Abstract
RNA templates of 33 nucleotides containing the brome mosaic virus (BMV) core subgenomic promoter were used to determine the promoter elements recognized by the BMV RNA-dependent RNA polymerase (RdRp) to initiate RNA synthesis. Nucleotides at positions −17, −14, −13, and −11 relative to the subgenomic initiation site must be maintained for interaction with the RdRp. Changes to every other nucleotide at these four positions allow predictions for the base-specific functional groups required for RdRp recognition. RdRp contact of the nucleotide at position −17 was suggested with a template competition assay. Comparison of the BMV subgenomic promoter to those from other plant and animal alphaviruses shows a remarkable degree of conservation of the nucleotides required for BMV subgenomic RNA synthesis. We show that the RdRp of the plant-infecting BMV is capable of accurately, albeit inefficiently, initiating RNA synthesis from the subgenomic promoter of the animal-infecting Semliki Forest virus. The sequence-specific recognition of RNA by the BMV RdRp is analogous to the recognition of DNA promoters by DNA-dependent RNA polymerases.
Viral RNA replication, a process fundamental to pathogenicity, requires specific recognition of RNA features by proteins. RNA-dependent RNA polymerase (RdRp) is a complex composed of viral and cellular proteins that directs viral RNA synthesis from infecting RNA templates (1). Many viral RdRp proteins have been sequenced and analyzed (2); however, a comprehensive mechanism describing RNA synthesis is lacking. Consequently, general knowledge of RdRps is significantly less than that of other RNA and DNA polymerases.
To investigate the mechanism of RNA-directed RNA synthesis, we study brome mosaic virus (BMV), type member of the bromovirus group of plant viruses in the alphavirus-like superfamily of (+)-strand RNA viruses (3). The BMV genome is comprised of three RNAs designated 1, 2, and 3, and a subgenomic RNA4 which is initiated from (−)-strand RNA3. Enriched BMV RdRp preparations from infected barley can, in a highly specific manner, synthesize (−)-strand RNA from (+)-strand templates and subgenomic (+)-strand products from (−)-strand templates (4–6).
We have developed a system to study BMV subgenomic RNA initiation from minimal templates, designated proscripts, because they contain the promoter and template for (+)-strand RNA synthesis. We have shown that the 20 nt 3′ of the subgenomic initiation nucleotide, recognized as the subgenomic core promoter (7, 8), are sufficient to direct (+)-strand RNA synthesis (6), permitting the design of proscripts focusing only on the core promoter.
In this paper, we have used a functional assay to determine that nucleotides −17, −14, −13, and −11 relative to the subgenomic initiation site are required for RNA synthesis from the subgenomic core promoter. Moreover, at least one of these nucleotides, −17, was found likely to be contacted by the BMV RdRp. The resolution achieved in this study allows us to predict the base moieties contacted by RdRp and demonstrate sequence-specific recognition between an RdRp and its cognate template. The four required nucleotides identified within the BMV promoter are highly conserved in members of the alphavirus-like superfamily. In fact, the BMV RdRp could recognize and accurately synthesize RNA using the Semliki Forest virus (SFV) subgenomic promoter. This heterologous interaction provides evidence for a common mode of template recognition by the RdRps of the alphavirus-like superfamily.
MATERIALS AND METHODS
Synthesis of Proscripts.
PCR was used to generate cDNA copies of the (−)-strand BMV RNA3 encompassing the subgenomic promoter. Pairs of primers, one of which contained a T7 promoter and the other harboring discrete changes within the subgenomic promoter, were used in PCR with the cDNA clone of RNA3, pB3TP8 (9). Proscripts and size markers were generated using T7 RNA polymerase (Ampliscribe; Epicentre Technologies, Madison, WI) as described (6). RNAs were purified with Qiagen (Chatsworth, CA) columns using the manufacturer’s protocol to remove NTPs and proteins remaining from the T7 transcription reaction. RNAs were visually inspected by denaturing PAGE and quantified by ethidium bromide staining and/or UV absorbance.
RdRp Activity Assay and Product Analysis.
