Abstract
DNA polymerase δ (pol δ) plays an essential role in DNA replication, repair, and recombination. We have purified pol δ from Schizosaccharomyces pombe more than 103-fold and demonstrated that the polymerase activity of purified S. pombe pol δ is completely dependent on proliferating cell nuclear antigen and replication factor C. SDS/PAGE analysis of the purified fraction indicated that the pol δ complex consists of five subunits that migrate with apparent molecular masses of 125, 55, 54, 42, and 22 kDa. Western blot analysis indicated that the 125, 55, and 54 kDa proteins are the large catalytic subunit (Pol3), Cdc1, and Cdc27, respectively. The identity of the other two subunits, p42 and p22, was determined following proteolytic digestion and sequence analysis of the resulting peptides. The peptide sequences derived from the p22 subunit indicated that this subunit is identical to Cdm1, previously identified as a multicopy suppressor of the temperature-sensitive cdc1-P13 mutant, whereas peptide sequences derived from the p42 subunit were identical to a previously uncharacterized ORF located on S. pombe chromosome 1.
DNA replication in eukaryotes requires three distinct DNA polymerases, α, δ, and ɛ (1–6). DNA polymerase α, complexed with DNA primase, initiates DNA synthesis by catalyzing the formation of pre-Okazaki fragments (7, 8). A direct role of DNA polymerase δ (pol δ) in the replication of simian virus 40 (SV40) DNA has been shown. This polymerase is involved in the maturation of pre-Okazaki fragments on the lagging strand and the synthesis of leading strand DNA (3). In contrast, no role for pol ɛ has been demonstrated in SV40 DNA synthesis. However, pol ɛ is essential for viability of Saccharomyces cerevisiae and it has been proposed that pol ɛ acts at the cellular replication fork and/or plays a role in DNA repair pathways (9). In contrast to pol α, pols δ and ɛ require two accessory protein factors, proliferating cell nuclear antigen (PCNA) and replication factor C (RFC, also called Activator 1), for processive action during strand elongation of low levels of primer ends (10–12). Complexed with a primed template, RFC recruits PCNA and assembles it onto DNA in the presence of ATP (13–15). PCNA, a ring-shaped homotrimeric protein, encircles DNA, acting as a sliding clamp that tethers the pols to the template DNA (16–18). Subsequent ATP hydrolysis is required for the DNA polymerase to enter the complex to initiate chain elongation in the presence of dNTPs (13, 19, 20–23).
Human (h) pol δ has been shown to be a heterodimer consisting of a 125-kDa subunit containing both the polymerase and 3′ to 5′ exonuclease domains and a 48-kDa subunit essential for the stimulation of polymerase activity by PCNA (24–28). In Saccharomyces cerevisiae (sc), pol δ includes subunits of 125 kDa (homologous to the h pol δ large subunit), 58 kDa (homologous to the h pol δ 48-kDa subunit), and other subunits that remain to be defined (2, 29). Studies of Schizosaccharomyces pombe (sp) have identified genes for the large catalytic subunit of pol δ, pol3+/cdc6+, and two additional genes, cdc1+ and cdc27+, and their respective proteins of 125, 51, and 43 kDa (30–33). The Pol3/Cdc6 and Cdc1 proteins are homologous to those isolated from S. cerevisiae, suggesting that the structure and function of pol δ in these two yeasts may be similar. Genetic and biochemical studies have shown that the p125 subunit of sp pol δ (Pol3/Cdc6) interacts with Cdc1, the homologue of the h pol δ 48-kDa subunit and the sc pol δ 58-kDa subunit. Furthermore, Cdc27 interacts with Cdc1, suggesting that Pol3, Cdc1, and Cdc27 form a complex that constitutes the pol δ holoenzyme (32).
We report here on the purification of sp pol δ and demonstrate that both Cdc1 and Cdc27 proteins copurify with the Pol3 catalytic subunit, confirming that these three subunits interact. In addition, we have detected two proteins of 42 and 22 kDa that copurify with sp pol δ, suggesting that the sp pol δ holoenzyme comprises five distinct subunits. In support of this, the sequence of the 22-kDa protein reported here is identical to the gene product Cdm1, which was previously identified as a multicopy suppressor of the S. pombe temperature-sensitive mutant cdc1-P13 (32).
Furthermore, we present peptide sequencing data indicating that the p42 subunit is encoded by a previously uncharacterized ORF present in a cosmid c9G1, which is present on chromosome 1 of S. pombe.
MATERIALS AND METHODS
Reagents and Enzymes.
Poly(dA)4500 and oligo(dT)12–18 were obtained from Life Sciences (St. Petersburg, FL) and Pharmacia LKB, respectively. [3H]dTTP was from Amersham. E. coli single-stranded DNA-binding protein (SSB) was from Pharmacia. The polyclonal antibody against the sp pol δ p125 subunit was a generous gift from G. Baldacci (30), and the antibodies for detection of Cdc1 and Cdc27 were as described previously (32).
Assays for sp PCNA, sp RFC, and sp Pol δ.
