Abstract
Forced expression of the retinoblastoma (RB) gene product inhibits the proliferation of cells in culture. A major target of the RB protein is the S-phase-inducing transcription factor E2F1. RB binds directly to the activation domain of E2F1 and silences it, thereby preventing cells from entering S phase. To induce complete G1 arrest, RB requires the presence of the hbrm/BRG-1 proteins, which are components of the coactivator SWI/SNF complex. This cooperation is mediated through a physical interaction between RB and hbrm/BRG-1. We show here that in transfected cells RB can contact both E2F1 and hbrm at the same time, thereby targeting hbrm to E2F1. E2F1 and hbrm are indeed found within the same complex in vivo. Furthermore, RB and hbrm cooperate to repress E2F1 activity in transient transfection assays. The ability of hbrm to cooperate with RB to repress E2F1 is dependent upon several distinct domains of hbrm, including the RB binding domain and the NTP binding site. However, the bromodomain seems dispensable for this activity. Taken together, our results point out an unexpected role of corepressor for the hbrm protein. The ability of hbrm and RB to cooperate in repressing E2F1 activity could be an underlying mechanism for the observed cooperation between hbrm and RB to induce G1 arrest. Finally, we demonstrate that the domain of hbrm that binds RB has transcriptional activation potential which RB can repress. This suggest that RB not only targets hbrm but also regulates its activity.
The retinoblastoma (RB) gene is often inactivated in a wide variety of human tumors (1). It encodes a protein that is regulated in a cell cycle-dependent manner (2). During the G1 phase of the cell cycle, RB is in an hypophosphorylated form, which is presumably the active form of the protein. At the end of the G1 phase, RB is phosphorylated by the cell cycle-dependent protein kinases cyclin D/cdk4 or cdk6 (3).
The RB protein plays a central role in the control of the G1/S transition (2) and can be inactivated by a variety of S-phase-inducing viral transforming proteins such as the adenovirus E1A protein (1, 4). Furthermore, ectopic expression of RB in some transformed cell lines induces a growth arrest into the G1 phase of the cell cycle (5). The domain of RB responsible for this growth inhibitory activity consists of the E1A interaction domain called the “pocket domain,” which RB shares with two other proteins targeted by E1A, the p107 and p130 proteins (6). In addition to the pocket, the extreme C terminus is also required for RB-induced growth arrest (5, 7).
RB is thought to inhibit cell proliferation, at least in part, through its effect on transcription (3, 8). One of the major targets of RB is the E2F transcription factor which is activated at the G1/S transition and transactivates genes whose products are required during the S phase of the cell cycle, such as the dehydrofolate reductase or the DNA polymerase α genes (9). E2F plays an important role in cell cycle control because deregulated expression of the E2F1 protein induces cell entry into S phase and can lead to transformation (10–12). The E2F transcription factor is composed of heterodimers between one of the five “E2F” proteins and one of the three “DP” proteins (13). RB targets E2F1-, E2F2- and E2F3-containing heterodimers, where as p107 and p130 target E2F4- and E2F5-containing heterodimers (14).
The RB protein contacts directly the E2F1 activation domain and silences it (15, 16). Furthermore, the E2F1/RB complex is able to bind to promoters bearing E2F sites and represses transcription mediated by these promoters (17). This repression is likely to be mediated by an inhibitor domain within the RB protein, because RB represses transcription when targeted to a promoter through an heterologous DNA binding domain (18, 19). In addition to repressing E2F polymerase II-regulated promoters, RB is also able to repress transcription mediated by the RNA polymerases I and III (20, 21).
Several lines of evidence indicate that repression of E2F-regulated promoters is responsible, at least in part, for RB-induced growth arrest. Domains of RB responsible for repressing E2F1 activity are indeed required for RB to inhibit cell proliferation (5), and overexpression of E2F1 relieves the cell cycle block induced by RB (22). Furthermore, an E2F1 mutant, which cannot activate transcription or bind RB, is oncogenic, presumably because it is unable to recruit RB to repress E2F-regulated promoters (23).
