Abstract
The causal agent of chrysanthemum chlorotic mottle (CChM) disease has been identified, cloned, and sequenced. It is a viroid RNA (CChMVd) of 398–399 nucleotides. In vitro transcripts with the complete CChMVd sequence were infectious and induced the typical symptoms of the CChM disease. CChMVd can form hammerhead structures in both polarity strands. Plus and minus monomeric CChMVd RNAs self-cleaved during in vitro transcription and after purification as predicted by these structures, which are stable and most probably act as single hammerhead structures as in peach latent mosaic viroid (PLMVd), but not in avocado sunblotch viroid (ASBVd). Moreover, the plus CChMVd hammerhead structure also appears to be active in vivo, because the 5′ terminus of the linear plus CChMVd RNA isolated from infected tissue is that predicted by the corresponding hammerhead ribozyme. Both hammerhead structures of CChMVd display some peculiarities: the plus self-cleaving domain has an unpaired A after the conserved A9 residue, and the minus one has an unusually long helix II. The most stable secondary structure predicted for CChMVd is a branched conformation that does not fulfill the rod-like or quasi-rod-like model proposed for the in vitro structure of most viroids with the exception of PLMVd, whose proposed secondary structure of lowest free energy also is branched. The unusual conformation of CChMVd and PLMVd is supported by their insolubility in 2 M LiCl, in contrast to ASBVd and a series of representative non-self-cleaving viroids that are soluble under the same high salt conditions. These results support the classification of self-cleaving viroids into two subgroups, one formed by ASBVd and the other one by PLMVd and CChMVd.
Keywords: satellite RNAs, RNA self-cleavage
The discovery of viroids in the early seventies (1) led to the modification of the paradigm that considered viruses as the smallest inciting agents of infectious diseases. Viroids, single-stranded circular RNAs of 246–375 nucleotides able to infect certain plants, are currently the lowest step of the biological scale (2). Viroids display singular functional properties: their genomes are not translated, and two of the 26 different “species” sequenced so far are able to self-cleave through hammerhead ribozymes (3), an aspect with important evolutive implications.
Although chrysanthemum chlorotic mottle (CChM) was one of the first diseases presumed to be induced by a viroid, the molecular structure of its causal agent has remained an enigma for more than 20 years. Biological evidence supporting the involvement of a small RNA with a size consistent with that expected for a viroid was presented (4), and in accordance with this hypothesis, electron microscope studies of thinly sectioned tissues and sap preparations failed to reveal virus-like particles (5). However, a differential RNA was never found in the zone of nondenaturing polyacrylamide gels where infectivity was located (6), and in cases where such an RNA was reported it turned out to be most probably a contamination of chrysanthemum stunt viroid (CSVd), another viroid already identified in the same plant. Furthermore, the causal agent of the CChM disease displayed some unusual properties when compared with typical viroids, particularly its high instability in crude leaf extracts and its insolubility in 2 M LiCl (4, 7), which raised intriguing questions about its structure. Finally, coinoculation experiments showed that members of the group of viroids, which include CSVd, citrus exocortis viroid (CEVd), and a mild and a severe strain of potato spindle tuber viroid, but exclude the agent of CChM disease, exhibited crossprotection in a variety of combinations. These results were interpreted as an indication that CSVd, CEVd, and potato spindle tuber viroid, but not the agent of CChM disease, affect a common biological process (8). These unique properties, which even led to the consideration of the possible involvement in CChM disease of a novel class of small pathogenic RNAs (9), created the challenge to characterize the causal agent of CChM disease. Here we report the molecular structure of this agent, which is a viroid (CChMVd) whose unusual features explain some of the above observations.
MATERIALS AND METHODS
Infectivity Bioassays.
A type isolate of the agent of CChM disease was used. Bioassays were performed on the natural host chrysanthemum, Dendranthema grandiflora Tzvelev. cv “Bonnie Jean” in growth chambers at 28°C with constant fluorescent illumination. Under these conditions the typical symptoms appeared approximately 8–10 days after inoculation. Mechanical inoculations were made by applying nucleic acid extracts with a tip to three expanding leaves of each plant previously dusted with carborundum. Inocula were made in borate buffer (0.2 M boric acid-NaOH, pH 9) (4). Each preparation was assayed on four plants.
RNA Analysis and Purification.
Crude extracts from chrysanthemum leaves were obtained with buffer-saturated phenol (10). Where indicated nucleic acids were made 2 M LiCl and left overnight at 4°C, and the soluble and insoluble fractions were collected by centrifugation at 8,000 g for 20 min; other details are described in Results. Polysaccharides were removed with methoxyethanol (11). RNAs were analyzed in nondenaturing 5% polyacrylamide gels (130 × 100 × 2 mm) with TAE buffer (40 mM Tris/20 mM sodium acetate/1 mM EDTA, pH 7.2) and in denaturing 5% polyacrylamide gels of the same size containing 8 M urea and 0.25 × TBE buffer (12). In some cases a combination of both systems was used (13). To improve the resolution RNAs were also analyzed in 5% long denaturing gels (370 × 200 × 1 mm) containing 8 M urea and 1 × TBE (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3). Gels were stained with ethidium bromide and silver for preparative and analytical purposes, respectively. Where indicated RNAs were eluted from gel sections (10) and bioassayed.
cDNA Synthesis, Cloning, and Sequencing.