The synthesis of poly(dT) during the elongation of multiply primed poly(dA):oligo(dT) DNA templates is dependent on the presence of pol δ, PCNA, and RFC. The activities of these proteins were measured in reaction mixtures (30 μl) containing 40 mM Tris⋅HCl (pH 7.8), 7 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 166 μg/ml BSA, 50 ng of poly(dA)4000·oligo(dT)12–18 (20:1), 33 μM [3H]dTTP (300 cpm/pmol), and 0.3 μg of E. coli SSB. For the assay of PCNA activity, reaction mixtures were supplemented with 0.15 unit of pol δ and 0.13 unit of RFC; for the assay of RFC, reaction mixtures contained 50 ng of PCNA and 0.15 unit of pol δ; for the assay of pol δ, reaction mixtures contained 50 ng of PCNA and 0.13 unit of RFC. Reactions were incubated at 37°C for 60 min, and the incorporation of [3H]dTMP into an acid-insoluble form was measured by liquid scintillation counting. One unit of pol δ or RFC supported the incorporation of 1 nmol of dTMP under the conditions specified above.
sp PCNA Isolation.
The plasmid construct for overexpression of His-tagged sp PCNA (34) was transformed into E. coli M15[pREP4] cells. Cell cultures (10 liters) were grown in 2× YT medium (containing 16 g of Bacto-tryptone, 10 g Bacto-yeast extract, 5 g of NaCl per liter) to an OD600 of 0.7 prior to induction with 1 mM IPTG for 3 hr. The resulting cells (18 g) were lysed for 1 hr at 0°C in 100 ml of buffer containing 50 mM Tris⋅HCl buffer (pH 8.0), 20 mM potassium phosphate buffer (pH 8.0), 0.5 M KCl, 3 mg/ml lysozyme, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM EGTA, 0.2 μg/ml aprotinin, 0.2 μg/ml leupeptin, and 0.1 μg/ml antipain. Lysed cells were centrifuged at 18,000 rpm for 30 min, and the supernatant (85 ml) was mixed with 15 ml of Ni2+/NTA beads for 2 hr at 4°C. The beads were then washed three times with 75 ml of buffer containing 50 mM Tris⋅HCl (pH 8.0), 20 mM potassium phosphate (pH 8.0), and 1 M KCl, followed by washing with the same buffer containing 0.25 M KCl. The beads were then packed into a column, and PCNA was eluted with a solution containing 0.5 M imidazole, 20 mM potassium phosphate (pH 8.0), 0.25 M KCl, and 10% glycerol. Fractions containing PCNA, as identified by SDS/PAGE, were pooled and dialyzed for 12 hr against buffer containing 25 mM Tris⋅HCl (pH 7.5), 25 mM KCl, 1 mM EGTA, 0.1 mM PMSF, 0.2 μg/ml aprotinin, 0.2 μg/ml leupeptin, and 0.1 μg/ml antipain. SDS/PAGE analysis indicated that PCNA had been purified to ≥95% homogeneity with a yield of 200 mg.
sp Pol δ Isolation.
The isolation of sp pol δ was carried out using a procedure modified from that described for the purification of DNA pol III from S. cerevisiae (4). S. pombe cells (1.2 kg) were resuspended in 1,200 ml of buffer containing 0.2 M Tris⋅HCl (pH 8.0), 2 mM EDTA, 2 mM DTT, 10% glycerol, 0.01% Brij 58, 2 μM pepstatin A, 10 mM NaHSO3, 0.5 mM PMSF, and 2 mM benzamidine and were broken with glass beads using the DynoMill (Impandex, Clifton, NJ). Saturated ammonium sulfate solution (saturated at 4°C) was added to 5%, and the mixture was centrifuged at 15,000 × g for 60 min. The supernatant was adjusted to 55% with solid ammonium sulfate (280 g/ml), and the pellet obtained after centrifugation at 15,000 × g for 90 min was dissolved in 300 ml of buffer A (50 mM potassium phosphate, pH 7.0/10% glycerol/2 mM EDTA/2 mM DTT/2 μM pepstatin A/10 mM NaHSO3/0.5 mM PMSF/2 mM benzamidine) and 0.2 M KCl. This pol δ-containing fraction was dialyzed against three changes of 4 liters of the same buffer over a 12-hr period. The dialyzed material was loaded onto a phosphocellulose column (5.5 × 23.5 cm, 500 ml) equilibrated with 16 liters of buffer A with 0.2 M KCl and eluted sequentially with 800 ml of buffer A containing 0.4 M KCl and 500 ml of buffer A containing 0.7 M KCl. The 0.4 M KCl fraction (330 ml) containing pol δ activity was diluted to 0.1 M KCl with 1,170 ml of buffer B (25 mM potassium phosphate, pH 7.0/10% glycerol/2 mM EDTA/1 mM DTT/2 μM pepstatin A/10 mM NaHSO3/0.5 mM PMSF/2 mM benzamidine) and loaded onto a Q Sepharose column (2.5 × 8.5 cm, 40 ml) and eluted with a 1-liter linear gradient of 0.1–0.4 M KCl in buffer B. The active fractions (peaking at 0.2 M) were pooled (225 ml), diluted to 0.15 M KCl with 75 ml of buffer B, and loaded onto a SP-Sepharose column (1.0 × 7.0 cm, 5 ml). A 200-ml linear gradient of 0.15–0.5 M KCl in buffer B was used to develop this column, and active fractions (peaking at 0.2 M KCl) were pooled (25 ml), diluted to 0.1 M KCl with 30 ml of buffer B, and loaded onto a single-stranded DNA cellulose column (1.0 × 2.8 cm, 2 ml) equilibrated with buffer B and 0.1 M KCl. A 50-ml linear gradient of 0.1–0.4 M KCl in buffer B was applied to this column, and active fractions peaking at 0.2 M KCl were pooled (16 ml) and loaded directly onto a hydroxylapatite column (1.0 × 1.4 cm, 1 ml). A 40-ml linear gradient of 0.025–0.3 M potassium phosphate, pH 7.0, in buffer B containing 0.2 M KCl was used to elute bound proteins, and the pol δ activity eluting at 0.1 M potassium phosphate was pooled (8 ml) and concentrated to 4 ml using a centrifugal filter device (Millipore). A portion of this fraction (0.2 ml) was applied to a 5-ml 15–35% glycerol gradient in buffer B containing 0.2 M KCl. After centrifugation at 45,000 rpm for 18 hr at 4°C, fractions (0.2 ml) were collected from the bottom of the tube. A summary of the purification procedure is presented in Table 1. The yield of sp pol δ after the glycerol gradient step has been calculated assuming that this procedure was carried out with the entire fraction isolated after hydroxylapatite chromatography.
