Abstract
Aged individuals suffer from multiple dysfunctions during skeletal muscle atrophy. The purpose of this study was to determine differential changes in gene expression in atrophied soleus muscle induced by hindlimb immobilization in young (3–4 months) and old (30–31 months) rats. The hypothesis was that differentially expressed mRNAs with age-atrophy interactions would reveal candidates that induce loss of function responses in aged animals. Each muscle was applied to an independent set of Affymetrix micoarrays, with 385 differentially expressed mRNAs with atrophy and 354 age-atrophy interactions detected by two-factor ANOVA (α of 0.05 with a Bonferroni adjustment). Functional trends were observed for 23 and 15 probe sets involved in electron transport and the extracellular matrix, respectively, decreasing more in the young than in the old. Other functional categories with atrophy in both ages included chaperones, glutathione-S-transferases, the tricarboxylic acid cycle, reductions in Z-line-associated proteins and increases in probe sets for protein degradation. Surprisingly, myosin heavy chain IIb and IIx mRNAs were suppressed in the atrophied soleus muscle of old rats as opposed to the large increases in the young animals (16- and 25-fold, respectively, with microarrays, and 61- and 68-fold, respectively, with real-time PCR). No significant changes were observed in myosin heavy chain IIb and IIx mRNA with micoarrays in the atrophied soleus muscles of old rats, but they were found to increase six- and fivefold, respectively, with real-time PCR. Therefore, deficiencies in pre-translational signals that normally upregulate myosin heavy chain IIb and IIx mRNAs during atrophy may exist in the soleus muscle of old animals.
In response to changes in contractile activity, skeletal muscle is extremely plastic (Booth & Baldwin, 1996), for example half its mass may be lost in 1 month when immobilized in a shortened position (Booth, 1977). The clinical outcome of atrophy is a weaker muscle that requires rehabilitation to regain mass and strength (Paulos et al. 1991). Adherence to physical activity interventions is low (van der Bij et al. 2002), which is particularly harmful in older populations where sarcopenic (age-induced wasting) muscles may set up a negative cycle of weakness causing loss of function and inactivity, which results in additional atrophy and weakness (Buchner & Wagner, 1992; Walston & Fried, 1999). Although the skeletal muscles of young and old rats show a comparable magnitude of atrophy (27–37 %) when limbs are immobilized, muscles from older animal limbs fail to regrow in conditions of normal cage activity, suggesting that differences in gene expression exist between the young and old, even though their losses in mass from immobilization do not (Chakravarthy et al. 2000; Zarzhevsky et al. 2001). One set of mRNAs expected to change with immobilization were myosin heavy chains (MHCs) IIb and IIx. MHC IIb and IIx mRNAs were previously shown to increase 2.6- and 24-fold, respectively, in the soleus muscle of 11-week-old rats after 1 week of hindlimb immobilization (Jänkälä et al. 1997), and thus could serve as positive controls for immobilization-induced changes in gene expression. The hypothesis of the current study was that multiple mRNAs would be expressed differentially between young and old rats after 10 days of immobilization-induced muscle atrophy, where mRNAs with age-atrophy interactions would reveal candidates that induce loss of function responses in aged animals.
To test the hypothesis, RGU34 A-C Affymetrix microarrays, containing ≈ 24 000 genes and expressed sequence tags (ESTs), were employed, which allowed a global, unbiased determination of mRNA expression in the soleus muscle of hindlimb-immobilized rats. Two previous reports used arrays to determine gene expression during the soleus muscle atrophy produced by 21 and 35 days of rat hindlimb suspension, respectively (Stein et al. 2002; Wittwer et al. 2002). However, fewer genes (8000 (Stein et al. 2002) and 1200 (Wittwer et al. 2002)) were assayed in the suspension experiments. In addition, the paper by Stein et al. (2002) failed to adjust P-values for multiple t tests so that an undefined number of false positives may be included in their list of significant differences. Finally, while hindlimb suspension and immobilization both remove mechanical loading, only immobilization removes isotonic contractions. For example, the EMG activity of the suspended and immobilized soleus muscles are reduced by 19 % (Alford et al. 1987) and 85–90 % (Fischback & Robbins, 1969; Hnik et al. 1985), respectively, indicating that differences in muscle activity exist between the two models of unloading. Thus, differential responses of some genes between the suspension and immobilization models of unloading atrophy are likely to differ. Knowledge of novel differences in muscle mRNA expression during limb immobilization with age would allow the development of more novel, scientifically based therapies to prevent immobilization-induced atrophy in the muscle of older individuals.
Methods
Animals
Fischer 344x Brown Norway Fl male rats were obtained from the National Institute of Aging (Harlan Labs, Indianapolis, IN, USA). Rats were sacrificed at the ages of 3–4 months (young, n = 10) and 30–31 months (old, n = 10). The rats were fed regular rat chow and water ad libitum, were housed 2–3 per cage, in filter-topped guinea-pig cages with rubber mat bottoms, and maintained on a 12:12 h light:dark cycle. While the atrophy data being presented have not been published previously, the data from the control soleus muscles described here were incorporated as part of a larger aging experiment in a previous publication (Pattison et al. 2003), which found 682 differentially expressed probe sets between young and old rats. The data was split into multiple publications due to its large size and the multiple interactions being studied.
Hindlimb immobilization
Prior to casting, animals were anaesthetized with an intraperitoneal injection of a cocktail containing ketamine (49 mg ml−1), xylazine (6.2 mg ml−1) and acepromazine (2.0 mg ml−1) at a concentration of 0.10 ml (100 mg body weight)−1. Once anaesthetized, both hindlimbs were fixed from the waist down in the plantar-flexed position with wire-mesh-reinforced plaster casts. Animals were maintained in the immobilized condition for 10 days. Four experimental groups were formed (young control, young immobilized, old control and old immobilized; the effects of aging between young and old controls have been presented in a previous publication; Pattison et al. 2003). Following 10 days of immobilization, rats were anaesthetized with 0.123 ml (100 mg body weight)−1 of ketamine/xylazine/acepromazine cocktail prior to muscle excision and exsanguination to produce death.
Sample processing
Soleus muscles were extracted, weighed, snap-frozen in liquid nitrogen and then powdered using a mortar and pestle cooled by liquid nitrogen. Both soleus muscles from a single rat formed a single observation, where soleus muscle RNA from a single rat was applied to an individual array. Total RNA was extracted from an aliquot of muscle powder put directly into a TRIzol solution (Invitrogen, Carlsbad, CA, USA) and homogenized on ice using a Polytron homogenizer (Kinematica, Lucerne, Switzerland) on setting 7 for three pulses, 15 s per pulse. Total RNA was purified using RNeasy columns (Qiagen, Valencia, CA, USA). Methods for sample preparation are described in detail in the Affymetrix Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA, USA) and are described only briefly here. Synthesis of cDNA was carried out using 10 µg of purified total RNA with a T7-(dT)24 primer (100 pmol µl−1), using components of the Superscript Choice kit (Invitrogen). All incubations were performed in a Mastercycler Gradient thermocycler (Eppendorf, Westbury, NY, USA). The resulting double-stranded cDNA was quantified using a PicoGreen kit (Molecular Probes, Eugene, OR, USA). A 2 µg aliquot of cDNA was used in the in vitro transcription reaction, utilizing biotinylated nucleotides from bioarray high-yield RNA transcript labelling kits (Enzo Diagnostics, Farmingdale, NY, USA). The resulting cRNA was further purified using RNeasy columns (Qiagen). The purified biotinylated cRNA was then fragmented and hybridized to Affymetrix rat genome U34A, B and C arrays and analysed with fluorescence intensity scanning according to Affymetrix protocols (Affymetrix Expression Analysis Technical Manual). All hybridization and scanning of the arrays was performed in the University of Missouri DNA Core Facility (Columbia, MO, USA).