BMV RdRp was prepared from infected barley as described (10). Standard assays consisted of 1 pmol of template RNA (unless stated otherwise) with 10 μl of RdRp in a 40-μl reaction containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM DTT, 0.5% (vol/vol) Triton X-100, 2 mM MnCl2, 200 μM ATP and UTP, 500 μM GTP, and 250 nM [α-32P]CTP (Amersham). Reactions were incubated at 30°C for 90 min and stopped by phenol/chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.4 M ammonium acetate. Products were separated by electrophoresis on 20% denaturing (8 M urea) polyacrylamide gels. Gels were wrapped in plastic and exposed to film at −80°C. Product bands were quantified using a PhosphorImager (Molecular Dynamics) and values were compared with the amount of product generated from the wild-type (WT) template to derive the relative percent activity of mutant templates. All values represent the mean of at least three independent experiments.
RESULTS
Accurate Subgenomic Initiation.
To determine the recognition elements contained within the subgenomic core promoter, a 33-nt proscript (−20/13) was constructed which contains the WT promoter sequence 20 nt 3′ of the subgenomic initiation start site in (−)-strand RNA3. This proscript directs the synthesis of a 13-nt product, the first 11 nt of which are BMV sequence followed by two guanylates added by T7 RNA polymerase to allow labeling of RdRp products with [α-32P]CTP. The BMV sequence within proscript −20/13 is complementary to the viral (+)-strand RNA3 from positions 1,222 to 1,252 and serves as the WT control (Fig. 1A).
Figure 1.
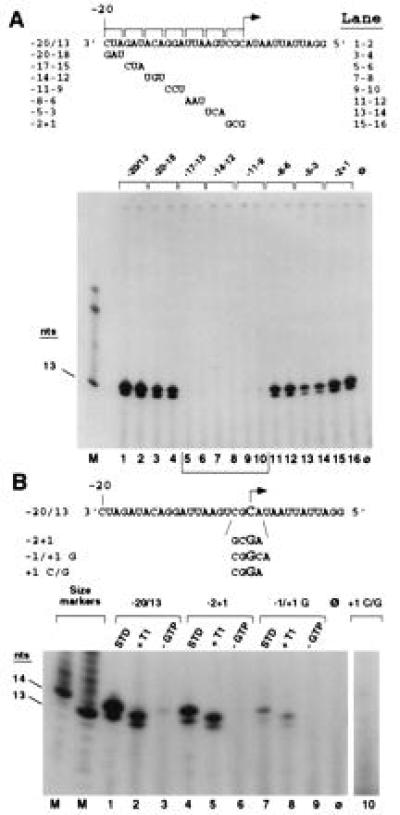
Regions in the BMV subgenomic promoter required for accurate and efficient initiation of RNA synthesis. (A) Triple transversion mutants. The sequence shown is proscript −20/13 containing the WT BMV core promoter directing synthesis of a 13-nt product. Nucleotide changes, in groups of three, in proscripts are shown with brackets and the mutant nucleotides are displayed below the WT sequence. Identity of input templates, in duplicate reactions, is indicated at the top of the gel. Reaction products were separated by denaturing PAGE (lanes 1–16) and visualized by autoradiography. The predominant RdRp product was 14 nt as judged by comparison to a 13-nt size marker (lane M) due to the nontemplated addition of one nucleotide, possibly by RdRp. Lane φ represents the products of a control reaction with no added template. Region of the subgenomic promoter required for activity is bracketed below the gel. (B) Sequence requirements for accurate initiation of RNA synthesis. The WT promoter sequence is shown on top. RNAs with altered initiation sequences are shown below. The initiating nucleotide (WT or mutant) is denoted with an arrow and larger size font. STD indicates products of a standard RdRp reaction (lanes 1, 4, and 7) and reactions lacking GTP are indicated by −GTP (lanes 3, 6, and 9). RNase T1 digestion of the RdRp products is indicated with +T1 (lanes 2, 5, and 8). Size markers (lane M) indicate size of the RdRp products; products from reactions with no added template are shown (lane φ).