To efficiently block cells into the G1 phase of the cell cycle, RB requires the presence of hbrm/BRG-1 activity (24). hbrm and BRG-1 are the two known human homologues of the yeast SWI2/SNF2 protein (25–27). They contain a helicase-like domain, which exhibits DNA-dependent ATPase activity, and a bromodomain located at the C terminus of the protein. These proteins belong to two distinct large multimolecular SWI/SNF complexes, which are able to remodel chromatin structure in vitro in the presence of ATP (28, 29). Through chromatin remodeling, it can assist transcription factors such as GAL4 to bind in vitro to their target DNA sequence on nucleosomal DNA (30, 31). The SWI/SNF complex possesses some properties of a specific coactivator, presumably by helping transcription factors to bind promoters, thereby removing the nucleosome-induced transcriptional repression (28). In addition, the yeast SWI/SNF complex is under certain conditions detected within the RNA polymerase II holoenzyme (32). However, recent studies on mammalian cells failed to detect colocalization or cofractionation of brm/BRG-1 with the RNA polymerase II (33). In mammalian cells, SWI/SNF is clearly involved in transcriptional activation by some members of the nuclear receptor superfamily, including the glucocorticoid receptor, as shown in transient transfection experiments (25, 27, 34). This activity requires several distinct domains on hbrm, including the ATP binding site and the N-terminal part of the molecule, but the bromodomain seems dispensable (25).
The hbrm/BRG-1 proteins possess some growth-inhibitory properties that are dependent on the presence of RB (24, 35). Conversely, a dominant-negative mutant of hbrm/BRG-1 relieves RB-induced growth arrest of SAOS2 cells, indicating that RB and hbrm/BRG-1 cooperate to block cells in G1 (24). This functional cooperation requires a physical interaction between RB and hbrm/BRG-1, mediated through a domain within hbrm/BRG-1 related to the RB binding site of the viral transforming protein E7 (24, 34).
In this paper, we show, by coimmunoprecipitation from transfected cells, that RB is able to target hbrm to E2F1. We also demonstrate that E2F1 and hbrm are present within the same complex in vivo. In transfection experiments, RB and hbrm cooperate to repress E2F1 activity, indicating that, besides the glucocorticoid receptor, E2F1 could be another common target for these two proteins. This cooperation requires several distinct functional domains on hbrm, including the E7-like RB binding domain. Taken together, our results show that hbrm can act as a corepressor for RB. Repressing E2F1 may be responsible, at least in part, for the functional cooperation between hbrm and RB to repress cell proliferation.
MATERIALS AND METHODS
Cell Culture, Transfections and Chloramphenicol Acetyltransferase (CAT) Assays.
SAOS2 and U2OS human osteosarcoma cells, C33A human epithelium cervical cells, and WI-38 cells were maintained in DMEM supplemented with 10% fetal calf serum and grown at 37°C (5% CO2). Cells were transfected overnight using the calcium phosphate coprecipitation technique. After 24-h incubations, extracts from transfected cells were used for CAT assays. Results were quantified by PhosphorImager.
In Vivo Expression Plasmids.
(E2F)3TK-CAT, CMV-E2F1, and CMV-DP1 have been described (36). CMV-RB and mutant are gifts from W. Kaelin (Harvard Medical School). CMV-HA-hbrm and mutants have been described (25). The reporter construct G5E1b-CAT was a gift from M. Green (University of Massachusetts Medical Center). GAL4-hbrm protein was expressed from a cytomegalovirus (CMV)-driven promoter and contains amino acids 1206 to 1304 from hbrm. GAL4-E2F1 was described previously (37).
Coimmunoprecipitation from Transfected Cells.