sp RFC Isolation.
For the isolation of sp RFC, S. pombe cells (600 g) were resuspended in buffer A (600 ml) and subjected to the same procedure described above for the isolation of sp pol δ through the phosphocellulose step. sp RFC activity eluted from the phosphocellulose column with buffer A containing 0.4 M KCl (260 ml, 4,100 units, 332 mg protein) and was dialyzed against two changes of 4 liters of buffer B containing 50 mM KCl over a 12-hr period. Following application to a Q Sepharose column (2.5 × 10.5 cm, 50 ml), bound proteins were eluted with a 500-ml linear gradient of 0.05–0.5 M KCl in buffer B. Active fractions (eluting at 0.2 M KCl) were pooled (80 ml, 8,080 units, 43 mg protein) and loaded onto a hydroxylapatite column (1.0 × 11.2 cm, 8 ml), which was developed with a 150-ml linear gradient of 0.025–0.4 M potassium phosphate, pH 7.0, in buffer B containing 0.2 M KCl. RFC-enriched fractions (eluting at 0.25 M potassium phosphate) were pooled (16 ml, 2,144 units, 8.8 mg protein), dialyzed against 2 liters of buffer C (25 mM Hepes, pH 7.5/10% glycerol/1 mM DTT/2 μM pepstatin A/2 mM NaHSO3/0.1 mM PMSF/1 mM benzamidine) with 0.2 M KCl, and mixed with 1 ml of sp PCNA-linked Affi-Gel 15 beads (containing 10 mg of PCNA per ml of beads) equilibrated with the same buffer in the presence of 2 mM ATP and 7 mM MgCl2 (35). After rocking for 4 hr at 4°C, the mixture was packed in a column and washed with the same buffer prior to elution of bound proteins with 25 ml of 0.5 M KCl in buffer C, and active fractions were pooled (5 ml, 732 units, 36 μg of protein). SDS/PAGE analysis indicated that the RFC complex was purified to near homogeneity.
Protein Sequence Analysis.
sp Pol δ containing glycerol gradient fractions was pooled (2.4 ml, ≈5 μg protein) and precipitated with 0.1 volume of 0.15% sodium deoxycholate and 0.1 volume of 72% trichloroacetic acid. The mixture was centrifuged at 15,000 rpm for 30 min at room temperature, and the pellet was dissolved in 20 μl of 2× SDS sample buffer, neutralized with 1 M Tris⋅HCl, pH 8.3, and then resolved by 12% SDS/PAGE and transferred to a nitrocelloluse membrane (0.2 μm, Schleicher & Schuell). The 42- and 22-kDa protein bands were excised from the Poinceau S-stained nitrocellulose membrane, subjected to in situ tryptic digestion (36, 37) prior to peptide resolution through a 1-mm RP-HPLC column (ODS 1 mm × 150 mm). Chemical sequencing and matrix-assisted laser-desorption ionization time-of-flight mass spectrometry were carried out as described (38–40).
RESULTS
Isolation of sp PCNA and sp RFC.
PCNA, RFC, and SSB all stimulate pol δ-catalyzed DNA synthesis of primed single-stranded DNA templates. To assay for sp pol δ activity during purification, PCNA and RFC were purified from S. pombe as described in Materials and Methods. It has previously been shown that h PCNA can substitute for sp PCNA in vivo (41). To test whether sp PCNA and sp RFC are able to substitute for h PCNA and h RFC, respectively, in supporting in vitro DNA synthesis, poly(dA):oligo(dT) assays were carried out in which the pol δ holoenzyme was reconstituted with h pol δ, h or sp RFC, and h or sp PCNA. sp RFC and sp PCNA supported h pol δ-dependent DNA synthesis to an extent similar to that observed with the homologous species pol δ-holoenzyme. Thus, for its isolation, sp RFC activity was assayed for its ability to stimulate h pol δ-dependent DNA synthesis in the presence of sp PCNA. Purified sp RFC migrated in SDS/PAGE as five distinct protein bands, one of 125 kDa and four with apparent molecular masses of 36–40 kDa, more closely resembling the h RFC subunit sizes (145 kDa and 36–40 kDa) than those of sc RFC (94 kDa and 36–40 kDa) (data not presented). The relationship of the sp RFC small subunits to the corresponding subunits of h RFC and sc RFC, however, remains to be elucidated.