First-strand CChMVd-cDNA was synthesized with avian myeloblastosis virus reverse transcriptase and random hexamers using linear CChMVd RNA as template. Second-strand cDNA was generated by the RNase H method (14), and the resulting double-stranded cDNA was cloned in the SmaI site of pUC 18. To identify CChMVd-specific cDNAs, the resulting inserts were sequenced with chain terminating inhibitors (15) and T7 DNA polymerase. Inserts with sequences corresponding to ribosomal RNA and ribulose-1,5-bisphosphate carboxylase oxygenase were discarded, and the rest, with sequences not deposited in the EMBL database, served as templates for random-primed synthesis of radioactive probes, which were used in Northern blot hybridizations of RNAs from healthy and CChM-affected plants. This approach identified a probe derived from an insert of 82 nt that hybridized with a band of the expected size present only in plants displaying the CChM disease symptoms. With the sequence of this 82-nt insert a pair of adjacent primers of opposite polarities PI (5′-TGGGACTGGCCCCATCTCCCTTCTCC-3′) and PII (5′-GTCGGTTCGCTCTCGTAGTCACAGCC-3′) was designed and then used for the synthesis of full-length CChMVd-cDNA by reverse transcription–PCR. For reverse transcription reaction 20 ng of purified circular CChMVd RNA and 500 ng of primer PI were annealed in 10 mM Tris⋅HCl, pH 8.5/20 mM KCl by boiling for 2 min and cooling rapidly on ice. First-strand cDNA was synthesized as before (16), precipitated with ethanol, and dissolved in 10 μl of water. PCR amplification was performed in 50 μl containing 5 μl of the cDNA solution, 0.5 μg each of primers PI and PII, 2.5 units of Pfu DNA polymerase, and the buffer recommended by the supplier (Stratagene). The PCR cycling profile (30 cycles) was as described (16), and the amplified products of the expected full length were eluted and cloned in the SmaI site of pUC18. cDNA inserts were sequenced in both orientations as already indicated.
Analysis of the in Vitro Self-Cleavage Reactions.
The EcoRI-BamHI fragments of the recombinant pUC18 plasmids containing the full-length inserts of CChMVd were subcloned in pBluescript II KS+ (Stratagene). CChMVd head-to-tail dimeric constructs were obtained by ligation of monomeric PCR-amplified products, and synthesized with the phosphorylated primers PI and PII, and subsequent cloning in the SmaI site of pBluescript II KS+. Radioactive and nonradioactive transcripts of both polarities were obtained with T3 and T7 RNA polymerases as described (17). Transcription products were separated in 5% polyacrylamide gels containing 8 M urea and 40% formamide and autoradiographed or scanned with a bioimage analyzer (Fuji BAS1500).
Self-cleavage of purified transcripts eluted from gels was performed by incubation at 40°C for 1 hr in the self-cleavage buffer (50 mM Tris⋅HCl, pH 8/5 mM MgCl2/0.5 mM EDTA) (17). The samples were previously heated in 1 mM EDTA (pH 6) at 95°C for 2 min and snap-cooled in ice. The extension of self-cleavage was analyzed as indicated.
Computer Analysis.
The secondary structure of lowest free energy for CChMVd RNA was determined by the circular version of the mfold program (Genetics Computer Group, Madison, WI) (18).
RESULTS
Detection of a Small RNA Associated with CChM Disease.
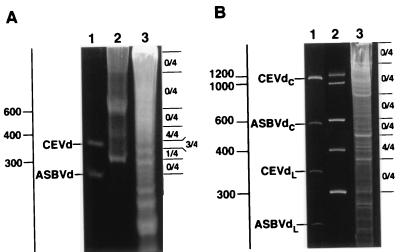
Total nucleic acids from healthy and infected chrysanthemum plants were partitioned in 2 M LiCl. Bioassays revealed that most of the infectivity was in the 2 M LiCl insoluble RNAs (data not shown), confirming previous observations (7). From here on, we continued with this fraction, which was analyzed by PAGE in 5% nondenaturing gels. After staining with ethidium bromide or silver, no differential band was detected in the material from infected plants (data not shown). To determine where the infectious RNA laid, the gel was cut into sections, and linear RNAs and the largest and the smallest known viroids (Fig. 1A) were taken as markers. The RNAs eluted from the sections were bioassayed and, in line with previous results (7), most of the infectivity was found in the region between the RNA markers of 300 and 400 nt (Fig. 1A).
Figure 1.