U2OS human osteosarcoma cells were transfected as described above. Twenty-four hours after transfection cells were lysed in 1 ml lysis buffer (50 mM Tris, pH 8/300 mM Kcl/30 mM MgCl2/10 mM EDTA/0.5% Nonidet P-40/0.1 mM phenylmethylsulfonyl fluoride, aprotinin, leupeptin, and pepstatin). The lysis mixture was incubated on ice for 20 min and cleared by centrifugation at 12,000 × g for 10 min at 4°C. Relevant antibodies were added to 1 ml of extract and incubated at 4°C for 2 h. Protein A-Sepharose/protein G-Sepharose (a 50/50 mix) was added and the mixture rotated slowly overnight at 4°C. The immune complexes were pelleted and washed three times with lysis buffer. Immunoprecipitates were eluted by competition with a hemagglutinin (HA) peptide (1 mM final) in 50 mM Tris (pH 8), 150 mM NaCl, 1 mM EDTA, and 0.2% Nonidet P-40. Immunoprecipitates were then analyzed by SDS/PAGE, transferred to a nitrocellulose membrane, and subjected to Western blot analysis with either RB antibody XZ55 (PharMingen) HA antibody 12CA5 (Boehringer Mannheim) or E2F1 antibody KH95 (PharMingen). Immunoreactive bands were detected with an ECL kit (Amersham) according to the manufacturer’s instructions.
Preparation of Anti-hbrm/BRG-1 Antibodies.
Polyclonal rabbit anti-hbrm/BRG-1 antibodies were generated using full-length HA-hbrm protein that was produced with a baculovirus system and immunopurified using and anti-HA monoclonal antibody. To produce polyclonal chicken anti-hbrm/BRG-1 antibodies, we inserted a fragment of the mouse mBRG-1 cDNA in pGEX2T in-frame with glutathione S-transferase. The fragment of mouse cDNA encoded amino acids 39–333 in the corresponding human sequence. The mouse and the human amino acid sequence are 92% identical in this region. The fusion protein was expressed in Escherichia coli and affinity purified on glutathione-Sepharose 4B under conditions recommended by the manufacturer (Pharmacia). The antibodies were collected from the egg yolk by PEG precipitation then affinity purified.
Cell Extracts and Immunoprecipitations.
WI-38 cells were grown to confluency, lysed in IP0.1 buffer (20 mM Hepes, pH 7.6/10% glycerol/25 mM MgCl2/0.1 mM EDTA/0.2% Nonidet P-40/0.1 M potassium acetate/2.25 mg/ml pepstatin/10 mg/ml leupeptin/10 mg/ml aprotinin/2 mM phenylmethylsulfonyl fluoride/0.1 mM DTT), and then sonicated for 5 min followed by vigorous vortex mixing. The extracts were finally cleared by centrifugation. For immunoprecipitations, 50 μl of rabbit anti-hbrm/BRG-1 antibody were prebound to 10 μl of protein A-Sepharose beads (Pharmacia). The beads were then washed extensively in IP0.1 buffer and added to precleared extract from one 10-cm plate. Washing and elution of the immunoprecipitate was performed as described (38). For Western blot analysis, the membrane was blocked with PBS/0.2% Tween 20/10% horse serum and incubated with chicken anti-hbrm/BRG-1 and mouse monoclonal anti-E2F antibodies. Horseradish peroxidase-coupled secondary antibodies were obtained from Biocytex (Marseille, France) (rabbit anti-chicken IgY) and Amersham (sheep anti-mouse). Enhanced chemiluminescence (ECL) reagents (Amersham) were used for detection.
RESULTS
RB can induce growth arrest, at least partly, by repressing the activation capacity of the S-phase-inducing transcription factor E2F1. RB has been shown to induce growth arrest in cooperation with hbrm (24). One possible explanation for this cooperativity is that RB and hbrm cooperate to repress E2F1 activity.
To investigate such a mechanism, we first sought evidence of an in vivo complex containing RB, hbrm, and E2F1. The RB protein has been shown to form independent complexes in vivo with either E2F1 (39, 40) or hbrm (24, 34). We used an indirect immunoprecipitation assay to establish whether RB is able to bind both E2F1 and hbrm simultaneously. This is potentially possible because hbrm and E2F1 possess distinct RB binding motifs.