GeneChip analysis
The rat genome U34 array set, which assays the relative abundance of ≈24 000 genes and EST clusters (based on UniGene Build 34), contained 26 388 probe sets. Each probe set consisted of 16 perfectly matched (complementary) 25-mers, corresponding to different regions along the length of an mRNA. Similarly, 16 mismatched pairs (containing a single mutated base) that do not perfectly complement a transcript's sequence were used as a measure of non-specific background binding. Microarray Suite 5.0 software (Affymetrix) was employed to determine significant differences between the perfectly matched and mismatched probe pairs, based on their fluorescence intensity. The software utilized a one-sided Wilcoxon's signed-rank test to calculate a P value reflecting the significance of differences between pairs. The resulting P-values were used as a qualitative measure of the ability to detect a given transcript, where values of P < 0.04 were called ‘Present’, those between 0.04 and 0.06 were called ‘Marginal’ and those of P > 0.06 were called ‘Absent’. Only those probe sets that were ‘Present’ or ‘Marginal’ in ≥ 60 % of the samples for an experimental group were carried on for statistical analyses: 12 485 probe sets were sufficiently detected in at least one of the experimental groups (i.e. young control, young immobilized, old control or old immobilized soleus muscles). Microarray Suite 5.0 software (Affymetrix) uses statistically based algorithms to determine transcript abundance based on the mean fluorescence intensity across the probe set (termed ‘Signal’). The Signal for each probe set was calculated as the one-step biweight estimate of the combined differences of all of the probe pairs in the probe set. The Signal value was used for all subsequent statistical analyses. Fold changes were calculated as the result of the mean immobilized Signal intensity/mean control Signal intensity (fold increase in atrophy) or mean control Signal intensity/mean immobilized Signal intensity (fold decrease with atrophy). Microarray analyses have been criticized as being ‘quite elusive about measurement reproducibility’ (Claverie, 1999). However, Bakay et al. (2002) have reported that experimental error among Affymetrix microarrays is not a significant source of unwanted variability in expression profiling experiments (R2 = 0.979). In the current experiment, duplicate arrays were also found to have small, unwanted inter-chip variability (R2 = 0.981). In addition, a recent publication concerning a single human patient, in which RNA was prepared from two distinct breast tumours and placed on duplicate U95A GeneChips (four chips in total), found a very low degree of experimental variability between microarrays (R2 = 0.995), and between the two tumours (R2 = 0.987; Unger et al. 2001).
Real-time PCR
Total RNA samples were DNase-treated with an RNase-free DNAse kit (Qiagen). The samples were then purified with an RNeasy column (Qiagen). The lack of DNA contamination was validated by PCR and real-time PCR. Samples were analysed in an ABI 7000 (Applied Biosystems, ABI) after a two-step real-time-PCR reaction. Total RNA was reverse transcribed by two different methods. First, 2 µg of total RNA was reverse transcribed using random hexamers for single-stranded cDNA synthesis (ABI). Alternatively, total RNA was reverse transcribed in double-stranded cDNA during previous cDNA synthesis for array sample processing with a T7-oligo(dT)24 primer. The resulting double-stranded cDNA was quantified using a PicoGreen kit (Molecular Probes). Complementary DNA from both preparations was then amplified by PCR in a reaction consisting of: 25 ng of each cDNA sample (run in duplicate) in a mix of 900 nm primers, 250 nm minor groove binder probe and Taqman Universal PCR Master Mix (ABI). Any samples that contained a range of > 0.2 cycle time (CT), were re-assayed. The primer/probe sequences are shown in Table 1 in Supplementary Material 1, available online at: DOI: 10.1113/jphysiol.2003.047233.
Table 1.
Functional categorization of atrophy main effects for selected probe sets identified as significant after Bonferroni adjustment
| Gene name | GenBank accession no. | Raw P value | Fold change |
|---|---|---|---|
| Amino acid metabolism | |||
| Ornithine decarboxylase 1 | J04791 | 6.400E-10 | −2.95 |
| X07944 | 4.887E-09 | −2.16 | |
| J04792 | 4.190E-08 | −2.03 | |
| Glutamic–oxaloacetic transaminase 2, mitochondrial(aspartate aminotransferase 2) | M18467 | 3.097E-07 | −1.60 |
| ESTs similar to mouse glutamate–cysteine ligase, modifier subunit | AI008385 | 9.011E-08 | −1.88 |
| Branched-chain alpha-ketoacid dehydrogenase subunit El alpha | J02827 | 2.757E-06 | 1.65 |
| Cytosolic cysteine dioxygenase 1 | AA942685 | 3.696E-06 | 1.94 |
| Chaperone | |||
| ESTs similar to mouse McKusick-Kaufman syndrome protein | AI104266 | 1.786E-06 | −1.69 |
| ESTs similar to mouse DnaJ (Hsp40) homologue, subfamily A, member 4 | AA799570 | 3.570E-09 | −3.01 |
| AA848268 | 3.653E-09 | −3.13 | |
| ESTs similar to mouse heat shock protein, 105 kDa | AI236601 | 1.233E-08 | −2.80 |
| Heat shock protein 60 (liver) | X54793 | 3.466E-08 | −1.76 |
| ESTs similar to human DnaJ (Hsp40) homologue, subfamily B, member 4 | AI008643 | 4.930E-08 | −2.46 |
| Heat shock 70 kDa protein 5 (glucose-regulated protein precursor (GRP 78)/ immunoglobulin heavy chain binding protein) | S63521 | 1.433E-07 | −2.43 |
| Testis-specific heat shock protein-related gene hst70 | X15705 | 1.930E-07 | −3.00 |
| Heat shock 70 kDa protein 4 | AF077354 | 6.643E-07 | −1.77 |
| Heat shock 10 kDa protein 1 | AI170613 | 2.102E-06 | −1.41 |
| Glutathione transferase | |||
| Glutathione S-transferase, mu 5 | U86635 | 7.809E-11 | 2.53 |
| Glutathione-S-transferase, mu type 2 (Yb2) | X04229 | 8.867E-10 | 2.03 |
| J03914 | 2.279E-09 | 3.82 | |
| J02592 | 6.327E-09 | 3.92 | |
| J02810 | 2.987E-08 | 1.54 | |
| Glutathione S-transferase, mu 5 | U86635 | 1.990E-08 | 2.04 |
| Glutathione S-transferase, mu type 3 (Yb3) | E01415 | 1.059E-07 | 1.86 |
| Microsomal glutathione S-transferase 1 | J03752 | 2.269E-06 | 1.91 |
| ESTs similar to human glutathione S-transferase theta 1 | AA819192 | 3.090E-06 | −1.43 |
| Protein degradation | |||
| ESTs similar to mouse autophagy 12-like (S. cerevisiae) | AI178979 | 1.320E-06 | 1.24 |
| For proteasomal ATPase (SUG1) (also known as protease (prosome, macropain) 26S subunit, ATPase 5) | AB000491 | 9.077E-07 | 1.77 |
| ESTs similar to mouse proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | AI172107 | 1.168E-06 | 1.46 |
| Proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 | AI175576 | 1.886E-06 | 2.00 |
| Proteasome (prosome, macropain) subunit, alpha type 7 | AI179950 | 2.441E-06 | 1.41 |
| ESTs similar to mouse proteasome (prosome, macropain) subunit, beta type 4 | AI172162 | 2.759E-06 | 1.73 |
| Cathepsin L | S85184 | 4.372E-11 | 4.66 |
| S85184 | 1.76E-07 | 2.11 | |
| Protein degradation continued Calpain 3 | J05121 | 2.215E-09 | −1.81 |
| AF052540 | 2.729E-09 | −1.85 | |
| AF061726 | 1.148E-07 | −1.95 | |
| Membrane-bound transcription factor protease, site 1 | AI101111 | 9.134E-09 | 1.64 |
| ESTs similar to mouse f-box and leucine-rich repeat protein 3a | AI145387 | 3.803E-09 | 2.10 |