Various mutations within the promoter sequence were created. Initially, transversions, in groups of three nucleotides, were synthesized to scan the entire promoter and determine which regions were required for RNA synthesis. Positions −17 to −9 were identified as containing essential nucleotides because mutations in this region of the promoter reduced the ability of the BMV RdRp to initiate synthesis of subgenomic RNA to about 2% of WT activity (Fig. 1A, lanes 5–10). Mutations at nucleotide positions −5 to −3 had a lesser effect, retaining 16% of WT activity (Fig. 1A, lanes 13 and 14). The three nucleotide transversions covering all other positions of the core promoter each only reduced synthesis by 50%. The predominant RdRp product from all templates was 14 nt due to the nontemplated addition of one nucleotide, a phenomenon common to all DNA-dependent RNA polymerases and the poliovirus RdRp (11).
Unexpectedly, subgenomic synthesis was relatively unaffected by replacement of nucleotides from −2 to the initiation site, +1 (Fig. 1A, lanes 15 and 16). Previous work has demonstrated that the identity of the initiating cytidylate for the subgenomic RNA4 must be maintained (6). The three nucleotide transversion in the −2+1 proscript places a cytidylate at position −1 and it is conceivable that the BMV RdRp is able to utilize this nucleotide for initiation of RNA synthesis directing a product of the same size as the WT proscript by either facilitating the use of a nontemplated guanylate (instead of the templated cytidylate at the +1 position) or bypassing the +1 position completely. To test whether the initiation position can be moved, two additional proscripts were made (Fig. 1B). Proscript +1 C/G specifically replaces the initiating cytidylate with a guanylate while leaving all other positions as WT sequence. Proscript −1/+1 G inserts a guanylate into the initiation site and moves the WT +1 cytidylate into the +2 position. This proscript encodes a 14-nt product (15 nt with nontemplated addition) if the inserted guanylate at the +1 position is recognized by the BMV RdRp. However, if the initiation site can be shifted to the cytidylate now occupying the +2 position, then a 13-nt product (14 nt with nontemplated addition) should be observed. Accurate initiation of the RdRp products can be verified by withholding GTP which is required only for initiation or by treatment with RNase T1 which specifically cleaves after guanylates. Thus, a correctly initiated RdRp product will decrease by 1 nt after RNase T1 digestion.
The RdRp products from proscripts −2+1, −1/+1 G, and WT −20/13 were all initiated with GTP as judged by the reactions lacking GTP and by RNase T1 treatment (Fig. 1B, lanes 1–9) and are the same size (13 and 14 nt) as T7 size markers. However, no product was synthesized from the +1 C/G proscript (Fig. 1B, lane 10). We therefore conclude that a cytidylate residue is required as the initiating nucleotide, conforming with earlier observations. It appears that the BMV RdRp can recognize a cytidylate at either one nucleotide 3′ or 5′ proximal to the original initiation site (Fig. 1B, lanes 4, 5, 7, and 8). All of the reactions demonstrate accurate initiation and validate the use of this system for characterization of the RdRp-subgenomic core promoter interaction. These experiments also confirm that the addition of the nontemplated nucleotide generating the 14-nt product occurs at the 3′ end of the (+)-strand product.
Nucleotides Critical for Initiation of Subgenomic RNA Synthesis.
Data from the three nucleotide transversions indicated that positions −17 to −9 contain nucleotides important for RNA synthesis. To identify the critical residues, proscripts that contain single nucleotide transversions at each position within this region and at positions −5 to −3 of the subgenomic core promoter were constructed (Fig. 2A). Positions −17, −14, −13, −11, −10, and −5 were important for synthesis because transversions significantly decreased RNA synthesis by the BMV RdRp (Fig. 2A, lanes 2, 5, 6, 8, 9, and 11). The most critical positions, −17, −14, −13, and −11, all had activities below 3% of the WT proscript. Changes at positions −10 and −5 were less severe, retaining 31% to 18% of WT synthesis. As a negative control, no synthesis was observed from a proscript, +1 C/G, which contains a transversion at position +1 (Fig. 2A, lane 14).
Figure 2.