Analysis of nucleic acids by PAGE under nondenaturing (A) and denaturing conditions (B). Gels were stained with ethidium bromide. Lanes 1, CEVd (371 nt) and ASBVd (247 nt), which in denaturing conditions are separated in two bands corresponding to the circular and linear forms indicated by the subscripts C and L, respectively. Lanes 2, ladder of RNA markers with their sizes in nt indicated at the left. Lanes 3, nucleic acid preparation of CChMVd-infected tissue. After electrophoresis nucleic acids were eluted from the regions delimited by the horizontal bars at the right and inoculated onto blocks of four chrysanthemum plants (the numerators of the fractions indicate the number of plants that developed symptoms).
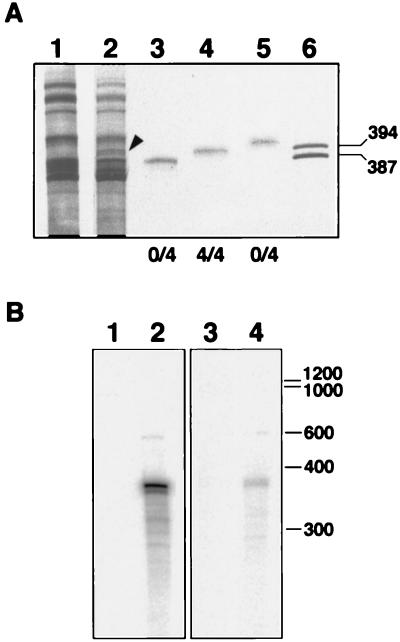
The same nucleic acid preparations then were analyzed by PAGE in 5% denaturing gels containing 8 M urea and 0.25 × TBE, in which circular and linear forms of viroids are considerably separated (12). The infectivity was recovered from the gel section around the linear RNA marker of 400 nt (Fig. 1B), but no symptoms were induced by the RNAs eluted from sections where viroid circular RNAs of 250–375 nt migrate. These results indicated that the RNA causing the CChM disease is a linear molecule of approximately 400 nt, although the coexistence of a circular form accumulating at very low levels was not ruled out. In an attempt to detect such a circular RNA, we separated nucleic acids by two consecutive PAGE steps with a protocol designed for identifying new viroid-like RNAs (13). After silver staining, no differential RNA could be detected in the zone where viroid circular forms migrate (data not shown), indicating that, if present, the levels of a circular CChM-specific RNA must be very low. At that point we considered that the linear RNA of approximately 400 nt causing the CChM disease could have remained undetected against a background of similarly sized cellular RNAs due to the limited resolution of the gels used. To examine this possibility, preparations enriched in the pathogenic RNA, obtained by eluting the nucleic acids from the section of a nondenaturing PAGE delimited by the linear RNA markers of 300–400 nt, were electrophoresed in long denaturing gels. Using this approach, an RNA with the expected size of the pathogenic agent was detected in preparations from CChM-infected plants, but not in parallel healthy controls (Fig. 2A). The differential RNA, and those from the adjacent regions of the gel, were eluted and inoculated. The typical symptoms were induced by the differential RNA (Fig. 2A) and, moreover, a correlation between the infectivity and the intensity of the band of this RNA was observed (data not shown).
Figure 2.
(A) Analysis of nucleic acids by PAGE in long denaturing gels stained with silver. Lanes 1 and 2, nucleic acid preparations from healthy and CChMVd-infected plants, respectively, with the arrowhead indicating the position of a differential band. Lanes 3, 4, and 5, nucleic acids eluted from the differential band and from the adjacent lower and upper regions, respectively. Aliquots of these preparations were inoculated onto blocks of four chrysanthemum plants and the number of them that developed symptoms is indicated by the numerator of the fractions shown in lowest part of the figure. Lane 6, two DNA markers of 394 and 387 nt used to delimit approximately the gel region containing the differential band with which the infectivity was associated. (B) Northern blot hybridizations of chrysanthemum nucleic acids after denaturing PAGE, using 82-nt RNA probes for plus (lanes 1 and 2) and minus (lanes 3 and 4) CChMVd sequences. Lanes 1 and 3, nucleic acids from uninfected leaves. Lanes 2 and 4, nucleic acids from CChMVd-infected leaves.
Nucleotide Sequence and Proposed Secondary Structure of the CChM RNA.
The eluted linear RNA inducing the CChM disease was reversely transcribed using random hexamers and cloned. As described in Materials and Methods, a cDNA clone of 82 nt specific for this RNA was obtained. When RNAs from CChM-affected plants were separated by denaturing PAGE and analyzed by Northern blot hybridization with two probes of opposite polarities derived from the specific clone of 82 nt, two signals were detected with both probes (Fig. 2B). One was generated by the linear CChM RNA used as a template and the other, considerably less intense, by a slow moving RNA that might be its circular counterpart. To confirm this hypothesis the presumed circular CChM RNA was eluted and amplified by reverse transcription–PCR using a pair of adjacent primers with opposing polarities (PI and PII), derived from the sequence of the 82-nt fragment: a product the same length as the CChM linear RNA was obtained (data not shown), which subsequently was cloned and sequenced.
