Abstract
Background
Psoriasis, a chronic inflammatory skin disease, affects approximately 2% of the population worldwide. Although the aetiology of psoriasis is poorly understood, patients with disease of early onset (Type I, age of onset ≤ 40 years) usually have a strong genetic component to the disease.
Objectives
The purpose of this study was to investigate the role of the protein tyrosine phosphatase nonreceptor type 22 (PTPN22) gene region in susceptibility to Type I psoriasis.
Patients and methods
Thirteen single nucleotide polymorphisms (SNPs) mapping to the PTPN22 region were genotyped in 647 patients with Type I psoriasis and 566 normal controls.
Results
The rs2476601 (R620W) SNP, widely associated with other inflammatory autoimmune diseases, showed no evidence of association with susceptibility to Type I psoriasis. Two SNPs (rs1217414 and rs3789604) demonstrated significant association with Type I psoriasis and were subsequently genotyped in a further 253 unrelated patients and 2024 normal controls. rs1217414 and rs3789604 were also significantly associated with Type I psoriasis in the combined datasets (P = 0·003 and P = 0·0002, respectively); furthermore carriage of both risk alleles was also significantly associated (P = 0·002).
Conclusions
This study demonstrates evidence of association of two SNPs (rs1217414 and rs3789604) in the PTPN22 region with Type I psoriasis, providing evidence for a role of this gene in Type I psoriasis that is not conferred by the R620W variant previously associated with a number of inflammatory diseases.
Keywords: Psoriasis, PTPN22, genetic association, linkage disequilibrium
Psoriasis is a chronic immune-mediated skin disease that affects approximately 2% of the population worldwide.1 The most common characteristics of the disease are red, well-demarcated and heavily scaled plaques on the knees, elbows and scalp but any skin surface can be affected. Most patients (75%) of chronic plaque psoriasis first present before the age of 40 years – known as early onset or Type I psoriasis. Late onset or Type II psoriasis presents after the age of 40 years.2 Family studies have provided strong evidence that there is an underlying genetic component to Type I psoriasis although the precise mode of inheritance is poorly understood. Genome-wide scans on patients with psoriasis have identified a number of key genomic regions linked to Type I disease.3–12 The best documented of these is psoriasis susceptibility region 1 (PSORS1) situated on chromosome 6p; here 35–50% of the heritability of psoriasis is believed to be attributable to this locus3 with a disease penetrance of 10–15%.13 The PSORS1 susceptibility region harbours the HLA-Cw6 allele which has a strong association with Type I psoriasis; as many as 85% of patients are HLA-Cw6 positive compared with 15% in Type II patients.2 Aside from PSORS1, there is also a significant contribution towards psoriasis susceptibility from non-HLA genes and the identification of these loci are key to understanding the full genetic component that underpins the pathogenesis of the disease. Potential candidate genes can be selected for investigation on the basis of their function or through overlap with related conditions. In particular recent attention has focused on the protein tyrosine phosphatase nonreceptor type 22 (PTPN22) gene.
The PTPN22 gene encodes the protein lymphoid tyrosine phosphatase (Lyp) which has an N-terminal phosphatase domain and a long, noncatalytic C-terminus with many proline-rich motifs. The R620W polymorphism in PTPN22 is associated with several immune-mediated diseases including rheumatoid arthritis,14–16 Type I diabetes mellitus (initial disease associated),17 juvenile idiopathic arthritis,16 systemic lupus erythematosus15 and autoimmune thyroid disease15 but not others, including psoriasis15,16,18,19 and psoriatic arthritis;16,20 however, speculation regarding the latter remains because of a more recent publication that detected a positive association with this marker.21 This polymorphism is located in the N-terminal proline-rich motif, binding to the src homology 3 binding domain Csk and results in a substitution of arginine with tryptophan. Studies have suggested that the W620 variant has reduced binding to Csk, down-regulating the binding of Lyp with Csk.16 The mechanism(s) underlying the reduced binding are not as yet fully understood, with more recent publications stating that the polymorphism is in fact a gain of function regulator of T cells22 as opposed to a loss of function variant which was the initial hypothesis.