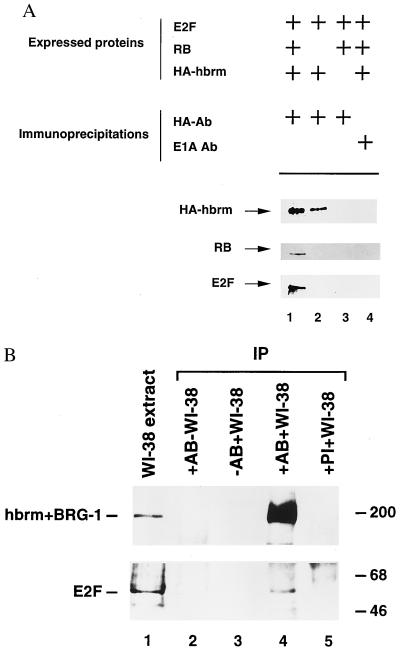
Fig. 1A shows that an interaction between E2F1 and hbrm is not readily detected in human cells lacking RB. Following transfection of U2OS cells with CMV-E2F1 and CMV-HA-hbrm, precipitation of hbrm by HA antibodies does not result in the indirect precipitation of E2F1, as detected by an E2F1-antibody Western blot (Fig. 1A, line 2). However, when a CMV-RB expression vector is included in the transfection, the E2F1 and RB proteins are readily detected in the hbrm immunoprecipitate (Fig. 1A, line 1). Omission of the CMV-HA-hbrm expression vector (Fig. 1A, line 3) or substitution of the HA antibody with an E1A-specific antibody (Fig. 1A, line 4) does not result in the precipitation of E2F1. This experiment indicates that RB can stimulate complex formation between the E2F1 and hbrm proteins.
Figure 1.
(A) RB can bind E2F1 and hbrm simultaneously. U2OS cells were transfected with 6 μg of the indicated expression vectors and 6 μg of DP1 expression vector. Whole-cell extracts were then immunoprecipitated using either a HA or a E1A antibody as indicated. Immunoprecipitates were assayed by Western blot analysis for the presence of hbrm (Top), RB (Middle), and E2F1 (Bottom) proteins. (B) hbrm/BRG-1 are associated with E2F1 in vivo. WI-38 cells were lysed in IP0.1 buffer and immunoprecipitated either in the absence (lane 3) or in the presence (lane 4) of anti-hbrm/BRG-1 antibodies from rabbit. Control immunoprecipitations were also performed in the presence of WI-38 extract and pre-immune serum (lane 5) and in the presence of anti-hbrm/BRG-1 antibodies, in the absence of WI-38 extract (lane 2). After analysis by 6% PAGE, proteins were transferred to nitrocellulose membrane and analyzed by Western blot with either anti-hbrm/BRG-1 antibodies from chicken (Upper) or an anti-E2F1 monoclonal antibody (Lower).
We next tried to establish whether E2F1 and hbrm exist in a single complex in vivo. We used an antibody that recognizes hbrm and the closely related protein BRG-1 in immunoprecipitation assays. Fig. 1B shows that the E2F1 protein is present in an immunoprecipitate of hbrm and BRG-1 from a normal human fibroblast extract, as shown by a Western blot using an E2F1-specific antibody (Fig. 1B Lower, line 4). This immunoprecipitate also contains the hbrm/BRG-1 proteins (Fig. 1B Upper, line 4). The interaction between E2F1 and hbrm/BRG-1 is specific because omission of the hbrm/BRG-1 antibody (Fig. 1B, line 3), or usage of a nonspecific antibody (Fig. 1B, line 5) does not result in the coprecipitation of E2F1. We estimate that ≈2.5% of endogenous E2F is complexed with hbrm. The interaction between E2F1 and hbrm is likely to be mediated by RB (Fig. 1A), but we cannot exclude the possibility that other proteins are involved in mediating or augmenting this interaction.
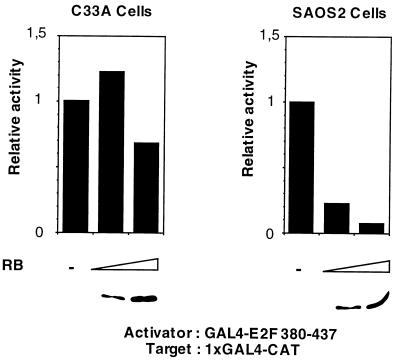
We next investigated the possibility that hbrm is involved in RB-induced repression of E2F1. We first tested whether RB would repress E2F1 in the absence of hbrm. Human epithelium cervical C33A cells contain no hbrm, present a low level of BRG-1, and lack functional pRB (25). The glucocorticoid receptor, whose activity is SWI/SNF-dependent, is largely inactive in these cells (25). As already described by others, when expressed at high levels, RB is able to repress E2F1 transcription in C33A cells (refs. 18, 41, and 42, and data not shown). However, as shown in Fig. 2, low, limiting levels of transfected RB were largely ineffective in repressing the activity of E2F1 activation domain in C33A cells (Fig. 2 Upper Left). However, similar amounts of RB as assayed by Western blot analysis (Fig. 2 Lower), repressed E2F1 activity in hbrm/BRG-1-expressing SAOS2 cells very efficiently (Fig. 2 Upper Right). This experiment suggested that hbrm could participate in E2F1 repression by RB.