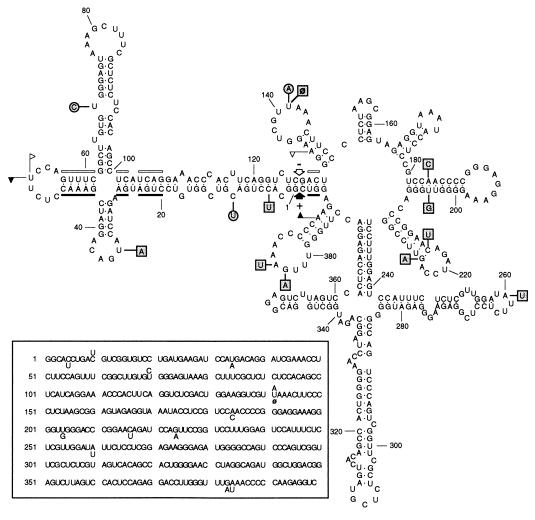
Fig. 3 (Inset) shows the primary structure of the CChM RNA, which is a circular molecule of 399 nt consisting of 112 G (28.1%), 109 C (27.3%), 87 A (21.8%), and 91 U (22.8%), therefore having a G+C content of 55.4%. Minor sequence heterogeneities were found in the indicated positions (Fig. 3, Inset). CChM RNA does not contain the characteristic core sequences of the central conserved region of several groups of non-self-cleaving viroids (2, 3). However, in both polarity strands the CChM RNA sequence does contain the conserved residues and the structural elements typical of the hammerhead structures found in the RNAs of ASBVd and PLMVd (19, 20), several satellite RNAs (21–24), the RNA form of a retroviroid-like element (25), and a transcript of the newt (26). This feature of the CChM RNA, together with the ability of the purified forms of this RNA to incite the CChM disease, and the absence of any detectable viral particles in CChM-affected plants (5), which could act as a helper virus, has led us to conclude that CChM RNA is a viroid: the chrysanthemum chlorotic mottle viroid (CChMVd).
Figure 3.
Predicted secondary structure of lowest free energy for the plus strand of the reference sequence variant of CChMVd. Plus and minus self-cleavage domains are delimited by flags, the 13 nucleotides conserved in most natural hammerhead structures are indicated by bars, and the self-cleavage sites are shown by arrows. Solid and open symbols refer to plus and minus polarities, respectively. Sequence heterogeneities found in two other variants, indicated within circles and squares, do not alter the proposed secondary structure. The pair of adjacent primers PI (antisense) and PII (sense) used for reverse transcription and PCR reactions cover the regions between positions 294–269 and 295–320, respectively. The same numbers are used in the minus and in the plus polarity. (Inset) Primary structure of the reference sequence variant of CChMVd (pCM5) with the polymorphism detected in the two other variants (pCM20 and pCM1) indicated below and above the reference sequence, respectively. ø indicates deletion.
The secondary structure of lowest free energy obtained for CChMVd is a branched conformation formed by a series of stem-loops with a free energy at 37°C of −516.6 kJ/mol. In the predicted secondary structure 68.7% of the residues are paired with a distribution of 55.5% G·C, 35% A·U, and 9.5% G·U bps. A close inspection of this secondary structure revealed that in the hairpins delimited by positions 181–207, 210–230, and 374–394 the stems are very stable, because they are long, are rich in G·C pairs, and have stretches of 3–4 consecutive pairs of this particular class. Moreover, the sequence polymorphism observed in the three variants does not affect the proposed secondary structure, and in particular the three aforementioned hairpins, either because the changes are found in loops or because when affecting a bp the substitutions are compensatory (Fig. 3). Therefore, it is likely that a least some portions of the computer-predicted structure could correspond to biologically significant domains.
Hammerhead Structures of CChMVd RNAs and Their Peculiarities.
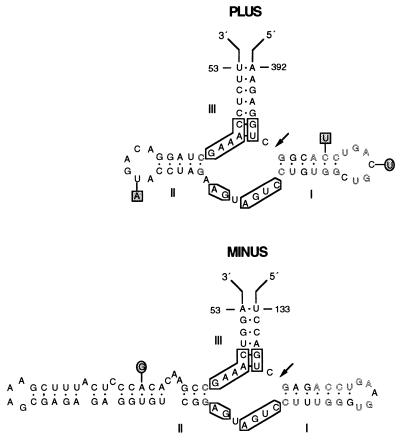
Fig. 4 shows that plus and minus strands of CChMVd can form hammerhead structures with the 11 conserved residues and the adjacent helices, which previously have been found around the in vitro self-cleavage sites of a group of small RNAs. Plus and minus hammerhead structures of CChMVd have stable helices III and short loops closing helices I and II (Fig. 4) (see ref. 27 for nomenclature), resembling the hammerhead structures of PLMVd more closely than those of ASBVd (19, 20). However, helix II of the minus CChMVd hammerhead structure is unusually long and composed of shorter helices separated by bulging residues. Moreover, when compared with other natural hammerhead structures, the plus CChMVd hammerhead structure has an extra A residue after the conserved A9, a situation previously reported only in the plus hammerhead structures of the satellites of the lucerne transient streak virus (28) and arabis mosaic virus (29), although in these cases the extra residue was a U.