Isolation of sp Pol δ.
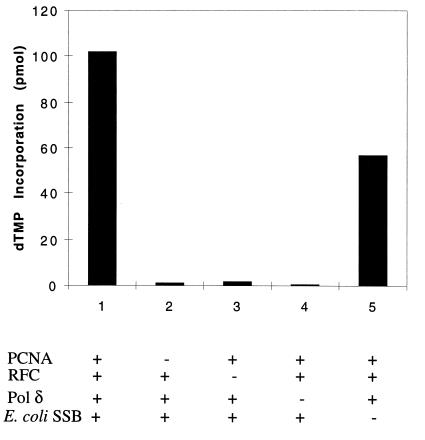
A summary of the purification procedure utilized for the isolation of sp pol δ is presented in Table 1. sp Pol δ was purified more than 1,000-fold with a 4% yield compared with the polymerase activity detected after phosphocellulose chromatography. Whereas the activity of the pol δ present in relatively crude fractions (the phosphocellulose fraction) was markedly stimulated by PCNA, RFC and pol δ cofractionate until the hydroxylapatite column and, thus, addition of RFC to these fractions does not stimulate pol δ-catalyzed DNA synthesis further. However, following hydroxylapatite fractionation, RFC and pol δ separate and pol δ activity becomes completely dependent on the addition of both PCNA and RFC (Fig. 1). In the poly(dA):oligo(dT) assay, poly(dT) synthesis was only stimulated 2-fold by E. coli SSB. In contrast, in reactions in which singly primed single-stranded M13 DNA was used in place of poly(dA):oligo(dT), DNA synthesis was completely dependent on the presence of an SSB (E. coli, h, or sp SSB; data not presented).
Figure 1.
Effects of PCNA, RFC, and E. coli SSB on pol δ activity with the poly(dA)⋅oligo(dT) template. Reaction mixtures (30 μl) contained 40 mM Tris⋅HCl (pH 7.8), 7 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 166 μg/ml BSA, 50 ng poly(dA)4000⋅oligo(dT)12–18 (20:1), and 33 μM [3H]dTTP (300 cpm/pmol). Where indicated, 0.13 unit of RFC, 50 ng PCNA, 0.3 μg E. coli SSB, and 0.15 unit of pol δ (hydroxylapatite fraction) were added. Reactions were incubated at 37°C for 60 min, and the incorporation of [3H]dTMP into an acid-insoluble form was measured by liquid scintillation counting.
Subunit Structure and Composition of sp Pol δ.
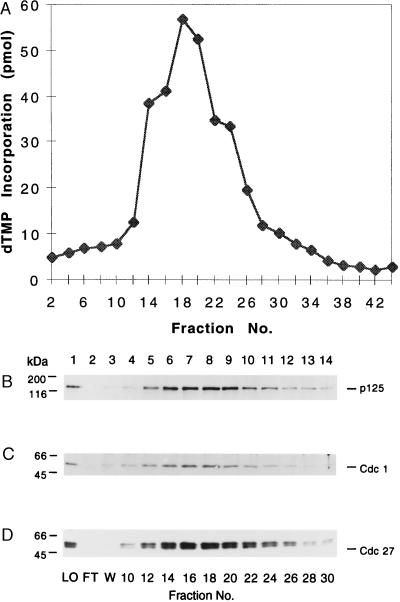
The availability of highly purified sp pol δ and antibodies specific for the p125 catalytic subunit, Cdc1, and Cdc27 prompted us to examine whether these subunits are all components of sp pol δ. For this purpose, we examined the elution profiles of both sp pol δ DNA synthetic activity and the p125, Cdc1, and Cdc27 subunits by Western blot analyses after hydroxylapatite chromatography. As shown in Fig. 2A, pol δ eluted between fractions 14 and 24 (0.1 M potassium phosphate), concomitant with the elution of the p125, Cdc1, and Cdc27 subunits as determined by Western blot analyses (Fig. 2 B–D, respectively). These three proteins were detected only in fractions containing polymerase activity, indicating their strong association.
Figure 2.
Elution profile of sp pol δ from a hydroxylapatite column. The elution procedure was carried out as described in Materials and Methods. (A) Elution profile of sp pol δ polymerase activity as determined by the poly(dA)⋅oligo(dT) assay. Aliquots (3.4 μl) of fractions eluting from the hydroxylapatite column were subjected to 10% SDS/PAGE and visualized by Western blotting with polyclonal antibodies against the p125 subunit (B), Cdc1 (C), and Cdc27 (D). LO, load on (single-stranded DNA cellulose fraction); FT, flow through; W, wash.