Figure 2.
RB represses less efficiently in C33A hbrm-negative cell line. C33A (Left) and SAOS2 cells (Right) were transfected with 1 μg of G1E1B-CAT, 2 μg of GAL4-E2F1, and with 25 and 50 ng (C33A) or 50 and 250 ng (SAOS2) of CMV-RB as indicated. Following a CAT assay, results were quantified using a PhosphorImager. The activity of the reporter in the absence of RB is normalized for both cell lines to a value of 1 (Upper). The level of expressed RB protein was assayed by immunoprecipitation followed by Western blot analysis using XZ55 antibody (Lower). Result of typical experiments is shown.
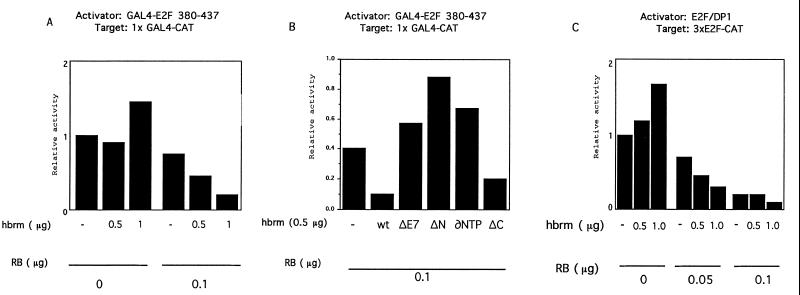
We next tried to establish whether exogenous expression of hbrm in C33A cells would restore normal repression by RB. Fig. 3A shows that transfection of an hbrm expression vector slightly stimulates the activity of a GAL4 E2F1 fusion protein. However, in the presence of low, limiting amount of exogenously expressed RB, the presence of hbrm results in the repression of GAL4 E2F1 activity. The repressive effect of RB is thus increased in the presence of hbrm, going, for example, from 1.5-fold in the absence of hbrm up to 7-fold in the presence of hbrm (Fig. 3A). This effect is not due to a change in the status of RB, because the amount of hypophosphorylated transfected RB remains constant whether or not hbrm is coexpressed, as assessed by Western blot analysis using the hypophosphorylated form-specific anti-RB antibody XZ55 (data not shown). This experiment shows that, in the presence of RB, hbrm is able to repress the activation capacity of GAL4–E2F1 fusion protein.
Figure 3.
hbrm cooperates with RB to repress the activation functions of E2F1. (A) C33A cells were transfected with 1 μg of G1E1B-CAT and 2 μg of GAL4-E2F1 (amino acids 380–437). Where indicated, 100 ng of CMV-RB and 0.5 or 1 μg of CMV-hbrm was added. The activity of the promoter in the absence of hbrm and RB is normalized to a value of 1. (B) Same as in A using 100 ng of CMV-RB and 0,5 μg of CMV-hbrm wild-type or mutant ΔE7 (deletion of the E7 homology domain), ΔN (deletion of the N-terminal domain), ∂NTP (NTP binding site mutant), and ΔC (deletion of the C-terminal domain). The activity of the promoter in the absence of hbrm and RB is normalized to a value of 1. (C) C33A cells were transfected with 1 μg of 3× E2F-CAT reporter vector and 100 ng of CMV-E2F1 and CMV-DP1. Where indicated, 100 ng of CMV-RB and 0.5 or 1 μg of CMV-hbrm was added. The activity of the promoter in the absence of hbrm and RB is normalized to a value of 1.