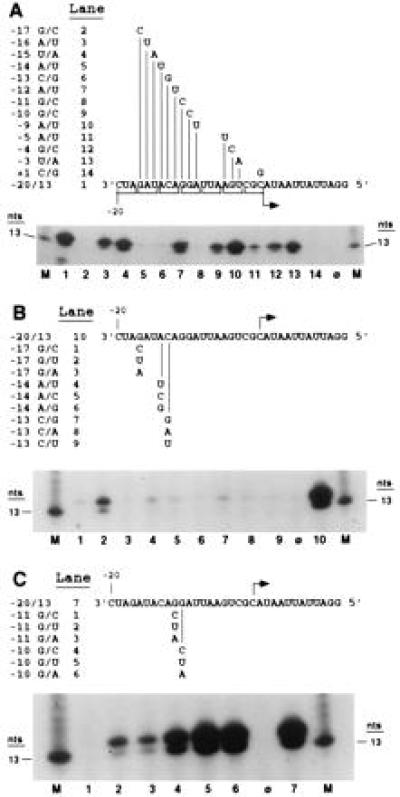
Mutational analysis of the BMV subgenomic core promoter. The lanes containing the RdRp reaction products from input proscripts are indicated adjacent to proscript names in each schematic. All products were separated on 20% denaturing polyacrylamide gels. Because these autoradiographs do not contain bands other than the RdRp products of the expected sizes, only the portions of the autoradiograph containing the RdRp products are shown. Lane φ indicates products made from reactions lacking input template whereas lane M contains a 13-nt size marker, generated by T7 RNA polymerase, of the identical sequence as the expected RdRp product. Products from proscripts supporting synthesis were accurately initiated as judged by comparison to size markers and the WT construct. (A) Single nucleotide transversion mutations in nucleotides −17 to −9, −5 to −3, and +1. Nucleotide sequences of the WT template (−20/13) and corresponding changes are shown with the original three nucleotide transversions bracketed. (B) All possible nucleotide replacements at positions −17, −14, and −13. Sequence of WT proscript (−20/13) is shown with appropriate changes indicated below. (C) All possible nucleotide replacements at positions −11 and −10. Schematic of WT proscript is shown and the identity of subsequent mutants is indicated below.
Proscripts containing all possible nucleotide replacements at the most critical positions, −17, −14, −13, and −11, were assayed to test the sequence specificity of these four positions. A change of the WT guanylate to uridylate (−17 G/U) at position −17 supported RNA synthesis at 13% of WT activity (Fig. 2B, lane 2). Other nucleotide substitutions at −17 (−17 G/C and −17 G/A) were unable to direct RNA synthesis (Fig. 2B, lanes 1 and 3). Nucleotides other than the WT sequence at positions −14 or −13 did not retain appreciable activity (Fig. 2B, lanes 4–9), demonstrating the importance of the adenylate at −14 and the cytidylate at −13. A change at position −11 of the WT guanylate to either uridylate or adenylate (−11 G/U or −11 G/A) retained about 8% activity, whereas the original −11 G/C proscript had only 1% of the activity of the WT proscript (Fig. 2C, lanes 1–3). To further demonstrate the specific requirements of the above four positions, the WT guanylate at position −10 was replaced with either a uridylate or adenylate without detrimental effects (Fig. 2C, lanes 5 and 6). In fact, both of these mutants, −10 G/U and −10 G/A, resulted in slightly better promoters than the WT proscript, generating 140% of the product of −20/13. These results demonstrate nucleotide specificity at positions −17, −14, −13, and −11 of the BMV subgenomic core promoter.
A summary of all the mutational data is presented in Fig. 3A. Examination of the functional moieties of the base in the replacement nucleotides which displayed partial activity suggests contact sites recognized by the BMV RdRp. To test this idea, competition experiments were performed to determine if proscripts −20/13, −17 G/C, and −17 G/U would affect synthesis from a second proscript with the WT promoter sequence directing synthesis of a 15-nt product (designated −20/15). The concentration of the competitor templates was varied from 0.1- to 10-fold molar excess of the −20/15 proscript. Competition was visualized by a decreasing amount of the 15-nt product with a concurrent increase of the 13-nt product (Fig. 3B). As expected, proscript −20/13 competed efficiently for synthesis because it also contains a WT subgenomic promoter (Fig. 3B, lanes 4–8). Synthesis from −20/15 was reduced 50% when −20/13 was present in an equimolar amount. In contrast, proscript −17 G/C was not an effective competitor, decreasing synthesis from −20/15 by only 15% when present at an equimolar ratio (Fig. 3B, lanes 9–13). The −17 G/U proscript inhibited synthesis from −20/15 at a reduced level (35%) from that observed with −20/13 (Fig. 3B, lanes 14–18). This level of inhibition is consistent with the result that synthesis from this mutant template was diminished but not abolished (Fig. 2B, lane 2). The result from this experiment is consistent with the idea that position −17, and most likely the other key nucleotides identified by mutagenesis, is contacted by the BMV RdRp.