| Ubiquitin C | D17296 | 7.469E-07 | 1.48 |
| TCA cycle | |||
| ESTs similar to mouse isocitrate dehydrogenase 3 (NAD+)beta | AI105469 | 5.725E-07 | −1.40 |
| Malate dehydrogenase-like enzyme | AF093773 | 1.239E-07 | −1.56 |
| ESTs weakly similar to mouse pyruvate dehydrogenase (lipoamide) beta | AI176117 | 2.037E-06 | −1.38 |
| ESTs highly similar to mouse isocitrate dehydrogenase 2 (NADP+), mitochondrial | AI172491 | 1.389E-11 | −2.24 |
| ESTs similar to human succinate dehydrogenase complex, subunit B, iron sulphur (Ip) | AI172320 | 2.166E-10 | −1.64 |
| Malate dehydrogenase mitochondrial | AI010480 | 2.723E-09 | −1.69 |
| ESTs similar to mouse succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | AA925554 | 1.268E-08 | −1.65 |
| AA925554 | 1.505E-07 | −1.87 | |
| Succinate–CoA ligase, GDP-forming, alpha subunit | J03621 | 6.303E-07 | −1.49 |
| Localized at or near Z-line | |||
| ESTs similar to human synaptopodin 2 (myopodin) | AI171209 | 1.918E-06 | −1.40 |
| ESTs similar to mouse cofilin 2, muscle | AI010742 | 3.842E-06 | −2.06 |
| Myozenin 2 (calsarcin-1) | AI104354 | 3.295E-07 | −1.56 |
| ESTs similar to mouse ankyrin repeat domain 2 (stretch-responsive muscle) | AI170842 | 1.452E-10 | −5.96 |
| ESTs similar to mouse LIM domain binding 3 (cypher) | AI175028 | 2.790E-07 | −1.89 |
| AA800216 | 9.527E-07 | −2.01 | |
| Cysteine-rich protein 3 | X81193 | 5.814E-07 | −3.09 |
| ESTs similar to mouse enigma homologue (R. norvegicus) | AA997341 | 7.486E-11 | −2.35 |
| ESTs similar to human enigma (LIM domain protein) | AI104117 | 4.209E-09 | −2.74 |
Under Raw P value, E-10 indicates × 10−10, E-09 indicates × 10−9, etc. The fold change, when positive, indicates that expression of a gene in soleus muscle from the immobilized groups increased compared to controls and, when negative, indicates that immobilized groups expressed the gene less than controls. A complete listing of probe sets can be found in Tables 2 and 3 in the Supplementary Material, available online at: DOI: 10.1113/jphysiol.2003.047233.
Only data from the oligo(dT)-primed cDNA are presented.
Data were analysed for relative changes in expression using the ΔΔCT method, according to User Bulletin no. 2 ABI PRISM 7700 Sequence Detection System. Relative efficiency plots were run to validate the use of the ΔΔCT method, where all slopes were < 0.1. All targets were normalized to p38. The p38 mRNA had an overall P value of 0.152. Finding an endogenous control gene to which to normalize targets required an exhaustive search of commonly used genes for normalization, as well as a couple taken from the microarray data itself. Microarray analyses showed that the most commonly used housekeeping genes changed significantly with age, atrophy and/or had an age-atrophy interaction, including: GAPDH, β-actin, β-glucuronidase, β-2-microglobulin, hypoxanthine guanine phosphoribosyltransferase1, phosphoglycerate kinase and transferrin receptor. Microarray data also suggested that β-tubulin (P = 0.064 age main effect) and cyclophilin A (P = 0.08 age main effect) were not ideal mRNAs to normalize to, although they were statistically non-significant for either an atrophy or an age effect. Furthermore, real-time PCR analysis performed on randomly primed total RNA showed that 18S and 28S changed significantly with atrophy (data not shown). In search of a potential normalization mRNA, we chose p38 because of its ability to be detected and failure to change in the microarray analyses. p38 mRNA, while not significantly different with microarray analysis for any of the treatments applied, surprisingly showed a significant change from real-time PCR analyses derived from randomly primed total RNA (data not shown). In response to the above findings, the hypothesis was tested that for the same RNA sample there might exist a difference between populations of reverse-transcribed cDNA with random priming (total RNA) and T7-oligo(dT)24-primer-amplified (polyA mRNA) RNA. The hypothesis was confirmed when p38 was found not to differ significantly (overall P = 0.152) when amplified from cDNA derived from an mRNA population with a T7-oligo(dT)24 primer. Thus, all real-time PCR data were obtained from cDNA amplified with a T7-oligo(dT)24 primer and normalized to p38 expression.
The differences in ΔCT values were analysed with a two-way ANOVA with P < 0.05 set as significant. Data are expressed as the calculated fold differences between different experimental groups (young control, young immobilized, old control and old immobilized).
Statistical methods
A 2 × 2 ANOVA (α = 0.05) was employed to compare the Signal values. The two factors were age (with levels of young and old) and atrophy (with levels of control and immobilized atrophy). Furthermore, a Bonferroni adjustment was used to determine statistical significance in order to correct for the multiple ANOVAs performed on the 12 485 probe sets that had been detected as Present (i.e. having sufficient hybridization as identified by the Affymetrix Microarray Suite 5.0 software). The microarray statistical analysis proceeded in two stages. In the first stage of analysis we looked at an overall model test to determine whether any of the factors had a significant effect. Genes for which the overall test was significant at a false discovery rate of < 0.001 were then considered at the second stage of analysis. At that point, the P-values for the main factor effects and interactions were examined. Those cases with a Bonferroni-adjusted P value of < 4.0 × 10−6 were considered significant. The Bonferroni adjustment minimizes the occurrence of false positives (Pattison et al. 2003). A 2 × 2 ANOVA was also employed to statistically compare differences in soleus mass with age and immobilization, where P < 0.05 was considered significant. For real-time PCR analysis, the ΔCT values were analysed with a 2 × 2 ANOVA with P < 0.05 set as significant for individual comparisons between different experimental groups (young control, young immobilized, old control and old immobilized).
Hierarchical clustering
Age-atrophy interactions were clustered hierarchically based on their relative Signal values. Prior to clustering, the Signal data were normalized for differences in abundance by setting the highest Signal intensity value in the four groups (young control, young atrophy, old control and old atrophy) to 100 %, the remaining groups were then set as a percentage of that figure. This normalization allowed similar graphical phenotypes to be clustered together. SAS version 8 (SAS Institute, Cary, NC, USA) software executed the clustering function on the normalized Signal values of all probe sets with significant age-atrophy interactions. One limitation of data clustering methods is that the clusters are based on sample data and, just like any statistical procedure, somewhat different values for mean responses and hence somewhat different clusters might be obtained if the whole experiment was to be repeated a second time. Also, clustering serves to model the sample data, where each clustering model will include some false positives (i.e. there will be some genes inappropriately clustered together due to the constraints of the model with true positives when they don't belong).