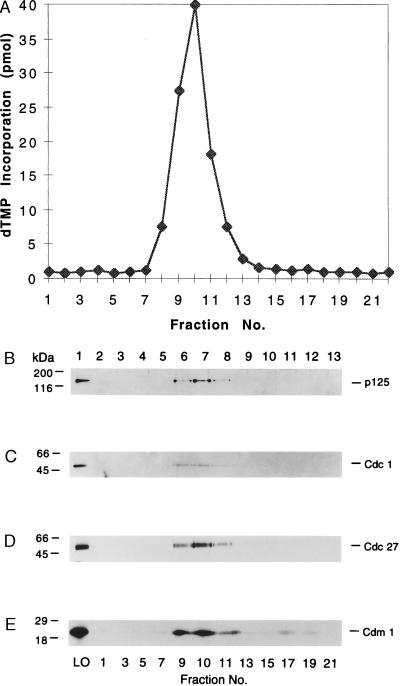
Glycerol gradient sedimentation of the sp pol δ-enriched hydroxylapatite fractions resulted in the appearance of DNA synthetic activity that peaked at fraction 10, between the catalase and aldolase marker proteins (Fig. 3A). Western blot analyses with antibodies against the p125, Cdc1, and Cdc27 subunits (Fig. 3 B–D, respectively) indicated that all three proteins cosedimented at a position coincident with sp pol δ activity (as seen following hydroxylapatite fractionation in Fig. 2). As observed after hydroxylapatite chromatography, these three proteins were detected only in fractions containing polymerase activity.
Figure 3.
Glycerol gradient sedimentation of sp pol δ. This step was carried out as described in Materials and Methods. (A) Glycerol gradient fractions were assayed for pol δ polymerase activity using the poly(dA):oligo(dT) assay. Aliquots (10 μl) of glycerol gradient fractions were subjected to Western blot analyses using polyclonal antibodies against the p125 subunit (B), Cdc1 (C), Cdc27 (D), and Cdm1 (E). LO, load on (hydroxylapatite fraction). In a separate gradient, sedimented under identical conditions, catalase and aldolase peaked at fractions 5 and 13, respectively.
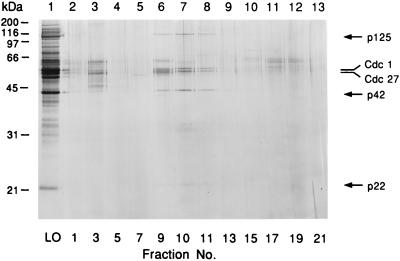
Examination of the glycerol gradient fractions by SDS/PAGE analysis followed by silver staining revealed that pol δ cosedimented as a complex containing five polypeptides with apparent molecular masses of 125, 55, 54, 42, and 22 kDa (Fig. 4). Western blot analysis confirmed that the 125-kDa (Pol3/Cdc6) polypeptide is the large subunit of pol δ, and that the protein bands of 55 and 54 kDa correspond to Cdc1 and Cdc27, respectively (as previously shown; refs. 32 and 42). Based on their amino acid composition, Cdc1 and Cdc27 are 51 and 43 kDa, respectively, suggesting that they migrate more slowly during SDS/PAGE than expected. Western blot analysis of Cdc27 (Figs. 2D and 3D) revealed the presence of two bands that migrated closely, most likely due to phosphorylation of Cdc27 (S.A.M., unpublished results). The comigration of the p125, Cdc1, and Cdc27 subunits with sp pol δ activity was evident in all chromatographic steps (summarized in Table 1) that were carried out with five different extract preparations. The previously unidentified proteins of 42 and 22 kDa were shown to cosediment with sp pol δ activity by SDS/PAGE analysis following hydroxylapatite chromatography and glycerol gradient sedimentation. When the two glycerol gradient fractions corresponding to the peak of pol δ activity were pooled, concentrated, and subjected to a second glycerol gradient centrifugation step, all five proteins continued to cosediment with sp pol δ activity. These observations suggest that all five protein subunits are associated with sp pol δ as a multiprotein complex.
Figure 4.
Polypeptide composition of sp pol δ. Aliquots (20 μl) of the glycerol gradient fractions were subjected to 12% SDS/PAGE analysis followed by silver staining. LO, load on.
Sequence Analysis of p42 and p22.
The two previously unidentified protein bands, p42 and p22, that copurified with sp pol δ were isolated and subjected to tryptic digestion as described in Materials and Methods. The sequences of three distinct tryptic peptides of the isolated 42-kDa band were obtained and are indicated in Fig. 5. No sequences homologous to these peptides were detected using the blast search program; however, a single cosmid, c9G1, was retrieved from the S. pombe genomic DNA sequencing project database at the Sanger Center, Cambridge, U.K. (http://www.sanger.ac.uk/projects/s_pombe/), which included sequences encoding all three peptides. These peptides are present within a single ORF of a predicted protein of 42 kDa. The nucleotide sequence and deduced amino acid sequence of p42 are presented in Fig. 5.
Figure 5.