Almost all studies of PTPN22 to date have focused on the R620W variant including those failing to find association with psoriasis. However, as activated T cells play a major role in the pathogenesis of psoriasis, PTPN22 remains a strong candidate gene for disease susceptibility.19 Therefore it is possible that polymorphisms in PTPN22 other than R620W may be important in psoriasis. A study of rheumatoid arthritis patients screened multiple single nucleotide polymorphisms (SNPs) across the gene and identified associations independent of R620W, illustrating the importance of systematic screening of this gene in autoimmune diseases.14 In this study we describe an investigation of SNPs covering the PTPN22 gene region in a case–control association study of U.K. patients with psoriasis.
Materials and methods
The study was a case–control candidate gene investigation of markers in the PTPN22 gene region carried out in a two-stage design.
Stage 1
Recruitment of patients with Type I psoriasis for the initial case cohort was coordinated through the Dermatology Centre, Hope Hospital, The University of Manchester, Manchester, U.K. All patients gave written informed consent and the study was approved by the Salford and Trafford Local Research Ethics Committee. In total 647 unrelated patients with Type I psoriasis (53·8% male; 46·2% female; mean age of onset 20 years; 50·9% patients HLA-Cw6 positive) were compared against 566 population-based controls of U.K. white ethnic origin.
Stage 2
A second cohort of 253 unrelated patients with Type I psoriasis (64·8% male; 32·4 female; mean age of onset 21 years) was provided by Guy's Hospital, London, U.K. A further 2088 controls were available from the 1958 British Birth Cohort study.23 These samples were used to evaluate further the SNPs found to be associated (P < 0·05 in both trend and genotypic association tests) in stage 1. The Oversight Committee for the Biomedical Assessment of the British 1958 Birth Cohort Study provided access to the DNA. Genotype data generated from the 64 nonwhite individuals included in the cohort was not included in the analysis, providing a total of 2024 controls (50·1% male; 49·9% female).23
Single nucleotide polymorphism selection
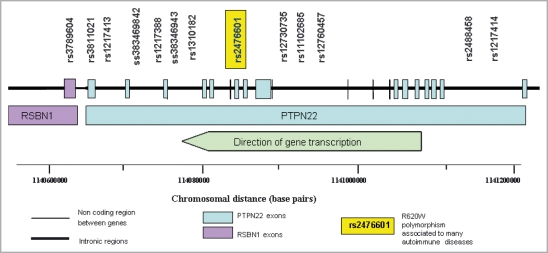
Seven haplotype-tagging SNPs across PTPN22 were augmented with three potentially functional SNPs, three SNPs with minor allele frequencies of less than 5% to increase the power of detecting association for rare SNPs and an additional SNP that had shown the most significant association to rheumatoid arthritis initially in an investigation by Carlton et al.14 who produced this particular SNP panel (Fig. 1). Assays were designed successfully for 13 of these markers.
Fig 1.
Map of the PTPN22 gene region and approximate location of genotyped SNPs.
Genotyping
Thirteen SNPs, including R620W, were genotyped in the PTPN22 region using Sequenom® MassArray™ technology according to the manufacturer's instructions using the iPLEX chemistry.
Statistical analyses
The statistical software package STATA version 8.2 (StataCorp Lp, College Station, TX, U.S.A.) was used to evaluate the differences in allele and genotype frequencies of each SNP between cases and controls and also to test the Hardy Weinberg equilibrium (HWE). Furthermore this software was used to perform stepwise logistic regression on the allelic and genotypic data of the combined dataset. Permutation testing of the data was performed using the PLINK statistical package,24 assessing the empirical significance level of the associations over 10 000 permutations.
Linkage disequilibrium
The genetic analysis software, HelixTree™ (Golden Helix Incorporated, Bozeman, MT, U.S.A.) was used to calculate the extent of linkage disequilibrium (LD) between the SNPs in the PTPN22 gene region. The parameters used for LD analysis are D′, which is a measure of the statistical significance of LD between two SNPs, and r2 which is the square of the correlation between two SNPs.