Figure 3.
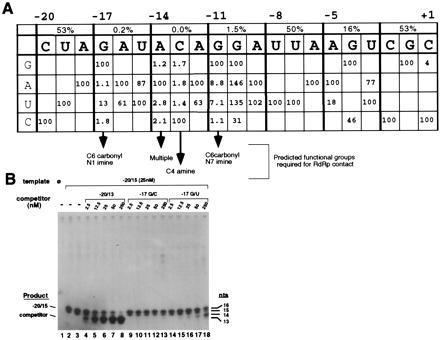
(A) Summary of the mutational analysis of the BMV subgenomic promoter. The WT sequence is in bold letters displayed horizontally with relative nucleotides positions marked above. Percentages in rectangles above the promoter sequence represent relative activity of the original triple transversion mutants. Key to nucleotide changes is at the left side. Values listed below the WT sequence represent the percent activity of the single nucleotide mutants compared with the WT proscript. The predicted functional groups required for the nucleotides at positions −17, −14, −13, and −11 are indicated below the arrows. (B) Potential binding contacts within the subgenomic core promoter. Competition between a WT promoter directing synthesis of a 15-nt product (−20/15) and one of three competitor templates encoding 13-nt products: −20/13, containing the WT BMV subgenomic promoter sequence; −17 G/C, a nonfunctional mutant template; −17 G/U, a partially functional template. The reaction in lane 1 was performed without any input templates whereas reactions in lanes 2–18 each contain 25 nM of the −20/15 template with the identity and quantity of competitor template indicated above the autoradiograph. The identities of the RdRp products and their sizes are denoted on each side of the autoradiograph.
Alphaviral Subgenomic Promoters.
A comparison of the BMV subgenomic promoter with those of other members of the alphavirus-like superfamily, infecting both plants and animals, reveals a striking conservation of the four nucleotides critical for synthesis by the BMV RdRp (Fig. 4A). In addition, all of these promoters contain a pyrimidine as the initiating nucleotide (uridylate in animal viruses and either a cytidylate or uridylate in plant viruses) with a highly conserved adenylate at the +2 position. This similarity implies that RdRps from members of the alphavirus-like superfamily recognize subgenomic promoters by a conserved mechanism.
Figure 4.
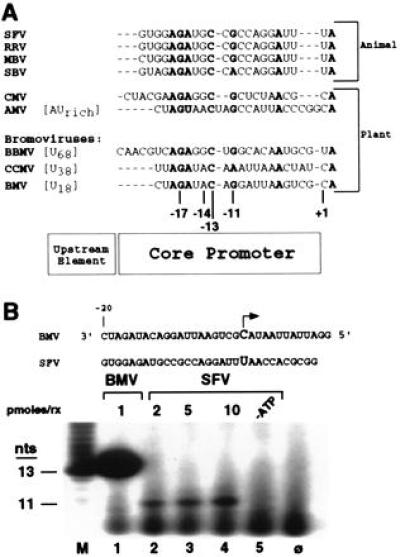
Conservation of subgenomic core promoters in the alphavirus-like superfamily. (A) Alignment of subgenomic promoters from both animal and plant viruses. Conserved nucleotides in the core promoter are shown in bold. Sequences were obtained from the GenBank database. Homopolymeric upstream elements, where present, are indicated with brackets. SFV, Semliki Forest virus; RRV, Ross River virus; MBV, Middelburg virus; SBV, Sindbis virus; CMV, cucumber mosaic virus; AMV, alfalfa mosaic virus; BBMV, broad bean mottle virus; CCMV, cowpea chlorotic mottle virus; BMV, brome mosaic virus. The alignment was generated using the clustalw software program. Positions −17, −14, −13, and −11 are noted below the BMV sequence; +1 denotes the initiation nucleotide. (B) BMV RdRp recognizes the SFV subgenomic promoter. A schematic for proscripts containing the WT subgenomic promoter for either BMV or SFV is shown with the initiating nucleotide indicated by the arrow. The 13-nt product from the BMV proscript −20/13 is shown in lane 1. The SFV proscript encodes an 11-nt product which requires an adenylate for correct initiation (lanes 2–5). Amounts of template (in pmol) used in an RdRp reaction are shown at the top of the gel; −ATP indicates absence of ATP in reaction. M denotes a 13-nt size marker produced by T7 RNA polymerase, and φ denotes a reaction with no added template.