Nucleotide and amino acid sequences of the sp pol δ p42 subunit. The three peptide sequences determined by chemical sequencing of tryptic peptides derived from the isolated 42-kDa subunit are underlined. These sequences were identical to the DNA sequence of the cosmid c9G1 retrieved from the S. pombe database (available from the Sanger Center, Cambridge, U.K.). The calculated molecular mass from the deduced amino acid sequence of the p42 subunit is 41,737 daltons, in keeping with the 42-kDa size observed following SDS/PAGE analysis.
Partial peptide sequences generated following tryptic digestion of the 22-kDa subunit were identical to the sequence of the S. pombe Cdm1 protein (Fig. 6). The cdm1+ gene was initially isolated as a multicopy suppressor of the temperature-sensitive cdc1-P13 mutant (32). Thus, p22 (Cdm1) has now been identified in association with the sp pol δ multisubunit complex by both genetic and biochemical approaches. The identity of this 22-kDa subunit was confirmed following immunoblot analysis of the sp pol δ containing glycerol gradient fractions using antibodies against the Cdm1 protein (Fig. 3E). In contrast to the other subunits, Western blot analysis indicated that a small portion of the Cdm1 subunit appeared to dissociate from the complex during glycerol gradient centrifugation. A detailed characterization of cdm1+ will be presented elsewhere (S.A.M., N. Reynolds, A. Watt, and P. A. Fantes, unpublished work).
Figure 6.
The complete amino acid sequence of the 22-kDa subunit deduced from clones of the cdm1 gene that rescued temperature-sensitive cdc1 mutants. The partial peptide sequences determined by chemical analysis of the 22-kDa subunit isolated as a part of the sp pol δ holoenzyme are underlined. Several of the peptides included sites at which no amino acid assignment could be made, and these are indicated with an asterisk above that position.
DISCUSSION
The previous demonstration that there is an interaction between the S. pombe Cdc1 and Pol3 subunits and between Cdc1 and Cdc27 suggested that these three proteins would be found in a complex. In this study, we have isolated and characterized S. pombe DNA polymerase δ and confirmed that Cdc1 and Cdc27 are associated with the catalytic 125-kDa subunit of pol δ and that these three proteins copurify with pol δ activity.
The Pol3 subunit, Cdc1, and Cdc27 have each been shown to be essential for cell cycle progression. S. pombe cells carrying temperature-sensitive mutations in the Pol3 subunit undergo cell cycle arrest in late S phase at the restricted temperature (43–45). Cells with temperature-sensitive mutations in either Cdc1 or Cdc27 undergo cell cycle arrest at the restricted temperature with a 2C DNA content and become hypersensitive to hydroxyurea at the permissive temperature. Cells harboring Cdc1 or Cdc27 mutations are highly elongated, contain a single nucleus, and do not enter mitosis. This cell cycle arrest is dependent on the rad/hus checkpoint pathway as demonstrated in cdc1 rad/hus or cdc27 rad/hus double-mutant cells, which fail to arrest prior to mitosis. Furthermore, Cdc1 or Cdc27 inactivation induces synthesis of the large transcript of suc22 (the small subunit of ribonucleotide reductase), which is associated with inhibition of DNA replication and DNA damage in S. pombe (32). Together, these results demonstrate that Pol3, Cdc1, and Cdc27 play important roles in replication, repair, and cell cycle progression (43–46).
The identification of two additional sp pol δ subunits of 42 and 22 kDa was based on their copurification with the p125, Cdc1, and Cdc27 subunits and with PCNA-dependent polymerase activity. Sequence data obtained from tryptic digests of the p42 and p22 protein bands indicated that they were not proteolyzed fragments of the p125, Cdc1, and/or Cdc27. The p42 subunit possesses a weak but significant sequence similarity with the budding yeast nuclear protein GCD14, which is of unknown function (47). The 22-kDa subunit was found to be identical to the Cdm1 protein, which was previously isolated by its ability to rescue cells harboring a temperature-sensitive cdc1 mutation (32). The significance of this interaction, as well as the role of the p22 and p42 subunits in governing the activity of sp pol δ, awaits further genetic and reconstitution studies.
Pol δ purified from mammalian cells has been shown to be a heterodimer of 125 and 48 kDa with no apparent Cdc27, p42, or Cdm1 homologues present. There are several possible explanations for this. Cdc27, p42, and Cdm1 may be unique to yeast, exhibit a lower affinity for the mammalian 125- and 48-kDa subunits, and thus separate from the heterodimer during purification or may only be required for the repair or checkpoint functions of pol δ and be dispensable for replication. Alternatively, exponentially growing fission yeast cells are predominantly in the G2 phase, and the association of p42, Cdc27, and Cdm1 with p125 and Cdc1 may be cell-cycle-dependent. Further studies with mammalian pol δ should distinguish between these possibilities.
The purified recombinant human p125-kDa subunit has catalytic and 3′ to 5′ exonuclease activities in vitro, suggesting that the 48-kDa subunit is not required for these activities. Recent studies have shown that polymerase activity of the human p125 subunit is not stimulated by PCNA (48); however, coexpression and subsequent purification of the p125- and 48-kDa subunits (homologous to Pol3 and Cdc1) yield a complex whose activity is stimulated by PCNA (28), suggesting that the 48-kDa subunit is responsible for mediating any interaction between pol δ and PCNA.