Results
Association testing: stage 1
In stage 1 of the study, two SNPs (rs1217414 and rs3789604) demonstrated significant genotypic association with Type I psoriasis (P ≤ 0·05 for both standard χ2 and χ2 test for trend, Table 1). No other SNPs, including R620W, showed evidence of association with Type I psoriasis by these tests (Table 1). The most significant difference in genotype frequencies of patients with Type I psoriasis and controls from stage 1 was observed for SNP rs1217414 (P = 0·0014), where risk was conferred by carriage of two copies of the minor allele (genotype TT) with an odds ratio (OR) = 2·39 (95% CI 1·43–4·11, P = 0·0005). However, the observed genotype frequencies in the controls deviated significantly from the expected number under the HWE for this SNP (P = 0·022). Nonetheless, the associations remained significant when comparing patient genotype frequencies with the expected HWE frequencies for controls. The genotype frequencies of rs3789604 were also significantly different when comparing patients with Type I psoriasis and controls in the initial analysis (Table 1). This association was based on a dominant model of inheritance for the major allele with OR = 1·43 (95% CI 1·13–1·80, P = 0·002) with the trend in frequencies also significant (Table 1) corresponding to an increase in major allele T frequency and TT genotype.
Table 1.
Genotype frequencies in cases and controls for Type I psoriasis in stage 1 of the study
| Patients | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype frequencies | Genotype frequencies | HWE in controlsa | Genotypic association | Trend test | ||||||
| SNP | 1_1 (%) | 1_2 (%) | 2_2 (%) | 1_1 (%) | 1_2 (%) | 2_2 (%) | exact P-value | χ2 | exact P-value | P-value |
| rs1217414 | 322 (50·79) | 253 (39·91) | 59 (9·31) | 309 (55·38) | 226 (40·50) | 23 (4·12) | 0·022 | 12·801 | 0·001 | 0·007 |
| rs2488458 | 369 (57·75) | 232 (36·31) | 38 (5·95) | 305 (55·56) | 215 (39·16) | 29 (5·28) | 0·304 | 1·121 | 0·564 | 0·663 |
| rs12760457 | 323 (50·71) | 265 (41·60) | 49 (7·69) | 258 (46·57) | 248 (44·77) | 48 (8·66) | 0·321 | 2·072 | 0·362 | 0·167 |
| rs11102685 | 523 (81·97) | 103 (16·14) | 12 (1·88) | 455 (81·40) | 102 (18·25) | 2 (0·36) | 0·212 | 6·691 | 0·031 | 0·701 |
| rs12730735 | 320 (50·79) | 262 (41·59) | 48 (7·62) | 245 (47·39) | 223 (43·13) | 49 (9·48) | 0·918 | 1·989 | 0·368 | 0·166 |
| rs2476601 | 408 (80·79) | 93 (18·42) | 4 (0·79) | 325 (82·70) | 64 (16·28) | 4 (1·02) | 0·552 | 0·799 | 0·687 | 0·549 |
| rs1310182 | 169 (34·00) | 246 (49·50) | 82 (16·5) | 101 (28·77) | 175 (49·86) | 75 (21·37) | 1·000 | 4·406 | 0·111 | 0·038 |
| ss38346943b | 487 (96·44) | 18 (3·56) | 0 (0·00) | – | – | – | – | – | – | – |
| rs1217388 | 373 (57·92) | 231 (35·87) | 40 (6·21) | 302 (54·32) | 223 (40·11) | 31 (5·58) | 0·266 | 2·309 | 0·312 | 0·399 |
| ss383469842 | 623 (96·89) | 20 (3·11) | 0 (0·00) | 535 (97·45) | 14 (2·55) | 0 (0·00) | 1·000 | 0·336 | 0·604 | 0·563 |
| rs1217413 | 395 (62·30) | 210 (33·12) | 29 (4·57) | 340 (62·85) | 178 (32·90) | 23 (4·25) | 1·000 | 0·087 | 0·972 | 0·797 |
| rs3811021 | 427 (67·14) | 192 (30·19) | 17 (2·67) | 358 (64·74) | 178 (32·19) | 17 (3·07) | 0·412 | 0·805 | 0·662 | 0·371 |
| rs3789604 | 445 (73·68) | 143 (23·68) | 16 (2·65) | 325 (64·61) | 160 (31·81) | 18 (3·58) | 0·887 | 10·646 | 0·005 | 0·002 |
Hardy Weinberg equilibrium (HWE) statistic calculated in STATA 8.2 from comparing genotype frequencies expected under HWE against the observed genotype frequencies.