Figure 4.
Hammerhead structures of plus and minus strands of the reference sequence variant of CChMVd. Arrows indicate the predicted self-cleavage sites. The conserved nucleotides present in all natural hammerhead structures (with the exception of those of plus satellite of barley yellow dwarf virus RNA and minus strand of a viroid-like RNA found recently in cherry, in which only 11 nucleotides are conserved) are boxed. Outlined fonts denote sequence identity between the CChMVd hammerhead structures. Sequence heterogeneities found in two other variants are indicated within circles and squares and do not alter the proposed hammerhead structures.
A cytidylate residue precedes the predicted self-cleavage sites of the CChMVd hammerhead structures, as in most other known hammerhead structures, and the residue between the conserved sequences CUGA and GA is a U in both CChMVd hammerhead structures, which also conforms to the situations observed in most natural hammerhead structures in which this residue is U, C, and exceptionally A. The domain formed by helix I and loop 1 shows extensive sequence identity between both CChMVd hammerhead structures (Fig. 4). Helices III of the plus hammerhead structures of CChMVd and PLMVd also share sequence similarities.
In Vitro and in Vivo Self-Cleavage of CChMVd RNAs.
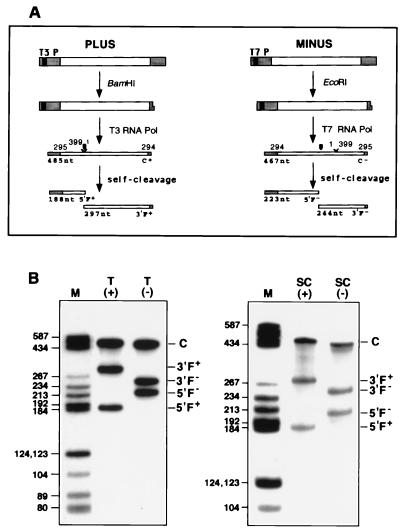
Monomeric plus and minus CChMVd RNAs transcribed in vitro from linearized plasmids with full-length CChMVd cDNA inserts, self-cleaved efficiently during transcription, and generated fragments with the sizes predicted by hammerhead structures (Fig. 5). The exact self-cleavage sites were confirmed by reverse transcription of the 3′F+ and 3′F− fragments with PI and PII primers, respectively, and comparison with the ladders obtained by sequencing the corresponding monomeric CChMVd cDNA clone using the same primers (data not shown). The extent of self-cleavage of plus and minus monomeric strands of CChMVd during transcription was 51% and 52%, respectively. When both full-length transcripts were purified and incubated under standard self-cleavage conditions (17), 54% of the plus and 68% of the minus CChMVd RNAs self-cleaved.
Figure 5.
In vitro synthesis and self-cleavage of monomeric plus and minus RNAs of the reference sequence variant of CChMVd. (A) Diagrams of plus and minus DNA templates and of the products generated by transcription with T3 and T7 RNA polymerases, respectively, after restriction with the indicated enzymes. Hatched boxes, vector sequences; filled boxes, RNA polymerase promoters; empty boxes, CChMVd sequence. The complete transcripts are C+ and C− and the cleavage fragments 5′F+, 3′F+, 5′F− and 3′F−. Positions in the CChMVd sequence are shown above the products and their expected sizes in nucleotides below. Self-cleavage sites are indicated by arrowheads. (B) Analysis by PAGE and autoradiography of the transcription (T) and of the self-cleavage (SC) reactions of purified monomeric products. Lanes M, radioactive DNA markers with their sizes in nucleotides indicated at left. The electrophoresis of the left panel was run for a shorter time than that of the right panel.
Evidence also was obtained to support the in vivo functioning of the CChMVd plus hammerhead structure. For this purpose the cDNA, obtained by reverse transcription of the linear CChMVd plus RNA with primer PI, was polyadenylated with terminal transferase, PCR-amplified with primers PI and a second one with a 3′ poly(T) tail, and cloned. Sequencing of the inserts revealed that the 5′ terminus of the linear CChMVd RNA was identical to that produced in the in vitro self-cleavage reaction (data not shown).
Infectivity of in Vitro Transcripts of CChMVd.
To confirm that the CChMVd RNA identified here was the causal agent of CChM disease, chrysanthemum plants were inoculated with the monomeric plus CChMVd RNA generated by self-cleavage in in vitro transcriptions from a plasmid with a dimeric head-to-tail CChMVd cDNA insert of the reference sequence variant. In two independent experiments the four inoculated plants became infected and developed the symptoms of the CChM disease concurrently with a set of control plants inoculated with a nucleic acid preparation from CChMVd-infected tissue. Northern blot hybridizations revealed that the onset of symptoms was accompanied by the appearance of the circular and linear monomeric CChMVd forms. The recombinant plasmid with the dimeric tandem repeat of CChMVd cDNA also was infectious and, although to a lesser extent, so was the monomeric CChMVd ds-DNA obtained by PCR amplification with the phosphorylated primers PI and PII.