It is interesting to note that as with pol δ there is a discrepancy in the subunit structure of pol ɛ from human or S. cerevisiae origins. Pol ɛ derived from human cells has been shown to include only two subunits of 215 and 55 kDa (49, 50), whereas sc pol ɛ contains subunits of >200, 80, 34, 30, and 29 kDa (5, 51, 52). Genes encoding the largest three of these sc subunits have been cloned, and those encoding the 200- and 80-kDa subunits have been shown to be essential. A single nonessential gene encodes both the 30- and 34-kDa subunits, suggesting that they may represent posttranslationally modified products of this gene (52).
The pol δ holoenzyme reconstituted from combinations of h and sp PCNA, RFC, and pol δ efficiently supported poly(dT) synthesis with a poly(dA):oligo(dT) template but showed variable efficiency in catalyzing nucleotide incorporation into a singly primed M13 DNA template. This difference was most evident in reactions containing sp pol δ, h PCNA, and sp RFC or h RFC. Other distinctions included a more marked salt sensitivity of reactions containing sp pol δ compared with those containing h pol δ (data not presented). Whether these effects are due to differences in the subunit structure and/or stability of sp pol δ and h pol δ is presently unclear.
Table 1.
Purification of S. pombe DNA polymerase
| Step | Protein, mg | Activity, units | Specific activity, units/mg of protein | Yield, % |
|---|---|---|---|---|
| Crude extract | 91,200 | — | — | — |
| Phosphocellulose | 930 | 19,050 | 21 | (100) |
| Q-Sepharose | 59 | 12,690 | 215 | 67 |
| Sp-Sepharose | 13 | 6,116 | 470 | 32 |
| SS-DNA cellulose | 5.4 | 4,132 | 765 | 22 |
| Hydroxylapatite | 0.28 | 1,378 | 4,921 | 7 |
| Glycerol gradient | 0.036 | 731 | 20,300 | 3.8 |
Acknowledgments
The authors are indebted to Dr. Z.-Q. Pan for helpful discussions and to Drs. Hediye Erdjument-Bromage and Paul Tempst of the Protein Center at Memorial Sloan–Kettering Cancer Center for their help in determining the peptide sequences reported here. Z.K. is a postdoctoral fellow of the Helen Hay Whitney Foundation. These studies were supported by the National Institutes of Health Grant GM 38559 to J.H., CA 54415 to T.S.-F.W., GM 38839 to M.O.D., and the Wellcome Trust (Grant 047406/Z/96) to S.A.M. J.H. is a Professor of the American Cancer Society.
ABBREVIATIONS
- pol
DNA polymerase
- RFC
replication factor C
- PCNA
proliferating cell nuclear antigen
- hSSB
human single-stranded DNA-binding protein (also called RPA)
- h
human
- sp
Schizosaccharomyces pombe
- sc
Saccharomyces cerevisiae
- PMSF
phenylmethylsulfonyl fluoride
References
- 1.Johnson A L, Barker D G, Johnston L H. Curr Genet. 1986;22:107–112. doi: 10.1007/BF00378201. [DOI] [PubMed] [Google Scholar]
- 2.Boulet A, Simon M, Faye G, Bauer G A, Burgers P M J. EMBO J. 1989;8:1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stillman B. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 4.Bauer G A, Heller H M, Burgers P M J. J Biol Chem. 1988;263:917–924. [PubMed] [Google Scholar]
- 5.Morrison A, Araki H, Clark A B, Hamatake R K, Sugino A. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 6.Wang T S-F. In: DNA Replication in Eukaryotic Cells. DePamphilis M, editor; DePamphilis M, editor. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1996. pp. 461–493. [Google Scholar]
- 7.Matsumoto T, Eki T, Hurwitz J. Proc Natl Acad Sci USA. 1990;87:9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T S-F. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- 9.Sugino A. Trends Biochem Sci. 1995;20:319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- 10.Lee S-H, Kwong A D, Ishimi Y, Hurwitz J. Proc Natl Acad Sci USA. 1989;86:4877–4881. doi: 10.1073/pnas.86.13.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsurimoto T, Stillman B. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 13.Lee S-H, Hurwitz J. Proc Natl Acad Sci USA. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsurimoto T, Stillman B. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 15.Burgers P M, Yoder B L. J Biol Chem. 1993;266:1961–1968. [Google Scholar]
- 16.Gogol E P, Young M C, Kubasek W L, Jarvis T C, von Hippel P H. J Mol Biol. 1992;224:395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- 17.Kong X-P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 18.Krishna T S R, Kong X-P, Gary S, Burgers P, Kuriyan J. Cell. 1994;79:1233–1244. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 19.Lee S-H, Pan Z-Q, Kwong A D, Burgers P M J, Hurwitz J. J Biol Chem. 1991;266:22707–22717. [PubMed] [Google Scholar]