Because RB and hbrm interact in vivo, their cooperation to repress E2F1 may depend on their ability to form such a complex. If this is so, then we would expect hbrm mutants lacking the RB binding site to be defective in E2F1 repression. We therefore tested a panel of hbrm mutants in the E2F1 repression assay. Fig. 3B shows that an hbrm mutant lacking the RB binding site (ΔE7) does not cooperate with RB to repress E2F1 activity. Instead, like the wild-type hbrm protein in the absence of RB, ΔE7 can slightly stimulate E2F1 activity. Two other mutants of hbrm were also defective in E2F1 repression. The ΔN mutant, which lacks the N terminus and ∂NTP that has a point mutation in the NTP binding site, were unable to cooperate with RB to repress E2F1. However, a mutant carrying a deletion of the hbrm bromodomain (ΔC) was still able to repress E2F1, although at slightly reduced capacity compared with wild-type hbrm. These results confirm that an interaction between hbrm and RB is important for E2F1 repression and indicate that other functions of hbrm, including its NTPase activity, play a role in the repression of E2F1.
We next tested whether hbrm would also cooperate with RB to repress the E2F transcription factor. For these experiments, we monitored the effect of RB and hbrm on the activity of the E2F1/DP1 complex as assayed on an E2F-site bearing promoter. Fig. 3C shows that as on GAL4–E2F fusion protein, hbrm has a small activating effect E2F1/DP1 by itself. However, under conditions of limiting RB concentrations (i.e., concentrations that will not fully repress E2F1 activity), hbrm is able to repress E2F1/DP1. Thus RB represses the activity of the E2F1/DP1 transcription factor more efficiently in the presence of hbrm.
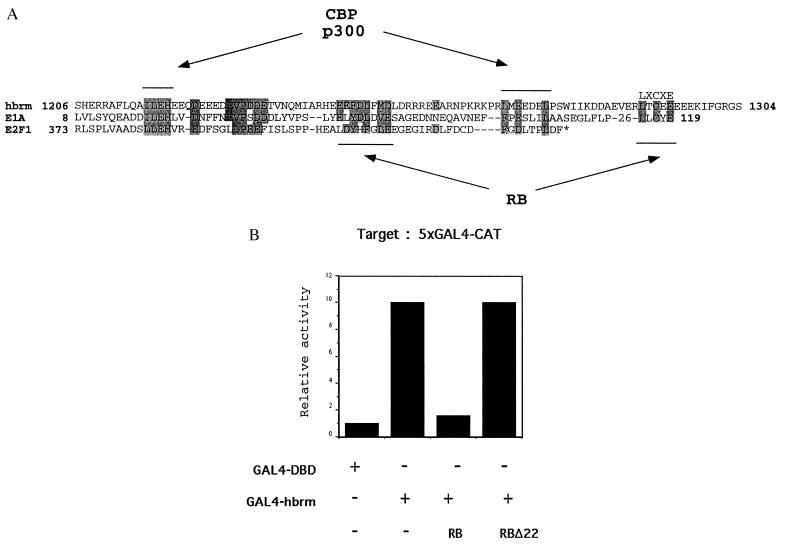
RB has the capacity to repress basal transcription when tethered to the promoter (18, 19). This result suggests that RB may negatively regulate the functions of a protein(s) present on most promoters. Given that hbrm may participate in activation of many genes, we hypothesized that one of the targets for RB-induced repression of promoter activity may be hbrm. This lead us to examine the sequences around the LXCXE motif in hbrm for any clues as to how RB may regulate the activity of hbrm. This lead to the observation that sequences directly N-terminal to the LXCXE motif possess similarity to sequences found in the related activation domains of E2F1 and E1A (37). Fig. 4A shows that the part of the sequence similarity overlaps the RB binding residues in E1A CR1, and in E2F1 as well as one of the p300/CBP binding sites characterized in E1A CR1. This observation raised the possibility that this region of hbrm may represent a transcription activation domain. To test this hypothesis, we took a 98-residue region of hbrm, which contains the E2F1/E1A homology and the characterized LXCXE-motif binding site for RB, and linked it to the GAL4 DNA binding domain to test for activation capacity. Fig. 4B shows that indeed this small region of hbrm has the ability to activate transcription of a GAL4-bearing promoter. Given that this region contains an LXCXE motif and possibly an additional RB binding motif related to that in E2F1, we tested the ability of RB to repress its activity. As seen in Fig. 4B, the activation capacity of hbrm is dramatically repressed by the presence of RB. This repressive capacity is dependent on an intact RB-pocket domain because a pocket mutant RB Δ22 is unable to repress hbrm. These results indicate that RB has the potential to repress transcriptional activation induced by the E7 domain of hbrm.