Unique Properties of CChMVd and PLMVd in High Salt Conditions.
Using the infectivity bioassay as a semiquantitative test, experiments reported previously (7) and confirmed in the present work (see above) have shown that most of the inciting agent of the CChM disease is insoluble in 2 M LiCl. This is an uncommon situation when compared with the behavior of typical viroids as potato spindle tuber viroid, CEVd, and CSVd, which remain essentially in the soluble fraction under the same conditions. To address this question more precisely, the solubility of a series of viroids in 2 M LiCl was monitored by Northern blot hybridization instead of by bioassay. To provide a uniform environment and discard the effects of nonspecific precipitations, we added circular forms of different viroids purified by two consecutive PAGE steps (13) to aliquots of a nucleic acid preparation from CChMVd-infected plants obtained by phenol extraction and chromatography on nonionic cellulose with a buffer containing 35% ethanol (10). The mixtures then were fractionated with 2 M LiCl, and the presence of each viroid in the soluble and insoluble fractions was examined by Northern blot hybridization using radioactive full-length RNA probes with the complementary sequences. Table 1 shows that the viroids lacking self-cleavage and ASBVd were predominantly found in the soluble fraction, whereas CChMVd and PLMVd were mostly detected in the precipitate.
DISCUSSION
By a combination of two consecutive PAGE steps, the first under nondenaturing conditions and the second in long denaturing gels, we have identified an RNA that induces the typical symptoms of the CChM disease when inoculated to chrysanthemum plants. These results, together with the lack of virus-like particles in the infected tissue (5), show that this RNA is a viroid, CChMVd. The very low levels at which the linear, and particularly the circular, forms of CChMVd accumulate, explain why previous attempts to identify this pathogenic RNA failed. Cloning and sequencing CChMVd revealed a size of 398–399 nt, the largest viroid characterized so far, excluding those with sequence duplications. Both polarity strands of CChMVd can adopt the hammerhead structures that have been found in all viroid-like plant satellite RNAs (21–24) and in the RNA form of a retroviroid-like element from carnation (25), but only in two of the 26 sequenced viroids, ASBVd and PLMVd (19, 20).
In terms of their stability and overall architecture, CChMVd hammerhead structures resemble those of PLMVd (20) but differ from those of ASBVd (19). The efficient in vitro self-cleavage of plus and minus monomeric CChMVd RNAs during transcription and after purification indicates that these reactions most probably occur through single-hammerhead structures as in PLMVd (20, 30), rather than through double-hammerhead structures like those proposed in ASBVd (31). Two findings support an in vivo role for at least the plus CChMVd hammerhead ribozyme: (i) the 5′ terminus of the plus linear strand of CChMVd is identical to that generated in the in vitro self-cleavage reaction, and (ii) the changes detected in the three CChMVd variants in the regions of both hammerhead structures do not affect their stabilities (Fig. 4). The hammerhead structures of CChMVd are more closely related to each other (Fig. 4) than to any other hammerhead structures. This is also the case for the hammerhead structures of PLMVd (20), satellite of the lucerne transient streak virus (28), and a viroid-like RNA, presumably of a satellite nature, recently characterized in cherry (32). However, the hammerhead structures of CChMVd, in contrast to those of PLMVd, satellite of the lucerne transient streak virus and the cherry viroid-like RNA, are similar only in the helix I-loop I region, whereas the rest of the self-cleaving domains are clearly different, in particular the helix II-loop II region. As suggested previously the intramolecular similarities might result from template switching by an RNA polymerase (28) and lead to very stable foldings in this region in the secondary structures of lowest free energy predicted for CChMVd plus (Fig. 3) and minus RNAs (data not shown). These foldings, in which the residues flanking the scissile bonds are in close proximity as a result of forming part of helices, could facilitate ligation as opposed to the hammerhead structures promoting self-cleavage.
The plus hammerhead structure of CChMVd is peculiar in having an A inserted between the conserved A9 and the quasi-conserved G10.1 or, alternatively, between the conserved G8 and A9 (Fig. 4). This A can be regarded as an extra residue because A9 and G10.1 are contiguous in all natural hammerhead structures with the exception of those of plus satellites of the lucerne transient streak virus and arabis mosaic virus, which contain an extra U in this position (28, 29). Although the extra A is compatible with an extensive in vitro self-cleavage (Fig. 5), it can be suspected of having some minor negative effects on this reaction because it is absent in most natural hammerhead structures. Moreover, because x-ray crystallography analysis of two hammerhead ribozymes has revealed that three non-Watson–Crick pairs involving A9 and G12, G8 and A13, and U7 and A14, (33, 34) are adjacent to the bp G10.1 C11.1, the extra A could either be accommodated as a bulging residue or to induce a rearrangement of this region of the ribozyme. One can speculate that the extra A, which holds a special position connecting two helices in the secondary structure of lowest free energy (Fig. 3), might be involved in some functional property of CChMVd other than self-cleavage and be preserved on this basis. Viroids are systems in which the genetic information is very compressed due to their small size, and therefore the implication of specific regions of the molecule in more than one function can be presumed.