Genotype failure in the controls.
SNP, single nucleotide polymorphism.
Stage 2 and combined analysis
Markers rs1217414 and rs3789604 were genotyped in additional case and control samples (n = 253 and n = 2024, respectively). Because of the small numbers of patients with psoriasis in the second cohort with an approximate 10 : 1 ratio of controls to patients, this additional genotype data was combined with data from stage 1. There was no increased evidence of association of rs1217414 with Type I psoriasis in the combined data sets although the difference in genotype frequency did remain significant (P = 0·003, Table 2) with risk again conferred by carriage of two copies of the minor allele with an odds ratio (OR) of 1·55 (95% CI 1·16–2·08, P = 0·003). Permutation testing of the data showed that the best model remained significant over 10 000 permutations for this marker (Table 2). In the combined data sets an increased evidence of association was obtained for SNP rs3789604 by both the standard χ2 and χ2 test for trend (P = 0·0002 and 0·0001, respectively; Table 2). Association for the dominant model showed an OR of 1·35 (95% CI 1·16–1·58, P = 0·002) for the major allele. Furthermore, the assortment of cases and controls concluded that the association had not occurred by chance following permutation testing (Table 2).
Table 2.
Genotype frequencies in cases and controls for Type I psoriasis in the combined study
| Combined analysis | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Genotype | Patients n (%) | Controls n (%) | HWE in controlsa | P-value | Trend test | Permutation (best model)b |
| rs1217414 | CC | 435 (49·54) | 1301 (54·14) | ||||
| CT | 360 (41·00) | 951 (39·58) | 0·2041 | 0·003 | 0·002 | 0·013 | |
| TT | 83 (9·46) | 151 (6·28) | |||||
| rs3789604 | TT | 613 (73·41) | 1621 (65·63) | ||||
| TG | 199 (23·83) | 765 (30·97) | 0·6457 | <0·001 | <0·001 | <0·001 | |
| GG | 23 (2·76) | 84 (3·40) | |||||
Hardy Weinberg equilibrium (HWE) statistic calculated in STATA 8.2 from comparing genotype frequencies expected under HWE against the observed genotype frequencies.
Corrected empirical P-value calculated for 10 000 permutations using PLINK software.
SNP, single nucleotide polymorphism.
HLA-Cw6 stratification
Stratification of patients for HLA-Cw6 status (51% of patients were HLA-Cw6 positive) was performed only at stage 1 of the analysis. Association for rs1217414 and rs3789604 was significant although reduced in the HLA-Cw6 positive patients with P-values of 0·024 and 0·030, respectively (Table 3), suggesting that association of these markers with psoriasis was not dependent on HLA-Cw6 status.
Table 3.
Genotype frequencies of PTPN22 region single nucleotide polymorphisms (SNPs) in HLA-Cw6 positive cases and controls
| Stage 1 | ||||||
|---|---|---|---|---|---|---|
| SNP | Genotype | Patients n (%) | Controls n (%) | HWE in controlsa | P-value | Trend test |
| rs1217414 | CC | 176 (51·9) | 309 (55·4) | |||
| CT | 134 (39·5) | 226 (40·5) | 0·0215 | 0·024 | 0·058 | |
| TT | 29 (8·6) | 23 (4·1) | ||||
| rs3789604 | TT | 240 (73·4) | 325 (64·6) | |||
| TG | 79 (24·2) | 160 (31·8) | 0·8869 | 0·030 | 0·010 | |
| GG | 8 (2·4) | 18 (3·6) | ||||
Hardy Weinberg equilibrium (HWE) statistic calculated in STATA 8.2 from comparing genotype frequencies expected under HWE against the observed genotype frequencies.