Database searching
The target sequences for the significantly different probe sets were analysed with nucleotide basic local alignment search tool (BLAST) analysis to identify known genes and to determine significant gene homologies with other species (http://www.ncbi.nlm.nih.gov/BLAST/). The target sequence is the region of a given gene or EST that was probed by the RG U34 arrays. Further information about a given sequence and its homologues/orthologues was obtained from the Locuslink, Homologene, OMIM, mouse genome, rat genome, NetAffx and Proteome databases (http://www.ncbi.nlm.nih.gov/LocusLink/, http://www.ncbi.nlm.nih.gov/HomoloGene/, http://www.ncbi.nlm.nih.gov/entrez/, http://www.informatics.jax.org/, http://rgd.mcw.edu/, https://www.affymetrix.com/analysis/query/interactive-query and https://www.incyte.com/proteome/databases, respectively). A gene's biological processes and molecular functions were determined based on the defined gene ontologies given in the aforementioned databases.
Results
A 10 day period of hindlimb immobilization decreased the wet weight of the soleus muscle by 37 % in young rats (mean ± s.e.m. 0.136 ± 0.004 g in control and 0.085 ± 0.003 g in immobilized; P < 0.001) and 28 % in old rats (0.174 ± 0.005 g in control and 0.125 ± 0.007 g in immobilized; P < 0.001). A two-way ANOVA on the soleus muscle mass data showed an age and an atrophy main effect, but no age-atrophy interaction (P = 0.86), indicating that young and old animals show comparable losses in muscle mass with 10 days of hindlimb immobilization-induced atrophy. Over the 10 days of immobilization, the young and old rats both experienced a reduction in body mass of 14 %, where young rats weighed 285.5 ± 5.8 g prior to casting and 244.9 ± 7.1 g following immobilization; while old rats weighed 531.1 ± 8.8 g before casting and 455.5 ± 14.1 g after immobilization.
Use of a two-way ANOVA on the Signal values of detectable probe sets yielded two sets of data. First, a set of mRNAs that had an immobilization-induced atrophy main effect that was similar regardless of age. A total of 385 probe sets were found to have been affected by atrophy; 203 increases and 182 decreases (Table 1; Supplementary Table 2). The second set of results was mRNAs whose expression showed an age-atrophy interaction, where the atrophy effect differed in magnitude or direction between young and old. A total of 354 probe sets demonstrated an age-atrophy interaction, where 159 increased and 215 decreased with atrophy in one or in both age groups, but at different magnitudes (Supplementary Table 3).
Table 2.
Microarray data versus real-time PCR data normalized to p38 mRNA expression
| YC vs YI | OC vs OI | |||||||
|---|---|---|---|---|---|---|---|---|
| Microarray | Real-time PCR | Microarray | Real-time PCR | |||||
| Gene name | Fold change | P value | Fold change | P value | Fold change | P value | Fold change | P value |
| MHCIIb | 16.08 | 3.19E-09 | 60.80 | <0.0001 | 2.37 | 3.35E-01 | 5.78 | 3.12E-02 |
| MHCIIx | 25.44 | 7.87E-10 | 68.45 | <0.0001 | 2.76 | 5.88E-04 | 5.44 | 5.00E-04 |
| Homer2 | −3.94 | 4.12E-11 | −9.53 | <0.0001 | −2.03 | 1.27E-06 | −1.95 | 2.53E-02 |
| −3.90 | 1.22E-09 | — | — | −1.73 | 1.60E-05 | — | — | |
| −7.22 | 1.65E-11 | — | — | −2.52 | 1.05E-06 | — | — | |
| −5.28 | 1.25E-12 | — | — | −2.44 | 1.84E-07 | — | — | |
| −4.94 | 1.18E-06 | — | — | −1.75 | 3.14E-02 | — | — | |
YC, young control; YI, young immobilized; OC, old control; OI, old immobilized. The fold change, when positive, indicates that expression of a gene in soleus muscle from the immobilized groups increased compared to controls and, when negative, indicates that immobilized groups expressed the gene less than controls. Under P value, E-10 indicates × 10−10, E-09 indicates × 10−9, etc. The five sets of results for Homer2 were obtained with different probe sets.
In order to detect similar patterns of gene expression for age-atrophy interactions, the data were clustered hierarchically (Supplementary Fig. 1). The four major age-atrophy interaction expression patterns are illustrated in Supplementary Fig. 1. Of the 354 probe sets with significant age-atrophy interactions, 185 showed a pattern where the absolute decrease with atrophy was greater in the soleus muscle of the young than in the old rats (Supplementary Fig. 2A). Conversely, only 21 probe sets showed a greater absolute decrease in the soleus muscle of old than young (Supplementary Fig. 2B). Thus, nine times as many probe sets decreased in the young compared to the old atrophy group. For those probe sets with significant age-atrophy interactions that increased with atrophy: 110 probe sets increased more in the soleus muscle of young than of old rats (Supplementary Fig. 2C), but only 20 increased more in old than young (Supplementary Fig. 2D and Tables 2 and 3). Thus, five times as many probe sets increased more in the young than in the old. The significance of the age differential response remains to be elucidated. A total of 18 probe sets exhibited patterns that did not conform to one of these classifications.
Figure 1. Failure of MHC IIb and IIx mRNAs to increase in the soleus muscle of old rats after 10 days of hindlimb immobilization.
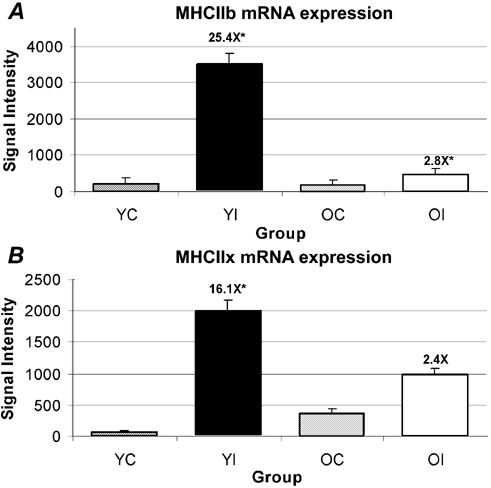
Failure of MHC IIb (A) and IIx (B) mRNAs to increase in the soleus muscle of old rats after 10 days of hindlimb immobilization (OI) when analysed by microarray analysis. Fold changes are shown above immobilized groups as the difference from the respective age-matched control. Values are the means ± s.e.m. of n = 5 per group. OI, old immobilized; OC, old control; YI, young immobilized; YC, young control. *Significant at P < 4.0 × 10−6.
Figure 2. An example of homer2 mRNA expression.
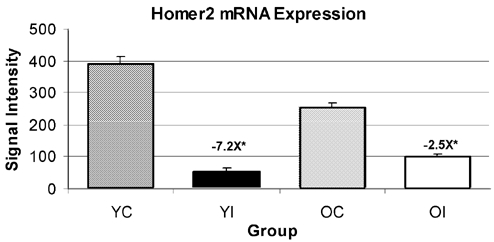
This example was selected from the five different probe sets for homer2 as detected from microarray analysis because this particular set was used for the design of primers and probes utilized in real-time PCR. The fold change is shown above the immobilized groups as the difference from respective age-matched controls. Values are the means ± s.e.m. of n = 5 per group. *Significant at P < 4.0 × 10−6.