The most stable secondary structure predicted for CChMVd is a clearly branched conformation that does not fulfill the rod-like or quasi-rod-like model proposed for the in vitro structure of most viroids. The only exception is PLMVd, whose predicted secondary structure is also branched and contains a left stem in which, like CChMVd, the conserved residues of both hammerhead structures and their corresponding self-cleavage sites are opposite each other (20). Moreover, in some PLMVd variants the sequences forming this left stem adopt a cruciform structure similar to that of CChMVd (S. Ambrós & R.F., unpublished data). Interestingly enough, when the solubility of a series of representative viroids in 2 M LiCl was investigated, CChMVd and PLMVd were found in the precipitate, whereas the others remained in the soluble fraction (Table 1). Because the solubility of similarly sized RNAs in high salt concentrations is presumably governed by conformation rather than by sequence, our results show that the tridimensional foldings of CChMVd and PLMVd are different from those of other viroids. The secondary structures of lowest free energy provide a likely explanation for this different behavior: viroids having a compact rod-like or quasi-rod-like structure, including ASBVd, are soluble in 2 M LiCl, whereas viroids with a branched conformation are insoluble under the same conditions. However, other explanations cannot be excluded: for example, CChMVd and PLMVd might be induced to adopt alternative conformations in 2 M LiCl, which, upon interacting with this highly ionic milieu, would be the ultimate cause of the insolubility of the two viroids. On the other hand, the high instability of CChMVd in crude extracts, when compared with CSVd (4), is also consistent with a more relaxed conformation and consequently more accessible to RNase digestion. On the other hand, our data also indicate that when a fractionation step with 2 M LiCl is used for isolating an unknown viroid, both fractions must be checked for the presence of such an RNA.
Chrysanthemum is the only known host for CChMVd (6), and in this respect it resembles PLMVd and ASBVd, which also have very restricted host ranges. Chrysanthemum is the only known herbaceous host of a self-cleaving viroid, and because the time elapsing between inoculation and onset of symptoms is short, this system is amenable for further site-directed mutagenesis studies aimed at mapping functional determinants in CChMVd.
The group of the self-cleaving viroids is presently formed by ASBVd, PLMVd, and CChMVd. Although no extensive sequence similarities exist between them, PLMVd and CChMVd are more closely related on the basis of their G+C content, predicted secondary structures of lowest free energy and morphology of their hammerhead structures, as well as on a physico-chemical basis such as their insolubility in 2 M LiCl. Because PLMVd is the type member of this subgroup we propose the name pelamoviroids for it, conforming to previous rules (35). In contrast to some typical non-self-cleaving viroids that are localized in the nucleus (36, 37), ASBVd accumulates mainly in the chloroplast (38, 39). If future work demonstrates that CChMVd and PLMVd are also chloroplastic, this will establish a cardinal difference between self-cleaving and non-self-cleaving viroids with major implications for their evolutive origin and replication.
Table 1.
Solubility in 2 M LiCl of different viroids
| Viroid | Fraction (%) in the supernatant* |
|---|---|
| ASSVd | 82 |
| CEVd | 98 |
| CSVd | 98 |
| CbVd1 | 94 |
| PBCVd | 93 |
| ASBVd | 98 |
| PLMVd | 4 |
| CChMVd | 2 |
ASSVd, apple scar skin viroid; CbVd1, coleus viroid 1; PBCVd, pear blister canker viroid.
Determined by scanning of Northern blot hybridization signals with a PhosphorImaging reader.
Acknowledgments
We thank Profs. R. K. Horst and P. Palukaitis for providing CChMVd inocula, A. Ahuir for excellent technical assistance, Dr. M. J. Rodrigo for help with computer programs, Drs. V. Pallás, C. Hernández and F. Di Serio for critical reading of the manuscript, and D. Donelland for English revision. This work was supported by Grants PB92–0038 and PB95–0139 from the Dirección General de Investigación Científica y Técnica de España and by contract CHRX-CT94–0635 from the European Commission (to R.F.). B.N. was a recipient of a predoctoral fellowship from the Ministerio de Educación y Ciencia de España.
ABBREVIATIONS
- CChM
chrysanthemum chlorotic mottle
- CChMVd
chrysanthemum chlorotic mottle viroid
- PLMVd
peach latent mosaic viroid
- ASBVd
avocado sunblotch viroid
- CSVd
chrysanthemum stunt viroid
- CEVd
citrus exocortis viroid
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the EMBL database (accession no. Y14700).