Figure 4.
hbrm activation domain shows homology to the activation domain of E2F1 and E1A 12S protein. (A) Alignment of amino acids 1,206–1,304 of the hbrm protein with the Ad12 E1A N terminus (amino acids 8–119) and the E2F1 activation domain (amino acids 373–437). Residues involved in RB and p300 binding in E1A and E2F1 are indicated. (B) RB binds and represses an activation domain in hbrm. C33A cells were transfected with 2 μg of G5E1B-CAT, 0.5 μg of GAL4 DNA binding domain, or GAL4-hbrm 1206–1304 fusion protein expression vectors and 0.5 μg of RB expression vectors as indicated. The activity of the promoter in the absence of hbrm and RB is normalized to a value of 1. Result of a typical experiment is shown.
DISCUSSION
In this report, we provide evidence that in the presence of RB, the hbrm protein has the capacity to repress the activation functions of the E2F1 transcription factor. This cooperativity is likely to be the result of the direct interaction between RB and hbrm for several reasons: (i) hbrm sequences containing the RB-binding LXCXE motif are required for the cooperation, (ii) RB has the capacity to contact both hbrm and E2F1 simultaneously, (iii) in vivo hbrm is found complexed with E2F1.
In the cell, hbrm is found in association with a set of other proteins that together make up the SWI/SNF chromatin remodeling complex. We do not know whether hbrm is part of this complex when it cooperates with RB to repress E2F1, or whether hbrm has functions outside SWI/SNF. Our results suggest that some of the properties of hbrm that are observed, while it is part of SWI/SNF, such as the NTPase activity, is required for its ability to repress E2F1. In addition, immunoprecipitation of hbrm can coimmunopurify other components of SWI/SNF. We therefore have to entertain the possibility that hbrm may be part of the SWI/SNF complex during its repression of E2F1 activity.
Coactivator functions are usually assigned to hbrm/BRG-1 and more generally to the SWI/SNF complex. However, we show here that hbrm can act as a specific corepressor of E2F1. Furthermore, the glucocorticoid receptor and E2F1 are paradoxically regulated in an opposite way by RB and hbrm. RB and hbrm collaborate to enhance activation by the glucocorticoid receptor (34) and for repressing E2F1 activity (this paper).
One explanation for this paradox could be that hbrm is part of distinct complex that represses transcription when it interacts with E2F1-RB. Consistent with this hypothesis is the fact that different SWI/SNF complexes exist in mammalian cells (43). Interestingly, RB has been shown to be capable of repressing transcription when tethered to the promoter via a GAL4 DNA binding domain (18, 19). These intrinsic repressive functions suggest that RB may negatively regulate some activity which is present on most promoters. This activity may be that of hbrm, when it is part of SWI/SNF. The SWI/SNF complex is required for the activity of a number of promoters in yeast and is found under certain conditions in the holoenzyme (32). These data suggest that SWI/SNF may be part of the “promoter complex.” Therefore, it is a possibility that RB can repress promoters by repressing the activity of hbrm. The fact that RB binds and represses an activation domain within hbrm is consistent with such a scenario.
Alternatively, the opposite functions of hbrm on the glucocorticoid receptor and on E2F1/RB could both reflect an increased binding to DNA of these factors, due to the nucleosome remodelling activity of the SWI/SNF complex. It is known that the SWI/SNF complex can help transcription factors such as GAL4 to bind in vitro to nucleosomal DNA (30). That mechanism has been proposed to explain the coactivator functions of the SWI/SNF complex observed in vivo on activating sequence-specific transcription factors, such as the glucocorticoid receptor (25). The E2F1/RB transcription factor is a repressor of transcription (17). By helping it to bind DNA, the SWI/SNF complex would be a corepressor of E2F1, as we observed in cotransfection experiments (Fig. 3). If that hypothesis was correct, RB would inhibit the transcription machinery by a mechanism independent of the SWI/SNF complex, but the presence of the SWI/SNF complex would be required for a stronger binding of RB to the promoter.