To examine the possibility that the mode of template recognition is conserved, we constructed a proscript containing the subgenomic promoter from SFV (12, 13) and tested its ability to be recognized by the BMV RdRp (Fig. 4B). So that the RNA product can be visually distinguished from the 13-nt product generated from the BMV proscript, the SFV proscript was designed to direct synthesis of an 11-nt product, the first 9 nt of which are WT SFV sequence followed by two guanylates added by T7 RNA polymerase to allow labeling of RdRp products with [α-32P]CTP. The SFV proscript directs the BMV RdRp to synthesize a product which is dependent upon ATP, used only as the initiating nucleotide for subgenomic synthesis from this template (Fig. 4B, lanes 2–5). The amount of synthesis was only 0.25% of the synthesis from an equimolar amount of WT −20/13 but significantly above background. This result demonstrates a heterologous interaction between an RdRp from a plant-infecting virus with an RNA template containing the subgenomic promoter from an animal-infecting virus.
DISCUSSION
Although initiation of subgenomic RNA synthesis has been studied in other RNA viruses (12–16), a detailed characterization of the promoter sequences directing synthesis has not been determined. More than 30 mutations in the BMV subgenomic core promoter were constructed to define the elements required for recognition by the BMV RdRp. Of the 20 nt comprising the core promoter, 4 (−17, −14, −13, and −11) were demonstrated to be essential. Moreover, we demonstrated that the BMV RdRp can use a cytidylate one position 3′ or 5′ of the WT initiation site, as well as a uridylate in the case of the SFV promoter. Competition experiments show that a proscript unable to direct RNA synthesis is also unable to inhibit synthesis from a WT proscript, indicating that changes at the four identified positions interfere with the BMV RdRp’s ability to bind these mutant templates.
In 1985, Miller et al. (5) demonstrated in vitro synthesis of full-length subgenomic RNA4 using a (−)-strand BMV RNA3 template with its 3′ end 20 nt from the initiating cytidylate. Subsequent studies reported that additional sequences flanking the promoter were required for high levels of subgenomic RNA synthesis (7, 8). Recent work from our laboratory (6) has shown that accurate and efficient RNA synthesis can occur using only the 20 nt comprising the core promoter when directing synthesis of a (+)-strand product fewer than 26 nt in length. Furthermore, a template with its 3′ end at position −17 was the smallest promoter capable of directing accurate RNA synthesis (6), correlating nicely with this study’s demonstration of the importance of this position.
Examination of the promoter mutants which partially enabled proscripts to be recognized by the BMV RdRp allow us to predict base-functional groups required for contact by the BMV RdRp (Fig. 3A). The C6 carbonyl and perhaps the N1 imine of the guanosine at position −17 are implicated as contact sites. Substitution with a uridylate (containing spatially equivalent carbonyl and imine groups) could partially restore this promoter’s ability to direct synthesis, as well as partially inhibit synthesis from a WT promoter (Fig. 2B, lane 2, and Fig. 3B, lanes 14–18). The lack of restoration of synthesis with a cytidylate to uridylate change at position −13 suggests the unique amine group at the cytosine C4 position may be required. The guanylate at position −11 could be changed to either an uridylate or adenylate and weakly direct RNA synthesis. These results indicate that the C6 carbonyl group and the N7 imine group both contribute to the interaction with the BMV RdRp. For position −14, several contact sites may be needed. The stringent requirements for these nucleotides may be largely responsible for the template specificity exhibited by the BMV RdRp.