References
- 1.Diener T O. Virology. 1971;45:411–428. doi: 10.1016/0042-6822(71)90342-4. [DOI] [PubMed] [Google Scholar]
- 2.Diener T O. FASEB J. 1991;5:2808–2813. doi: 10.1096/fasebj.5.13.1717335. [DOI] [PubMed] [Google Scholar]
- 3.Flores R, Di Serio F, Hernández C. Semin Virol. 1997;8:65–73. [Google Scholar]
- 4.Romaine C P, Horst R K. Virology. 1975;64:86–95. doi: 10.1016/0042-6822(75)90081-1. [DOI] [PubMed] [Google Scholar]
- 5.Dimock A W, Geissinger C M, Horst R K. Phytopathology. 1971;61:415–419. [Google Scholar]
- 6.Horst R K. In: The Viroids. Diener T O, editor; Diener T O, editor. New York: Plenum; 1975. pp. 291–295. [Google Scholar]
- 7.Kawamoto S O, Horst R K, Wong S M. Acta Horticult. 1985;164:333–340. [Google Scholar]
- 8.Niblett C L, Dickson E, Fernow K H, Horst R K, Zaitlin M. Virology. 1978;91:198–203. doi: 10.1016/0042-6822(78)90368-9. [DOI] [PubMed] [Google Scholar]
- 9.Palukaitis P F, Kurath G, Boccardo G. In: Viroids and Satellites: Molecular Parasites at the Frontier of Life. Maramorosch K, editor; Maramorosch K, editor. Boca Raton: CRC; 1991. pp. 59–77. [Google Scholar]
- 10.Pallás V, Navarro A, Flores R. J Gen Virol. 1987;68:2095–2102. [Google Scholar]
- 11.Bellamy A R, Ralph R K. Methods Enzymol. 1968;XII:156–160. [Google Scholar]
- 12.Sänger H L, Ramm K, Domdey H, Gross H J, Henco K, Riesner D. FEBS Lett. 1979;99:117–122. [Google Scholar]
- 13.Flores R, Durán-Vila N, Pallás V, Semancik J S. J Gen Virol. 1985;66:2095–2102. [Google Scholar]
- 14.Gubler U, Hoffman B J. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daròs J A, Flores R. RNA. 1995;1:734–744. [PMC free article] [PubMed] [Google Scholar]
- 17.Forster A C, Davies C, Hutchins C J, Symons R H. Methods Enzymol. 1990;181:583–607. doi: 10.1016/0076-6879(90)81153-l. [DOI] [PubMed] [Google Scholar]
- 18.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 19.Hutchins C J, Rathjen P D, Forster A C, Symons R H. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernández C, Flores R. Proc Natl Acad Sci USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prody G A, Bakos J T, Buzayan J M, Schneider I R, Bruening G. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 22.Bruening G. Methods Enzymol. 1989;180:546–558. doi: 10.1016/0076-6879(89)80123-5. [DOI] [PubMed] [Google Scholar]
- 23.Symons R H. Trends Biochem Sci. 1989;14:445–450. doi: 10.1016/0968-0004(89)90103-5. [DOI] [PubMed] [Google Scholar]
- 24.Miller W A, Hercus T, Waterhouse P M, Gerlach W L. Virology. 1991;183:711–720. doi: 10.1016/0042-6822(91)91000-7. [DOI] [PubMed] [Google Scholar]
- 25.Daròs J A, Flores R. Proc Natl Acad Sci USA. 1995;92:6856–6860. doi: 10.1073/pnas.92.15.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Epstein L M, Gall J G. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- 27.Hertel K J, Pardi A, Uhlenbeck O K, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach W L, Hodgson R, Symons R H. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forster A C, Symons R H. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- 29.Kaper J M, Tousignant M E, Steger G. Biochem Biophys Res Comm. 1988;154:318–325. doi: 10.1016/0006-291x(88)90687-0. [DOI] [PubMed] [Google Scholar]
- 30.Beaudry D, Bussière F, Lareau F, Lessard C, Perreault J-P. Nucleic Acids Res. 1995;23:745–752. doi: 10.1093/nar/23.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forster A C, Davies C, Sheldon C C, Jeffries A C, Symons R H. Nature (London) 1988;334:265–267. doi: 10.1038/334265a0. [DOI] [PubMed] [Google Scholar]
- 32.Di Serio F, Daròs J A, Ragozzino A, Flores R. J Virol. 1997;74:6603–6610. doi: 10.1128/jvi.71.9.6603-6610.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 34.Scott W G, Finch J T, Klug A. Cell. 1994;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 35.Elena S F, Dopazo J, Flores R, Diener T O, Moya A. Proc Natl Acad Sci USA. 1991;88:5631–5634. doi: 10.1073/pnas.88.13.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harders J, Lukacs N, Robert-Nicoud M, Jovin J M, Riesner D. EMBO J. 1989;8:3941–3949. doi: 10.1002/j.1460-2075.1989.tb08577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonfiglioli R G, Webb D R, Symons R H. Plant J. 1996;9:457–465. [Google Scholar]
- 38.Bonfiglioli R G, McFadden G I, Symons R H. Plant J. 1994;6:99–103. [Google Scholar]
- 39.Lima M I, Fonseca M E N, Flores R, Kitajima E W. Arch Virol. 1994;138:385–390. doi: 10.1007/BF01379142. [DOI] [PubMed] [Google Scholar]