Microarray analysis showed that MHC IIb and IIx transcripts were significantly increased by 25- and 16-fold, respectively, in the soleus muscle from immobilized limbs of young, but not old, rats (Fig. 1). The differential MHC IIb and IIx mRNA changes between young and old were confirmed by real-time PCR (Table 1). Real-time PCR analysis showed that MHC IIb transcripts were significantly increased 61- and sixfold in the soleus muscle from the immobilized young and old rats, respectively, as compared to their age-matched controls (Fig. 1A). With real-time analysis, MHC IIx transcripts were significantly increased by 68- and fivefold in the soleus muscle from the immobilized young and old rats, respectively, as compared to their age-matched controls (Fig. 1B). It should be noted that control values found for MHC IIx in real-time analysis were 5.14-fold higher in the old than in the young), which factors into the amplitude of the fold changes listed above (Supplementary Fig. 3). Although the control levels of MHC IIb and IIx mRNA increased with aging, as assessed with real-time PCR, the differences in MHC IIb and IIx mRNA with atrophy still showed that the amplitude of change in old was < 10 % of that of the young (Supplementary Fig. 3).
Figure 3. Hierarchical clustering and probe-set expression patterns.
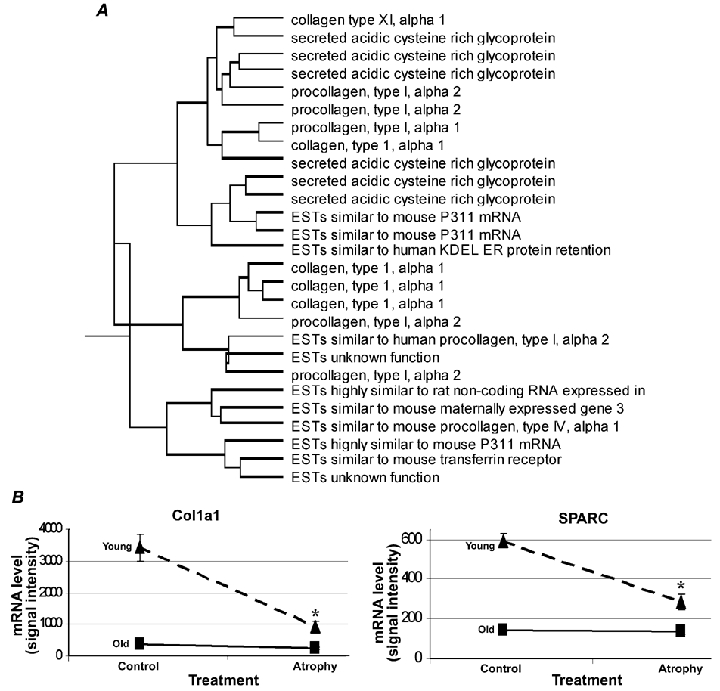
A, a cluster is shown that is composed predominantly of extracellular matrix genes obtained with hierarchical clustering analysis of expression profiles for age-atrophy interactions. B, representative examples of probe-set expression patterns from the shown hierarchical cluster of age-atrophy interactions.
Microarray analysis revealed that in soleus muscles from immobilized limbs, homer2 mRNA decreased in the range −3.9- to −7.2-fold (for five different probe sets) in the young and in the range −1.73- to −2.52-fold in the old rats as compared to age-matched controls (Table 2, illustrated in Fig. 2). The real-time PCR data showed a −9.5- and a −2-fold decrease for homer2 mRNA in the immobilized soleus muscles of young and old rats, respectively, relative to their age-matched controls.
A large number of probe sets exhibited similar directional changes by functional category in the atrophied soleus muscle of both young and old rats, examples of which are: decreases (10 probe sets whose proteins function as chaperones, nine in the tricarboxylic (TCA) cycle and nine associated with the Z-disc) and increases in expression (eight were glutathione-S-transferases and 11 with protein degradative functions; Table 1). Some of the functional families that showed age-atrophy interactions included amino acid metabolism (six probe sets increasing with atrophy in the young, but not in the old), cell cycle (four increasing with atrophy only in the young; another five showed an atrophy main effect by increasing in both ages), protein degradation (four increased only in young atrophy) and the TCA cycle (four decreased only in the young). The two largest functional families with age-atrophy interactions were electron transport (23 decreasing with atrophy only in the young; six decreased in both ages) and the extracellular matrix (15 only decreasing with atrophy in the young towards the already lower control values found in the old; Supplementary Tables 1 and 2). Probe sets for both the extracellular matrix and electron transport decreased to a greater degree in young than in old soleus muscle after 10 days of hindlimb immobilization. The preponderance of extracellular matrix probe sets decreasing in the young soleus muscle were for collagen isoforms.
In order to obtain further information from the age-atrophy interaction data, the data were clustered hierarchically according to their normalized signal intensities. This style of mathematical clustering grouped together probe sets that had similar graphical expression patterns. Interestingly, probe sets that were clustered together with similar patterns of expression often had similar biological functions (Supplementary Fig. 1). Figure 3 shows an example cluster where most of the probe sets clustered together by their relative expression pattern were involved in common biological roles in the extracellular matrix. Interestingly, the extracellular matrix probe sets revealed a trend where young muscle atrophy decreased extracellular matrix (ECM) mRNA levels to that of old control muscle, but atrophy had no further decrement in ECM mRNA levels in old muscle (Fig. 3).
Discussion
Losses in muscle strength with age are associated with increased mortality (Metter et al. 2002). The decrement in strength is partially attributable to reduced muscle mass and physical activity. Loss of muscle mass by limb immobilization is a model that mimics rapid losses of muscle strength in humans (Cartee, 1995). As the rate of muscle atrophy affects mortality in men after the age of 60 years more than the loss of absolute mass (Metter et al. 2002), it is important to consider both atrophy and age-atrophy interactions to better understand the biological mechanisms by which physical inactivity leads to muscle wasting. The hypothesis of the study was that multiple mRNAs would be expressed differentially between young and old rats after 10 days of immobilization-induced muscle atrophy. After a two-factor ANOVA and Bonferroni adjustment for multiple testing, 730 significant probe sets were identified that had either an atrophy main effect (385 probe sets) or an age-atrophy interaction (345 probe sets). These significant probe sets were then analysed using three approaches: (1) by individual probe set functions, (2) by the categorization of probe sets into functional groups based on their designated ontologies and (3) by hierarchically clustering together probe sets of similar expression patterns, with the aid of SAS statistical programs. Some of the identified differential responses with potentially important functions in immobilization-induced atrophy will be discussed.