The nucleotides identified as crucial for RNA synthesis in the BMV subgenomic promoter are highly conserved in the subgenomic promoters from other members of the alphavirus-like superfamily (Fig. 4A), implying a common mode of RdRp subgenomic promoter recognition. Accordingly, a proscript containing the animal-infecting SFV subgenomic promoter was able to direct accurately initiated RNA products using the BMV RdRp. Our study has demonstrated the heterologous recognition between an RdRp and RNA template from plant- and animal-infecting viruses.
A sequence-specific mode of recognition is strikingly similar to template recognition by the DNA-dependent RNA polymerases (DdRp). DdRps primarily interact with their respective templates via sequence-specific contacts (17). For example, the Escherichia coli RNA polymerase holoenzyme recognizes the identities of nucleotides at the −10 and −35 regions relative to the transcriptional start site (18). The single polypeptide bacteriophage polymerases (T7, T3, and SP6) contact specific nucleotides within a 17-nt consensus promoter primarily at nucleotides −11 to −9 (19, 20). The results of this study suggest that RdRps may discriminate between templates in a fashion analogous to that of DdRps.
Current dogma posits that protein and RNA interact primarily through a structure-specific mode of recognition allowing protein contact with one or more unstructured nucleotides (21). For example, the bacteriophage R17 coat protein recognizes a hairpin containing the ribosome binding site for the phage replicase (22). Also, viral RdRps replicating full-length products from linear templates have been shown to require various structures on the 3′ end of the RNA template to initiate accurate synthesis (23–26). One fundamental difference between genomic and subgenomic RNA synthesis is that subgenomic initiation occurs internally in a manner analogous to DdRps. Therefore, it is quite plausible that template selection by RdRp may proceed in a manner similar to that of DNA recognition by DdRps. Proteins recognizing unstructured RNA in a sequence-specific manner have been reported, including the translational repressor RegA of bacteriophage T4 which specifically recognizes an unstructured 12-nt sequence (27). RdRp sequence specificity has been suggested previously with the (−)-strand RNA influenza virus (28). Three nucleotides near the 3′ end of the (−)-strand RNA were found to be required for synthesis, although accurate initiation was not demonstrated. Subsequently, these nucleotides were shown to be directly contacted by the influenza RdRp (29). However, additional data demonstrated the need for sequences located at the 5′ end of the influenza RNA which form a partially double-stranded structure recognized by the RdRp (30, 31).
Although secondary structure may facilitate efficient subgenomic synthesis in full-length (−)-strand RNA3, several lines of evidence suggest that it plays a minor role in the BMV RdRp recognition of the subgenomic core promoter. The predicted hairpin structure in the WT proscript has a low free energy calculation, −0.2 kcal/mol (Fig. 5). This hairpin has a predicted melting temperature of 20°C and would most likely be denatured under our standard reaction conditions of 30°C. The −5 U/A mutant which strengthens the predicted structure had an adverse effect upon synthesis. Conversely, mutants that altered or obliterated the hairpin had either no (−10 G/U and −9 U/A) or only a slight (−8-6 triple mutant) effect on synthesis. Enzymatic studies could not determine a stable structure in the WT proscript (R.W.S., unpublished data). A minimal proscript 22 nt in length, lacking any predicted structure, supported accurate initiation (6). Furthermore, the computer-predicted structure shown in Fig. 5 differs in the location of the initiation nucleotide from the predicted structure of longer proscripts containing additional WT sequences which also directed accurate RNA synthesis (6). Finally, the specific nucleotides required to direct accurate RNA synthesis reside in a single-stranded region of the predicted structure of the WT (−20/13) proscript. Taken together, these results strongly argue that the BMV RdRp–core promoter interaction occurs primarily by the recognition of specific sequences.
Figure 5.