Linkage disequilibrium analysis
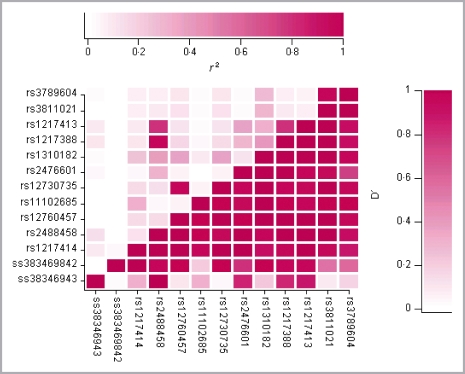
Analysis of the control cohort using the HelixTree™ software produced pairwise LD across the PTPN22 region (Fig. 2). D′ and r2 were measured as parameters for LD structure. LD was detected across the gene with D′ > 0·95 for all SNPs excluding ss38346943 and ss383469842. However, the r2 value showed that there were only five combinations of SNPs strongly correlated (r2 > 0·8, Fig. 2). SNPs rs1217414 and rs3789604, the two associated SNPs, demonstrated high D′ (0·88) but low r2 (0·06) with each other. A comparison between case and control LD plots showed no major differences in LD structure between the subgroups.
Fig 2.
Pairwise linkage disequilibrium pattern across the PTPN22 region. The genetic analysis software, HelixTree™ (Golden Helix Incorporated) was used to calculate the extent of LD between both the SNPs in the PTPN22 region. The parameters used for LD analysis are D′, which is a measure of the statistical significance of LD between two SNPs reflecting historical recombination; and r2, which is the square of the correlation between two SNPs thus measuring the correlation between alleles at two loci. The extent of LD between any two SNPs in this figure corresponds to the colour scale shown where no LD (D′ = 0 / r2 = 0) is white and complete LD (D′ = 1 / r2 = 1) is red.
Stepwise logistic regression
Stepwise logistic regression was conducted on both associated SNPs. The TT genotype of rs3789604 was the only significant variable in the regression model regardless of SNP order; OR = 1·39 (95% CI 1·15–1·68; P = 0·001).
Carriage of both associated risk alleles (TT) was associated with a significantly increased risk of psoriasis; OR = 1·16 (95% CI: 1·02–1·33; P = 0·002). Interestingly, carrying neither risk allele (CG) was found to have a protective effect; OR = 0·79 (95% CI: 0·67–0·93; P = 0·0003).
Discussion
The PTPN22 R620W polymorphism was not associated with Type I psoriasis; this replicated the findings of previous case–control studies.15,16,18,19 However, the investigation did detect an association between psoriasis and two other markers, namely rs1217414 and rs3789604, located in intron 1 of PTPN22 and exon 1 of the round spermatid basic protein 1 (RSBN1) gene downstream of PTPN22, respectively. SNP rs3789604 showed the strongest association with the disease in the combined analysis (P = 0·0002 and 0·0001 for χ2 and the χ2 trend test, respectively), which remained significant after permutation testing. Although the evidence for the association of rs1217414 was not increased in the larger sample size, this remained significant even after permutation testing. LD analysis demonstrated that although D′ LD was high between the alleles, LD correlation (r2) between rs1217414 and rs3789604 was low. SNP rs1217414 was excluded from stepwise logistic regression with rs3789604, indicating that there is no additional significant association with Type I psoriasis with this marker conditional on SNP rs3789604. A closer examination of these two markers revealed that 96% of individuals homozygous for the minor allele (TT) for SNP rs1217414 were also homozygous for the major allele (TT) for rs3789604 leading to the masking of rs1217414 by rs3789604 (but not reversed) in the effect on the disease. Carriage of both associated risk alleles (T allele for both rs1217414 and rs3789604) revealed a significantly increased risk of disease while carriage of the opposing alleles (C for rs1217414 and G for rs3789604) demonstrated an even greater protective effect against development of Type I psoriasis.