Repression of E2F-containing promoters by RB is considered to be one of the key mechanisms by which RB induces G1 arrest. The ability of hbrm to cooperate with RB in the repression of E2F1 may therefore be an underlying mechanism for the observed cooperation between RB and hbrm in the induction of G1 arrest. The LXCXE motif RB binding region of hbrm is also important for hbrm G1-arrest functions (24, 35). Indeed, in yeast cells there is a highly related hbrm homologue, SNF2, that is lacking the LXCXE motif, consistent with the fact that yeast cells do not possess RB or E2F1 proteins.
Acknowledgments
This work was supported by Medical Research Council (G930179 3MB) and Cancer Research Campaign (2081/0302) grants. C.L.C. was supported by a European Community Training and Mobility of Researchers fellowship.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- CMV
cytomegalovirus
- HA
hemagglutinin
References
- 1.Weinberg R A. Science. 1991;254:1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 3.Taya Y. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 4.Dyson N, Guida P, Munger K, Harlow E. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin X, Chittenden T, Livingston D M, Kaelin W G. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 6.Ewen M E, Xing Y G, Lawrece J B, Livingstone D M. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 7.Welch P J, Wang J. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 8.Kouzarides T. Semin Cancer Biol. 1995;6:91–98. doi: 10.1006/scbi.1995.0012. [DOI] [PubMed] [Google Scholar]
- 9.Adams P D, Kaelin W G. Semin Cancer Biol. 1995;6:99–108. doi: 10.1006/scbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 10.Johnson D G, Schwartz J K, Cress W D, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 11.Johnson D G, Cress W D, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1994;91:12823–12827. doi: 10.1073/pnas.91.26.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh P, Wong S H, Hong W. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam E, Lathangue N B. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 14.Slansky J E, Farnham P J. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 15.Helin K, Wu C L, Fattaey A R, Lee J A, Ngwu B D D, Harlow E. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 16.Flemington E K, Speck S H, Kaelin W G. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weintraub S J, Prader C A, Dean D C. Nature (London) 1992;328:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 18.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weintraub S J, Cjow K N, Luo R X, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 20.Cavanaugh A H, Hempel W M, Rogalsky L J T, Todorov G, Rothblum L. Nature (London) 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 21.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Nature (London) 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 22.Qin X Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krek W, Xu G, Livingston D M. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 24.Dunaief J L, Strober B S, Guha S, AKhavari P, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 25.Muchardt C, Yaniv M. EMBO J. 1993;11:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. Nature (London) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 27.Chiba H, Muramatsu M, Nomoto A, Kato H. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson C L, Tamkun J W. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L. EMBO J. 1996;15:5370–82. [PMC free article] [PubMed] [Google Scholar]
- 30.Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 31.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M E. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 32.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 33.Reyes, J. C., Muchardt, C. & Yaniv, M. (1997) J. Cell Biol. 137, in press. [DOI] [PMC free article] [PubMed]
- 34.Singh P, Coe J, Hong W. Nature (London) 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 35.Strober B E, Dunaief J L, Guha S, Goff S P. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin K, Trouche D, Hagemeier C, Sorensen T, Lathangue N B, Kouzarides T. Nature (London) 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 37.Trouche D, Kouzarides T. Proc Natl Acad Sci USA. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muchardt C, Sardet C, Bourachot B, Onufyk C, Yaniv M. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helin K, Lee J A, Vidal M, Dyson N, Harlow E, Fattaey A. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 40.Kaelin W K, Krek W, Sellers W R, Decaprio J A, Ajcenbaum F, Fuchs C S, Chittenden T, Farnham P J, Blanar M A, Livingston D M. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 41.Helin K, Harlow E, Fattaey A. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiebert S W, Chellapan S P, Horowitz J M, Nevins J R. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]