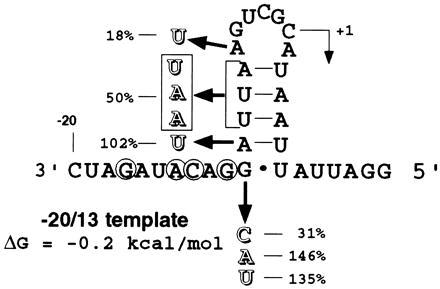
Correlation between the structure of proscript −20/13 and the ability of mutant BMV proscripts to direct RNA synthesis. Computer-predicted structure of the WT template (−20/13) is shown and positions −17, −14, −13, and −11 determined to be critical for RNA synthesis are circled. Mutations generated in this study which should affect the predicted secondary structure are indicated along with the relative activity of that mutant proscript. The initiation nucleotide (+1) is indicated with an arrow.
Acknowledgments
We thank the Indiana University Cereal Killers for helpful discussions. Funding was provided by National Science Foundation Grant MCB9507344. R.W.S. is supported by a National Institutes of Health Genetics Training Grant to the Indiana University Biology Department.
ABBREVIATIONS
- BMV
brome mosaic virus
- SFV
Semliki Forest virus, RdRp, RNA-dependent RNA polymerase
- DdRp
DNA-dependent RNA polymerase
- WT
wild type
References
- 1.Buck K W. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishihama A, Barbier P. Arch Virol. 1994;134:235–258. doi: 10.1007/BF01310564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldbach R, LeGall O, Welink J. Semin Virol. 1991;2:19–25. [Google Scholar]
- 4.Hardy S F, German T L, Loesch-Fries L S, Hall T C. Proc Natl Acad Sci USA. 1979;76:4956–4960. doi: 10.1073/pnas.76.10.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller W A, Dreher T W, Hall T C. Nature (London) 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 6.Adkins S, Siegel R W, Sun J-H, Kao C C. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh L E, Dreher T W, Hall T C. Nucleic Acids Res. 1988;16:981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.French R, Ahlquist P. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janda M, French R, Ahlquist P. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 10.Sun J-H, Adkins S, Faurote G, Kao C C. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 11.Neufeld K L, Galarza J M, Richards O C, Summers D R, Ehrenfeld E. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou J-H, Rice C M, Dalgarno L, Strauss E G, Strauss J H. Proc Natl Acad Sci USA. 1982;79:5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levis R, Schlesinger S, Huang H V. J Virol. 1990;64:1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Der Vossen E A G, Notenboom T, Bol J F. Virology. 1995;212:663–672. doi: 10.1006/viro.1995.1524. [DOI] [PubMed] [Google Scholar]
- 15.Johnston J C, Rochon D M. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 16.Kim K-H, Hemenway C. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- 17.Seeman N, Rosenberg J M, Rich A. Proc Natl Acad Sci USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sienbenlist U, Simpson R B, Gilbert W. Cell. 1980;66:269–280. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 19.Schick C, Martin C T. Biochemistry. 1995;34:666–672. doi: 10.1021/bi00002a034. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Ho H H, Maslak M, Schick C, Martin C T. Biochemistry. 1996;35:3722–3727. doi: 10.1021/bi9524373. [DOI] [PubMed] [Google Scholar]
- 21.Draper D E. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 22.Bernardi A, Spahr P F. Proc Natl Acad Sci USA. 1972;69:3033–3037. doi: 10.1073/pnas.69.10.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreher T W, Hall T C. J Mol Biol. 1988;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 24.Philipenko E V, Poperechny K V, Maslova S V, Melchers W J, Slot H J, Agol V I. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 25.Brown D, Gold L. Biochemistry. 1995;34:14775–14782. doi: 10.1021/bi00045a019. [DOI] [PubMed] [Google Scholar]
- 26.Song C, Simon A E. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 27.Webster K R, Spicer E K. J Biol Chem. 1990;265:19007–19014. [PubMed] [Google Scholar]
- 28.Seong B L, Brownlee G G. J Gen Virol. 1992;73:3115–3124. doi: 10.1099/0022-1317-73-12-3115. [DOI] [PubMed] [Google Scholar]
- 29.Fodor E, Seong B L, Brownlee G G. J Gen Virol. 1993;74:1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 30.Tiley L S, Hagen M, Matthews J T, Krystal M. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fodor E, Pritlove D C, Brownlee G G. J Virol. 1995;69:4012–4019. doi: 10.1128/jvi.69.7.4012-4019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