The results of this study are supported by the findings of a German study.18 In that study SNP rs2476601 was tested initially in an exploratory case–control study of 375 independent patients who were then combined with an additional 418 patients; there was no association with the R620W polymorphism (P = 0·22 and P = 0·19, respectively). However, the same study revealed psoriasis susceptibility in or near PTPN22 from haplotype analysis of four LD blocks across the region on 1p13·2.18 The common haplotype in three of the four LD blocks was found to be associated with psoriasis with only two remaining significant following Bonferroni correction (P = 0·02).18 Interestingly, the LD blocks containing these associated haplotypes spanned the region containing both the PTPN22 and RSBN1 genes; therefore the possibility remains that the associations detected in this study could be due to LD with other SNPs across the region. Furthermore, the rs3789604 marker has also been associated with rheumatoid arthritis independently of the well-characterized association with R620W.14 Interaction studies between the PTPN22 risk haplotypes identified by Huffmeier et al.18 and HLA-Cw6 returned negative estimates for both associated haplotypes thereby excluding a direct interaction between HLA-Cw6 and the causative allele within PTPN22 for susceptibility to psoriasis. The current study provides further evidence of lack of interaction between PTPN22 and HLA-Cw6.
SNP rs3789604 is located in a predicted transcription factor binding site in the 5′ end and exonic region (exon 1) of the RSBN1 gene. This marker is 1496 base pairs downstream of PTPN22 where the polymorphism results in either a synonymous mutation or putative transcription binding site.14 The rs3789604 polymorphism lies within a predicted transcription factor binding site for Pax-5, Pax-4, Nrf-1 and c-Myb.14 The minor allele G is believed to alter the binding activity for Pax-4 and Pax-5, which could regulate PTPN22 expression.14 Indeed, there is evidence of regulatory regions lying in adjacent genes, best described in the case of the sonic hedgehog gene (SHH) where a mutation residing approximately 1 Mb away from the gene called sasquatch (SSQ, now C6orf26) interrupts a long-range cis-acting regulator that disrupts SHH regulation and is thought to be responsible for preaxial polydactyly disease (a common limb malformation) in humans.25
The PAX genes are a family of transcription factors which are of primary importance during organogenesis. Furthermore PAX5 encodes the B-cell specific activator protein required for B lymphoid lineage.14 In the absence of PAX5, pro-B cells have the potential to differentiate into functional macrophages, osteoclasts, dendritic cells, granulocytes or natural killer cells depending on the cytokines inducing them,26 with a more recent publication determining that PAX5 functions to determine B- vs. T-cell developmental fate.27 Therefore it is conceivable that the rs3789604 polymorphism determines or initiates T-cell activation at the site of plaques of psoriasis. The SNP rs1217414 found to be associated with Type I psoriasis in this study has not been identified as associated with antibody-associated autoimmune diseases and its exact function is therefore unknown. Located in intron 1 of PTPN22 gene, it may have an effect on gene transcription.
This study confirms that the R620W polymorphism is not associated with Type I psoriasis thereby replicating existing studies. Evidence of association of two SNPs in the PTPN22 gene region with Type I psoriasis susceptibility may suggest that altered levels in PTPN22 transcription may influence T-cell function and thereby influence susceptibility to psoriasis.
Acknowledgments
This project was supported by an unrestricted Stiefel PhD studentship, awarded to Rhodri Llywelyn Smith (Stiefel Laboratories, Maidenhead, U.K.). We also acknowledge use of specimens from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
References
- 1.Griffiths CEM, Barker JNWN. Pathogenesis and clinical features of psoriasis. Lancet. 2007;370:263–71. doi: 10.1016/S0140-6736(07)61128-3. [DOI] [PubMed] [Google Scholar]
- 2.Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13:450–6. doi: 10.1016/s0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- 3.Trembath RC, Clough RL, Rosbotham JL, et al. Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet. 1997;6:813–20. doi: 10.1093/hmg/6.5.813. [DOI] [PubMed] [Google Scholar]