As expected, the microarray results showed a 16-fold increase in MHC IIb mRNA on the 10th day of hindlimb-immobilization-induced atrophy of the soleus muscle in young rats (Fig. 1A). In contrast, and unexpectedly, microarray data for the old rats exhibited no increase in MHC IIb mRNA in the immobilized soleus muscle. A similar differential trend occurred for MHC IIx mRNA in the soleus muscle, increasing 25-fold in array data in young rats, while no significant change was observed in the old muscle following 10 days of immobilization (Fig. 1B). The failure of MHC IIb and IIx mRNAs to increase with immobilization in the old as they did in the young soleus muscle is striking because it is well established that the mechanical unloading of young soleus muscles induces multi-fold increases in MHC IIx and IIb mRNAs (Haddad et al. 1998; Stevens et al. 1999a, b; Baldwin & Haddad, 2001; Cros et al. 2001; St-Amand et al. 2001). The dysregulation of MHCs IIb and IIx mRNA levels was validated by real-time PCR analysis (see Table 2 for comparisons). Real-time PCR analysis showed larger-amplitude fold increases for MHC IIb and IIx mRNAs (61- and 68-fold increases, respectively) with young atrophy than were observed from microarray analyses. We speculate that the differences between the microarray and real-time analyses are probably the result of the greater sensitivity of real-time PCR to detect even lower levels of the MHC IIb and IIx mRNA levels in controls than are possible with microarray analysis, resulting in larger fold differences. Supporting our speculation is that the real-time PCR analysis showed six- and five-fold significant increases between old controls and old immobilized groups for MHC IIb and IIx mRNAs, respectively, which were not significantly detected in microarray analyses. Nonetheless, the upregulation of MHC IIb and IIx mRNA levels seen in the atrophying soleus muscle of young rats was markedly suppressed in the aged animals, which permits the postulation that a shift in the pre-translational control of MHC IIb and IIx has occurred.
The aging defect in MHC IIx mRNA expression extends the observation of its failure to upregulate when older humans undergo skeletal muscle hypertrophy. Three months of resistance exercise training increased the transcript levels of skeletal muscle MHC I by 85 %, but decreased the transcript levels of MHC IIa and MHC IIx in both middle-aged (47–60 years) and old human subjects (> 65 years; Balagopal et al. 1997). Balagopal et al. (1997) concluded that ‘a decline in the synthesis rate of MHC implies a decreased ability to remodel this important muscle contractile protein and likely contributes to the declining muscle mass and contractile function in the elderly’. We speculate that the failure of MHC IIb and IIx mRNAs to upregulate in the old atrophied soleus muscle from a model that successfully forced a greater increase of their expression in younger muscles has uncovered a potential defect in type IIb and IIx MHC gene expression in old muscles. Although we do not have the data to support the following speculation, if MHC IIb and IIx mRNAs also failed to upregulate with age in faster phenotype muscles, the dysregulation of fast MHC expression could underlie why type IIb fibres are preferentially lost in the muscles of old rodents (Kugelberg, 1976). Another important mechanism for the loss of type II fibres in the aged is the loss of α motor nerves (Ansved & Larsson, 1990).
The striking inability of the aged animals to upregulate the mRNA expression of MHC IIx and IIb when compared to young animals, suggests that the aged animals lack the ability to activate the necessary molecular signalling mechanism(s) to increase the mRNA expression of MHC IIx or IIb in atrophied soleus muscle. Typically, the soleus muscle expresses both type I and type IIa MHC mRNA with more than 75 % of total MHC mRNA being accounted for by type I MHC, and no IIb MHC mRNA and only a marginal amount of type IIx MHC mRNA, accounting for less than 3 % of total MHC mRNA (Huey et al. 2001). During stimuli that induce atrophy of the soleus muscle there is typically a large upregulation of the IIx and IIb mRNA (Huey et al. 2001). The increase in MHC IIb mRNA has been termed in the past as de novo synthesis (di Maso et al. 2000; Huey et al. 2001). If the large increase in MHC IIb mRNA in atrophied muscle is mostly due to de novo synthesis, then the failure to increase MHC IIb mRNA in the old animals could be interpreted as a failure to increase the transcriptional activation of the endogenous MHC IIb and IIx promoters. Recently, Wheeler et al. (1999) and Allen et al. (2001) reported that MyoD binding to a proximal E-box regulated the activation of the MHC IIb promoter in C2C12 myotubes. Thus, one could expect that the failed increase in MHC IIb mRNA with age might be due to a failure of MyoD to be activated or to bind to the IIb promoter. However, MyoD and myogenin mRNA levels were unchanged in the array analyses. Finding a dysfunctional signalling pathway with age for MHC IIb and IIx is intriguing because other muscle-specific genes may also be regulated by similar defective mechanisms, which might explain why other muscle properties are altered during the aging/atrophy processes.
Proteins of the Homer family bind to ryanodine receptors (RyR1; Xiao et al. 2000) and dramatically enhance the Ca2+ responsiveness of RyR1 at the level of the single channel in skeletal muscle (Feng et al. 2002). The mRNA of another member of the homer family, homer 2a, decreased 2.5-fold (decrease of 40 %) in the heart as mice aged from 2 to 33 months (Bronikowski et al. 2003) and 40 % in old soleus muscle (Pattison et al. 2003). These observations have been extended here at the 10th day of immobilization by Homer2 mRNA decreasing more in young (range of −3.9- to −7.2-fold in five different probe sets) than in old (range of −1.73- to −2.52-fold) rats, as compared to their respective age-matched control values (Fig. 2). We propose a new hypothesis, that the downregulation of homer 2 mRNA with inactivity is a part of a more generalized adaptation to lower Ca2+ regulatory mechanisms in muscle with disuse. By analogy, it has been shown that cardiomyocytes isolated from non-exercised rats are associated with a decrease in the Ca2+ sensitivity of the contractile apparatus (Diffee & Nagle 2003).
Immobilization of limbs in a shortened position alters Z-disc morphology (Wanek & Snow, 2000), and differentially expressed mRNAs whose proteins are associated with the Z-line included: main effects – cysteine-rich protein 3 (also known as Csrp3, or muscle LIM protein) decreased threefold, ankyrin repeat domain 2 (also known as Ankrd2 or Arpp) decreased sixfold, enigma decreased 2.74-fold and elfin decreased 3.8- and 1.5-fold (multiple probe sets); interactions – small muscle protein (X-linked, Smpx, Csl, chisel) decreased 1.9- to 2.5- and 1.3- to 1.6-fold (multiple probe sets) in the soleus muscle of young and old rats, respectively. We propose the hypothesis that proteins of the Z-disc are signalled to downregulate by the lack of muscle stretch, as stretch receptors have been identified at this locale (Epstein & Davis, 2003).
Csrp3 mRNA was decreased threefold in the soleus muscle at the end of 10 days of immobilization-induced atrophy in both young and old muscles. Interestingly, the directional change in Csrp3 mRNA in the soleus muscle of 7 day hindlimb-suspended rats was the opposite, increasing two- to threefold (Willman et al. 2001), which might be explained by the fact that only the limb immobilization model lowers contractile activity, whereas EMG activity is not decreased in the hindlimb suspension model (Fischback & Robbins, 1969; Alford et al. 1987). Csrp3 may contribute to the maintenance of muscle cell differentiation via effects on the cytoarchitecture though their interactions and colocalization with α-actinin, zyxin and βI-spectrum (Khurana et al. 2002) and to promote myogenesis by binding to the transcription factor MyoD, enhancing MyoD activity (Kong et al. 1997). Targeted ablation of Csrp3 leads to severe myofibrillar disarray with subsequent dilated cardiomyopathy, putatively from the increased distension of the heart. In this context, the absence of stretch signals in skeletal muscle and the decrease in Csrp mRNA would contribute to its observed atrophy (Arber et al. 2002).
Hindlimb immobilization resulted in a decrease in 15 probe sets with roles in the extracellular matrix, all of which showed a trend of only decreasing with atrophy in young towards the already lower control values found in the old. Atrophied muscle in immobilized limbs exhibits a reorganization and distribution of collagen proteins, which Jarvinen et al. (2002) concluded probably contributes to the deteriorated function and biomechanical properties of the immobilized skeletal muscle. A pre-translational origin of the change in collagen proteins with disuse is suggested by the widespread decrease in collagen gene expression on the microarrays.