- 20.Burgers P M J. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 21.Hurwitz J, Dean F B, Kwong A D, Lee S-H. J Biol Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 22.Kuriyan J, O’Donnell M. J Mol Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 23.Herendeen R H, Kelly T J. Cell. 1996;84:5–8. doi: 10.1016/s0092-8674(00)80069-0. [DOI] [PubMed] [Google Scholar]
- 24.Byrnes J J, Downey K M, Black V C, So A G. Biochemistry. 1976;15:2817–2823. doi: 10.1021/bi00658a018. [DOI] [PubMed] [Google Scholar]
- 25.Chung D W, Zhang J, Tan C-K, Davie E W, So A G, Downey K M. Proc Natl Acad Sci USA. 1991;88:11197–11201. doi: 10.1073/pnas.88.24.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C L, Chang L-S, Zhang P, Hao H, Zhu L, Toomey N L, Lee M Y W T. Nucleic Acids Res. 1992;20:735–745. doi: 10.1093/nar/20.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Frugulhetti I, Jiang Y, Holt G, Condit R C, Lee M Y W T. J Biol Chem. 1995;270:7993–7998. doi: 10.1074/jbc.270.14.7993. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J-Q, He H, Tan C-K, Downey K M, So A G. Nucleic Acids Res. 1997;25:1094–1099. doi: 10.1093/nar/25.6.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugimoto K, Sakamoto Y, Takahashi O, Matsumoto K. Nucleic Acids Res. 1995;23:3493–3500. doi: 10.1093/nar/23.17.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pignede G, Bouvier D, Recondo A-M, Baldacci G. J Mol Biol. 1991;222:209–218. doi: 10.1016/0022-2836(91)90207-m. [DOI] [PubMed] [Google Scholar]
- 31.Park H, Francesconi S, Wang T S-F. Mol Biol Cell. 1993;4:145–157. doi: 10.1091/mbc.4.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacNeill S A, Moreno S, Reynolds N, Nurse P, Fantes P A. EMBO J. 1996;15:4613–4628. [PMC free article] [PubMed] [Google Scholar]
- 33.Iino Y, Yamamoto M. Mol Gen Genet. 1997;254:93–97. doi: 10.1007/s004380050395. [DOI] [PubMed] [Google Scholar]
- 34.Arroyo M P, Downey K M, So A G, Wang T S-F. J Biol Chem. 1996;271:15971–15980. doi: 10.1074/jbc.271.27.15971. [DOI] [PubMed] [Google Scholar]
- 35.Gerik K J, Gary S L, Burgers P M J. J Biol Chem. 1997;272:1256–1262. doi: 10.1074/jbc.272.2.1256. [DOI] [PubMed] [Google Scholar]
- 36.Tempst P, Link A J, Riviere L R, Fleming M, Elicone C. Electrophoresis. 1990;11:537–553. doi: 10.1002/elps.1150110704. [DOI] [PubMed] [Google Scholar]
- 37.Lui M, Tempst P, Erdjument-Bromage H. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 38.Elicone C, Lui M, Geromanos S, Erdjument-Bromage H, Tempst P. J Chromatogr. 1994;676:121–137. doi: 10.1016/0021-9673(94)00089-1. [DOI] [PubMed] [Google Scholar]
- 39.Erdjument-Bromage H, Lui M, Sabatini D M, Snyder S H, Tempst P. Protein Sci. 1994;3:2435–2446. doi: 10.1002/pro.5560031227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geromanos S, Casteel P, Elicone C, Powell M, Tempst P. In: Techniques in Protein Chemistry V. Crabb J W, editor; Crabb J W, editor. San Diego: Academic; 1994. pp. 143–150. [Google Scholar]
- 41.Waseem N H, Labib K, Nurse P, Lane D P. EMBO J. 1992;11:5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes D A, MacNeill S A, Fantes P A. Mol Gen Genet. 1992;231:401–410. doi: 10.1007/BF00292709. [DOI] [PubMed] [Google Scholar]
- 43.Francesconi S, Park H, Wang T S-F. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francesconi S, DeRecondo A M, Baldacci G. Mol Gen Genet. 1995;246:561–569. doi: 10.1007/BF00298962. [DOI] [PubMed] [Google Scholar]
- 45.Uchiyawa M, Galli I, Griffiths D J F, Wang T S-F. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Congley M J, Pierce A J, Modrich P. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 47.Cziepluch C, Kordes E, Pujol A, Jauniaux J C. Yeast. 1996;12:1471–1474. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1471::AID-YEA30%3E3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Zhou J Q, Tan C-K, So A G, Downey K M. J Biol Chem. 1996;271:29740–29745. doi: 10.1074/jbc.271.47.29740. [DOI] [PubMed] [Google Scholar]
- 49.Syvaoja J, Linn S. J Biol Chem. 1989;264:2489–2497. [PubMed] [Google Scholar]
- 50.Kesti T, Fanti H, Syvaoja J. J Biol Chem. 1993;268:10238–10245. [PubMed] [Google Scholar]
- 51.Hamatake R K, Hasegana H, Clark A B, Bebenek K, Kunkel T A, Sugino A. J Biol Chem. 1990;265:4072–4083. [PubMed] [Google Scholar]
- 52.Araki H, Hamatake R K, Johnston L H, Sugino A. Proc Natl Acad Sci USA. 1991;88:4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]