- 4.Nair RP, Henseler T, Jenisch S, et al. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet. 1997;6:1349–56. doi: 10.1093/hmg/6.8.1349. [DOI] [PubMed] [Google Scholar]
- 5.Lee YA, Ruschendorf F, Windemuth C, et al. Genome wide scan in German families reveals evidence for a novel psoriasis-susceptibility locus on chromosome 19p13. Am J Hum Genet. 2000;67:1020–4. doi: 10.1086/303075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veal CD, Clough RL, Barber RC, et al. Identification of a novel psoriasis susceptibility locus at 1p and evidence of epistasis between PSORS1 and candidate loci. J Med Genet. 2001;38:7–13. doi: 10.1136/jmg.38.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang XJ, He PP, Wang ZX, et al. Evidence for a major psoriasis susceptibility locus at 6p21 (PSORS1) and a novel candidate region at 4q31 by genome-wide scan in Chinese Hans. J Invest Dermatol. 2002;119:1361–6. doi: 10.1046/j.1523-1747.2002.19612.x. [DOI] [PubMed] [Google Scholar]
- 8.Tomfohrde J, Silverman A, Barnes R, et al. Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science. 1994;264:1141–5. doi: 10.1126/science.8178173. [DOI] [PubMed] [Google Scholar]
- 9.Samuelsson L, Enlund F, Torinsson A, et al. A genome-wide search for genes predisposing to familial psoriasis by using a stratification approach. Hum Genet. 1999;105:523–9. doi: 10.1007/s004399900182. [DOI] [PubMed] [Google Scholar]
- 10.Matthews D, Fry L, Powles AV, et al. Evidence that a locus for familial psoriasis maps to chromosome 4q. Nat Genet. 1996;14:231–3. doi: 10.1038/ng1096-231. [DOI] [PubMed] [Google Scholar]
- 11.Enlund F, Samuelsson L, Enerback C, et al. Psoriasis susceptibility locus in chromosome region 3q21 identified in patients from southwest Sweden. Eur J Hum Genet. 1999;7:783–90. doi: 10.1038/sj.ejhg.5200365. [DOI] [PubMed] [Google Scholar]
- 12.Karason A, Gudjonsson JE, Jonsson HH, et al. Genetics of psoriasis in Iceland: evidence for linkage of subphenotypes to distinct loci. J Invest Dermatol. 2005;124:1177–85. doi: 10.1111/j.0022-202X.2005.23703.x. [DOI] [PubMed] [Google Scholar]
- 13.Bowcock AM, Krueger JG. Getting under the skin: the immunogenetics of psoriasis. Nat Rev Immunol. 2005;5:699–711. doi: 10.1038/nri1689. [DOI] [PubMed] [Google Scholar]
- 14.Carlton VE, Hu X, Chokkalingam AP, et al. PTPN22 genetic variation: evidence for multiple variants associated with rheumatoid arthritis. Am J Hum Genet. 2005;77:567–81. doi: 10.1086/468189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Criswell LA, Pfeiffer KA, Lum RF, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–71. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinks A, Barton A, John S, et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–9. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 17.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–8. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 18.Huffmeier U, Steffens M, Burkhardt H, et al. Evidence for susceptibility determinant(s) to psoriasis vulgaris in or near PTPN22 in German patients. J Med Genet. 2006;43:517–22. doi: 10.1136/jmg.2005.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nistor I, Nair RP, Stuart P, et al. Protein tyrosine phosphatase gene PTPN22 polymorphism in psoriasis: lack of evidence for association. J Invest Dermatol. 2005;125:395–6. doi: 10.1111/j.0022-202X.2005.23802.x. [DOI] [PubMed] [Google Scholar]
- 20.Huffmeier U, Reis A, Steffens M, et al. Male restricted genetic association of variant R620W in PTPN22 with psoriatic arthritis. J Invest Dermatol. 2006;126:932–5. doi: 10.1038/sj.jid.5700179. [DOI] [PubMed] [Google Scholar]
- 21.Butt C, Peddle L, Greenwood C, et al. Association of functional variants of PTPN22 and tp53 in psoriatic arthritis: a case–control study. Arthritis Res Ther. 2006;8:R27. doi: 10.1186/ar1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–19. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 23.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lettice LA, Horikoshi T, Heaney SJ, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A. 2002;99:7548–53. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nutt SL, Heavey B, Rolink AG, et al. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 27.Cotta CV, Zhang Z, Kim HG, et al. Pax5 determines B- versus T-cell fate and does not block early myeloid-lineage development. Blood. 2003;101:4342–6. doi: 10.1182/blood-2002-10-3139. [DOI] [PubMed] [Google Scholar]