Mitochondrial protein concentrations in rat skeletal muscle decrease with age (Farrar et al. 1981; Holloszy et al. 1991). The physical inactivity of limb immobilization resulted in a decrease of 29 nuclear mitochondrial probe sets in the atrophying soleus muscle of young rats (only six of these decreased significantly in old), which would seem to suggest that atrophy-induced inactivity causes a muscular adaptation to downregulate the mRNAs involved in energy generation, possibly denoting mitochondrial dysfunction mimicking that of older muscles. Another potential contributor to mitochondrial dysfunction in sedentary skeletal muscle is if oxidative damage causes an accumulation of DNA mutations and deletions (Short & Nair, 2001). The increases in eight glutathione-S-transferase mRNAs in the atrophied muscles of the current data set suggests that these antioxidants are elevated to buffer the increases in oxidative stress of muscles in immobilized limbs, as reported previously (see Koesterer et al. 2002 for references.).
In summary, over 700 differentially expressed mRNAs were identified with a conservative Bonferroni adjustment. Among the identified genes, the failure of MHC IIb and IIx mRNAs to upregulate as vigorously in the soleus muscle of old, as compared to young, rats during a procedure that is well documented to cause a shift in MHC expression suggests a potential innate defect in the regulation of these genes that could contribute to the functional losses observed in aged animals. Future work will be necessary to determine the mechanism underlying this aging-associated failure to upregulate MHC IIb and IIx mRNAs during muscle disuse.
Acknowledgments
We thank Dr Gary Allen, who provided access to the Bioinformatics Consortium at the University of Missouri, and Dr Mark McIntosh for his leadership in establishment of the Affymetrix Core facility at the University of Missouri. We also thank Aaron Wheeler for his assistance and Dr Barry Prior for helpful discussions. The research was supported by NIH grant AG18881 and NASA grant NAG2–1372.
Supplementary material
The online version of this paper can be found at: DOI: 10.1113/jphysiol.2003.047233 and contains material entitled: Complete lists of probe sets identified as significant with an atrophy main effect, an age-atrophy interaction and results of hierarchical clustering of age-atrophy interactions.
References
- Alford EK, Roy RR, Hodgson JA, Edgerton VR. Electromyography of rat soleus, medial gastrocnemius, and tibialis anterior during hind limb suspension. Exp Neurol. 1987;96:635–649. doi: 10.1016/0014-4886(87)90225-1. [DOI] [PubMed] [Google Scholar]
- Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. J Biol Chem. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- Ansved T, Larsson L. Quantitative and qualitative morphological properties of the soleus motor nerve and the l5 ventral root in young and old rats. Relation to the number of soleus muscle fibers. J Neurol Sci. 1990;96:269–282. doi: 10.1016/0022-510x(90)90138-d. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 2002;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Bakay M, Chen YW, Borup R, Zhao P, Nagaraju K, Hoffman EP. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics. 2002;3:4. doi: 10.1186/1471-2105-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal P, Rooyackers OE, Adey DB, Ades PA, Nair KS. Effects of aging on in vivo synthesis of skeletal muscle myosin heavy-chain and sarcoplasmic protein in humans. Am J Physiol. 1997;273:E790–800. doi: 10.1152/ajpendo.1997.273.4.E790. [DOI] [PubMed] [Google Scholar]
- Baldwin KM, Haddad F. Effects of different activity and inactivity paradigms on myosin heavy chain gene expression in striated muscle. J Appl Physiol. 2001;90:345–357. doi: 10.1152/jappl.2001.90.1.345. [DOI] [PubMed] [Google Scholar]
- Booth FW. Time course of muscular atrophy during immobilization of hindlimbs in rats. J Appl Physiol. 1977;43:656–661. doi: 10.1152/jappl.1977.43.4.656. [DOI] [PubMed] [Google Scholar]
- Booth FW, Baldwin KM. Muscle plasticity: energy demand/supply processes. In: Rowell LB, Shepherd JT, editors. Handbook of Physiology, Section 12: Integration of Motor, Circulatory, Respiratory, and Metabolic Control during Exercise. New York: Oxford University Press; 1996. pp. 1075–1123. [Google Scholar]
- Bronikowski AM, Carter PA, Morgan TJ, Garland TJR, Ung N, Pugh TD, Weindruch R, Prolla TA. Lifelong voluntary exercise in the mouse prevents age-related alterations in gene expression in the heart. Physiol Genomics. 2003;12:129–138. doi: 10.1152/physiolgenomics.00082.2002. [DOI] [PubMed] [Google Scholar]
- Buchner DM, Wagner EH. Preventing frail health. Clin Geriatr Med. 1992;8:1–17. [PubMed] [Google Scholar]
- Cartee GD. What insights into age-related changes in skeletal muscle are provide by animal models? J Gerontol A Biol Sci Med Sci. 1995;50:137–141. doi: 10.1093/gerona/50a.special_issue.137. [DOI] [PubMed] [Google Scholar]
- Claverie JM. Computational methods for the identification of differential and coordinated gene expression. Hum Mol Genet. 1999;8:1821–1832. doi: 10.1093/hmg/8.10.1821. [DOI] [PubMed] [Google Scholar]
- Cros N, Tkatchenko AV, Pisani DF, Leclerc L, Leger JJ, Marini JF, Dechesne CA. Analysis of altered gene expression in rat soleus muscle atrophied by disuse. J Cell Biochem. 2001;83:508–519. doi: 10.1002/jcb.1248. [DOI] [PubMed] [Google Scholar]
- Diffee GM, Nagle D. Regional differences in effects of exercise training on contractile and biochemical properties of rat cardiac myocytes. J Appl Physiol. 2003;95:35–42. doi: 10.1152/japplphysiol.00951.2002. [DOI] [PubMed] [Google Scholar]
- Di Maso NA, Haddad F, Zeng M, McCue SA, Baldwin KM. Role of denervation in modulating IIb MHC gene expression in response to T(3). plus unloading state. J Appl Physiol. 2000;88:682–689. doi: 10.1152/jappl.2000.88.2.682. [DOI] [PubMed] [Google Scholar]
- Epstein ND, Davis JS. Sensing stretch is fundamental. Cell. 2003;112:47–50. doi: 10.1016/s0092-8674(03)00037-0. [DOI] [PubMed] [Google Scholar]
- Farrar RP, Martin TP, Ardies CM. The interaction of aging and endurance exercise upon the mitochondrial function of skeletal muscle. J Gerontol. 1981;36:642–647. doi: 10.1093/geronj/36.6.642. [DOI] [PubMed] [Google Scholar]
