen and in various cell lines. The role of α4–C in various cells and tissues remains to be determined. The wide distribution of α4, its binding to phosphatases, and the sensitivity of the binding to rapamycin raise the promise that further investigation of the function of α4 will prove interesting and worthwhile.
Acknowledgments
We thank Dr. Kim Arndt for sharing results prior to publication, Dr. Gregg Gunderson for providing anti-PP1C antibody, Dr. Ernest Y. C. Lee for providing recombinant PP1Cα, Dr. Thomas W. Sturgill for providing MAP kinase, and Dr. S. J. Parsons for the FLAG expression vector. We thank Lin Chen for phosphatase assays, Heekyoung Chung for preparation of the two-dimensional gel immunoblot, and Chris Todd for the panel of mouse tissue extracts. K.M. was supported by a postdoctoral fellowship from American Heart Association. This research was supported in part by a grant from the National Science Foundation (MCB9507357).
ABBREVIATIONS
- PP2A
protein phosphatase 2A
- GST
glutathione S-transferase
- phos a
phosphorylase a
- PKC
protein kinase C
- PKA
cAMP-dependent protein kinase
- MAP kinase
mitogen-activated protein linase
- MBP
myelin basic protein
- C subunit
catalytic subunit
References
- 1.Mumby M C, Walter G. Physiol Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- 2.Wera S, Hemmings B A. Biochem J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamibayashi C, Estes R, Slaughter C, Mumby M C. J Biol Chem. 1991;266:13251–13260. [PubMed] [Google Scholar]
- 4.Kamibayashi C, Lickteig R L, Estes R, Walter G, Mumby M C. J Biol Chem. 1992;267:21864–21872. [PubMed] [Google Scholar]
- 5.Kamibayashi C, Estes R, Lickteig R L, Yang S I, Craft C, Mumby M C. J Biol Chem. 1994;269:20139–20148. [PubMed] [Google Scholar]
- 6.Mayer-Jaekel R E, Hemmings B A. Trends Cell Biol. 1994;4:287–291. doi: 10.1016/0962-8924(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 7.McCright B, Rivers A M, Audlin S, Virshup D M. J Biol Chem. 1996;271:22081–22089. doi: 10.1074/jbc.271.36.22081. [DOI] [PubMed] [Google Scholar]
- 8.Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 9.Noronha C M C de, Takaori-Kondo A, McEntee M, Greene W C. Mol Biol Cell. 1996;7:355a. (abstr.). [Google Scholar]
- 10.Tung H Y L, Derocquigny H, Zhao L J, Cayla X, Rogues B P, Ozon R. FEBS Lett. 1997;401:197–201. doi: 10.1016/s0014-5793(96)01470-6. [DOI] [PubMed] [Google Scholar]
- 11.Kawabe T, Muslin A J, Korsmeyer S J. Nature (London) 1997;385:454–458. doi: 10.1038/385454a0. [DOI] [PubMed] [Google Scholar]
- 12.Andjelkovic N, Zolnierowicz S, Van Hoof C, Goris J, Hemmings B A. EMBO J. 1996;15:7156–7167. [PMC free article] [PubMed] [Google Scholar]
- 13.Arndt K T, Styles C A, Fink G R. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 14.Sutton A, Immanuel D, Arndt K T. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Como C J, Arndt K T. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 16.Kuwahara K, Matsuo T, Nomura J, Igarashi H, Kimoto M, Inui S, Sakaguchi N. J Immunol. 1994;152:2742–2752. [PubMed] [Google Scholar]
- 17.Inui S, Kuwahara K, Mizutani J, Maeda K, Kawai T, Nakayasu H, Sakaguchi N. J Immunol. 1995;154:2714–2723. [PubMed] [Google Scholar]
- 18.Usui H, Imazu M, Maeta K, Tsukamoto H, Azuma K, Takeda M. J Biol Chem. 1988;263:3752–3761. [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Kleiner U, Brautigan D L. Biochemistry. 1996;35:13858–13864. doi: 10.1021/bi961669e. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Parsons S, Brautigan D L. J Biol Chem. 1994;269:7957–7962. [PubMed] [Google Scholar]
- 22.Brautigan D L, Shriner C L. Methods Enzymol. 1988;159:339–346. doi: 10.1016/0076-6879(88)59034-1. [DOI] [PubMed] [Google Scholar]
- 23.Erickson A K, Payne D M, Martino P A, Rossomando A J, Shabanowitz J, Weber M J, Hunt D F, Sturgill T W. J Biol Chem. 1990;265:19728–19735. [PubMed] [Google Scholar]
- 24.Boyle W J, Van Der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 25.Jelinek T, Weber M J. BioTechniques. 1993;15:629–630. [PubMed] [Google Scholar]
- 26.Bastians H, Ponstingl H. J Cell Sci. 1996;109:2865–2874. doi: 10.1242/jcs.109.12.2865. [DOI] [PubMed] [Google Scholar]
- 27.Luke M M, Della Seta F, DiComo C J, Kobayashi R, Arndt K T. Mol Cell Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]