- Feng W, Tu J, Yang T, Vernon PS, Allen PD, Worley PF, Pessah IN. Homer regulates gain of ryanodine receptor type 1 channel complex. J Biol Chem. 2002;277:44722–44730. doi: 10.1074/jbc.M207675200. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Robbins N. Changes in contractile properties of disused soleus muscles. J Physiol. 1969;201:305–320. doi: 10.1113/jphysiol.1969.sp008757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad F, Qin AX, Zeng M, McCue SA, Baldwin KM. Interaction of hyperthyroidism and hindlimb suspension on skeletal myosin heavy chain expression. J Appl Physiol. 1998;85:2227–2236. doi: 10.1152/jappl.1998.85.6.2227. [DOI] [PubMed] [Google Scholar]
- Hnik P, Vejsada R, Goldspink DF, Kasicki S, Krekule I. Quantitative evaluation of electromyogram activity in rat extensor and flexor muscles immobilized at different lengths. Exp Neurol. 1985;88:515–528. doi: 10.1016/0014-4886(85)90067-6. [DOI] [PubMed] [Google Scholar]
- Holloszy JO, Chen M, Cartee GD, Young JC. Skeletal muscle atrophy in old rats: differential changes in the three fiber types. Mech Ageing Dev. 1991;60:199–213. doi: 10.1016/0047-6374(91)90131-i. [DOI] [PubMed] [Google Scholar]
- Huey KA, Roy RR, Baldwin KM, Edgerton VR. Temporal effects of inactivity on myosin heavy chain gene expression in rat slow muscle. Muscle Nerve. 2001;24:517–526. doi: 10.1002/mus.1035. [DOI] [PubMed] [Google Scholar]
- Jänkälä H, Harjola VP, Petersen ME, Harkonen M. Myosin heavy chain mRNA transform to faster isoforms in immobilized skeletal muscle: a quantitative PCR study. J Appl Physiol. 1997;82:977–982. doi: 10.1152/jappl.1997.82.3.977. [DOI] [PubMed] [Google Scholar]
- Jarvinen TA, Jozsa L, Kannus P, Jarvinen TL, Jarvinen M. Organization and distribution of intramuscular connective tissue in normal and immobilized skeletal muscles. An immunohistochemical, polarization and scanning electron microscopic study. J Muscle Res Cell Motil. 2002;23:245–254. doi: 10.1023/a:1020904518336. [DOI] [PubMed] [Google Scholar]
- Khurana T, Khurana B, Noegel AA. LIM proteins: association with the actin cytoskeleton. Protoplasma. 2002;219:1–12. doi: 10.1007/s007090200000. [DOI] [PubMed] [Google Scholar]
- Koesterer TJ, Dodd SL, Powers S. Increased antioxidant capacity does not attenuate muscle atrophy caused by unweighting. J Appl Physiol. 2002;93:1959–1965. doi: 10.1152/japplphysiol.00511.2002. [DOI] [PubMed] [Google Scholar]
- Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E. Adaptive transformation of rat soleus motor units during growth. J Neurol Sci. 1976;27:269–289. doi: 10.1016/0022-510x(76)90001-0. [DOI] [PubMed] [Google Scholar]
- Metter EJ, Talbot LA, Schrager M, Conwit R. Skeletal muscle strength as a predictor of all-cause mortality in healthy men. J Gerontol A Biol Sci Med Sci. 2002;57:B359–365. doi: 10.1093/gerona/57.10.b359. [DOI] [PubMed] [Google Scholar]
- Pattison JC, Folk LC, Madsen RW, Childs TE, Booth FW. Transcriptional profiling identifies extensive down regulation of extracellular matrix gene expression in sarcopenic rat soleus muscle. Physiol Genomics. 2003;15:34–43. doi: 10.1152/physiolgenomics.00040.2003. [DOI] [PubMed] [Google Scholar]
- Paulos LE, Wnorowski DC, Beck CL. Rehabilitation following knee surgery. Recommendations. Sports Med. 1991;11:257–275. doi: 10.2165/00007256-199111040-00005. [DOI] [PubMed] [Google Scholar]
- Short KR, Nair KS. Does aging adversely affect muscle mitochondrial function? Exerc Sport Sci Rev. 2001;29:118–123. doi: 10.1097/00003677-200107000-00006. [DOI] [PubMed] [Google Scholar]
- Stein T, Schluter M, Galante A, Soteropoulos P, Tolias P, Grindeland R, Moran M, Wang T, Polansky M, Wade C. Energy metabolism pathways in rat muscle under conditions of simulated microgravity. J Nutr Biochem. 2002;13:471. doi: 10.1016/s0955-2863(02)00195-x. [DOI] [PubMed] [Google Scholar]
- Stevens L, Gohlsch B, Mounier Y, Pette D. Changes in myosin heavy chain mRNA and protein isoforms in single fibers of unloaded rat soleus muscle. FEBS Lett. 1999a;463:15–18. doi: 10.1016/s0014-5793(99)01596-3. [DOI] [PubMed] [Google Scholar]
- Stevens L, Sultan KR, Peuker H, Gohlsch B, Mounier Y, Pette D. Time-dependent changes in myosin heavy chain mRNA and protein isoforms in unloaded soleus muscle of rat. Am J Physiol. 1999b;277:C1044–1049. doi: 10.1152/ajpcell.1999.277.6.C1044. [DOI] [PubMed] [Google Scholar]
- Unger MA, Rishi M, Clemmer VB, Hartman JL, Keiper EA, Greshock JD, Chodosh LA, Liebman MN, Weber BL. Characterization of adjacent breast tumors using oligonucleotide microarrays. Breast Cancer Res. 2001;3:336–341. doi: 10.1186/bcr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Bij AK, Laurant MG, Wensing M. Effectiveness of physical activity interventions for older adults: a review. Am J Prev Med. 2002;22:120–133. doi: 10.1016/s0749-3797(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Walston J, Fried LP. Frailty and the older man. Med Clin North Am. 1999;83:1173–1194. doi: 10.1016/s0025-7125(05)70157-7. [DOI] [PubMed] [Google Scholar]
- Wanek LJ, Snow MH. Activity-induced fiber regeneration in rat soleus muscle. Anat Rec. 2000;258:176–185. doi: 10.1002/(SICI)1097-0185(20000201)258:2<176::AID-AR7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Wheeler MT, Snyder EC, Patterson MN, Swoap SJ. An E-box within the MHC IIB gene is bound by MyoD and is required for gene expression in fast muscle. Am J Physiol. 1999;276:C1069–1078. doi: 10.1152/ajpcell.1999.276.5.C1069. [DOI] [PubMed] [Google Scholar]
- Willmann R, Kusch J, Sultan KR, Schneider AG, Pette D. Muscle LIM protein is upregulated in fast skeletal muscle during transition toward slower phenotypes. Am J Physiol. 2001;280:C273–279. doi: 10.1152/ajpcell.2001.280.2.C273. [DOI] [PubMed] [Google Scholar]
- Wittwer M, Fluck M, Hoppeler H, Muller S, Desplanches D, Billeter R. Prolonged unloading of rat soleus muscle causes distinct adaptations of the gene profile. FASEB J. 2002;16:884–886. doi: 10.1096/fj.01-0792fje. [DOI] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Worley PF. Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol. 2000;10:370–374. doi: 10.1016/s0959-4388(00)00087-8. [DOI] [PubMed] [Google Scholar]
- Zarzhevsky N, Carmeli E, Fuchs D, Coleman R, Stein H, Reznick AZ. Recovery of muscles of old rats after hindlimb immobilisation by external fixation is impaired compared with those of young rats. Exp Gerontol. 2001;36:125–140. doi: 10.1016/s0531-5565(00)00189-3. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Davis BS, Booth FW. IGF-I restores satellite cell proliferative potential in immobilized old skeletal muscle. J Appl Physiol. 2000;89:1365–1379. doi: 10.1152/jappl.2000.89.4.1365. [DOI] [PubMed] [Google Scholar]
- St-Amand J, Okamura K, Matsumoto K, Shimizu S, Sogawa Y. Characterization of control and immobilized skeletal muscle: an overview from genetic engineering. FASEB J. 2001;15:684–692. doi: 10.1096/fj.00-0150com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


