Abstract
Classical Hodgkin lymphoma (HL) is a malignant disorder characterized by the presence of neoplastic mononucleated Hodgkin and multinucleated Reed-Sternberg cells. Here, we show that both the interleukin (IL)–21 receptor as well as IL-21 are expressed by HL cells. IL-21 activates signal transducer of activation and transcription 3 (STAT3) and STAT5 in HL cell lines and activated human B cells. Ectopic expression of constitutively active STAT5 in primary human B cells resulted in immortalized B cells that have lost the B-cell phenotype and strongly resembled HL cells, which could partially be rescued by ectopic expression of the B cell–determining transcription factor E47. Data from experiments using reporter assays and overexpression of constitutively active IKK2 support the hypothesis that the STAT5 and nuclear factor-κB (NF-κB) pathways colaborate in HL genesis.
Introduction
Hodgkin lymphoma (HL) is characterized by the presence of a malignant cell population consisting of mononucleated Hodgkin cells and multinucleated Reed-Sternberg cells.1,2 A large variety of secondary molecular aberrations have been defined for HL; however, no HL-specific primary transforming event has been identified. The origin of these cells was elucidated only recently when Küppers et al found that HL cells express rearranged immunoglobulin genes.3 This indicated that HL cells are of B-cell origin, although they often lack typical B-lineage markers such as CD20, cell surface B-cell receptor (BCR), and CD79a.1,4 The detection of somatic hypermutations and the presence of crippled immunoglobulin (Ig)5 indicate that germinal center (GC) B cells are the precursors of HL cells. It is now assumed that GC B cells that escaped negative selection and acquired survival and proliferative advantages are the precursor of HL cells.2 The B cell–specific transcription program appears to be silenced in HL cells, resulting not only in abolition of Ig gene expression but also in silencing of B cell–specific factors that play essential roles in the GC stages of B cell development and maturation.6–8 The abolition of Ig gene expression may also be caused by epigenetic silencing.9 Recent evidence has shown that basic helix-loop-helix transcription factor E47, encoded by the E2A gene, which is essential for B-cell development,10–12 is inactivated in HL cells due to a combination of down-regulation of E2A gene expression and up-regulation of the E2A antagonists Id2 and the transcriptional repressor ABF-1.11 E47 is involved in establishing B-cell identity by regulating expression of a number of B cell–specific proteins and BCR.10,11 A conspicuous feature of HL cells is the aberrant expression of markers that are specific for cell types other than B cells. For instance, these cells express the myeloid cell marker CD15,13 the activation marker CD30,14 and the T-cell lineage transcription factor GATA3.15
HL cells display constitutively nuclear localization and activation of nuclear factor-κB (NF-κB).16,17 The presence of mutations in NFKBIA, the gene encoding IκBα, in 30% of primary Hodgkin lymphomas and NF-κB/REL locus amplifications suggests that genetic alterations that lead to continued activation of NF-κB play a crucial role in the transformation process of classical HL.5,18 However, constitutive activation of NF-κB can also be observed in other B-cell malignancies, making it unlikely that constitutive activation of NF-κB is the sole cause of HL genesis.19 Other transcription factors that might be involved in HL are the signal transducers of activation and transcription (STAT) proteins. STAT3,20 STAT5,21 and STAT622 have been shown to be continuously activated in HL cells. A possible role of STATs, in particular STAT3 and STAT5, in genesis of B-cell tumors has generated interest because these factors have been shown to play a role in control of proliferation and differentiation in various stages of normal B-cell development.23–25 STAT3 appears to be involved in plasma cell differentiation by regulating Blimp-1, which is essential for plasma cell differentiation.24,26,27 Recently we found evidence that STAT5 is involved in human memory B-cell development and self-renewal.25 Interleukin (IL)–21, which was recently identified as a major B-cell growth and differentiation factor,28,29 activates both STAT3 and STAT5. Upon ligand binding, IL-21Rα associates with the γc chain, a property it shares with the receptors for IL-2, IL-4, IL-7, IL-9, and IL-1530 leading to activation of JAK1, JAK3, STAT1, STAT3, and STAT5.28,30 In view of the prominent B-cell growth activities of IL-21, we have examined whether IL-21 and its receptor are expressed in HL cells and addressed the question whether one or both of the downstream targets STAT3 and STAT5 are involved in the genesis of HL.
Methods
Patient material and B-cell isolation
Tumor material was obtained from the Department of Pathology of the AMC in Amsterdam, The Netherlands. All lymphomas were classified according to the WHO classification criteria31 and judged at least by 2 pathologists. B cells were obtained from tonsils or peripheral blood (PB) of adults. The use of human tissue was approved by the Medical Ethical Committee of the AMC and was contingent on informed consent obtained in accordance with the Declaration of Helsinki. The cell sorting was done as described25 using a FacsARIA (Becton Dickinson, San Jose, CA).
Retroviral constructs, production of recombinant retrovirus, and retroviral transduction
The mammalian expression vector LZRS containing CA-STAT5 and CA-STAT5bER was previously described.25,32 CA-IKK2 was obtained from J. J. Schuringa (University of Groningen, Groningen, The Netherlands). IκBαSR was obtained from M. E. Poynter (University of Vermont, Burlington, VT). We obtained wild-type (WT) human STAT3 from E. Caldenhoven (Erasmus University, Rotterdam, The Netherlands) and generated Lazarus (LZRS)-WT-STAT3-IRES-GFP. Human WT STAT3 was fused with C-terminus of estrogen receptor (ER). The pSUPER and pRS constructs for STAT5 were described previously.25 Retrovirus was made with Phoenix-A cell line. Transduction of primary B cells was performed as described previously.25
Immunoblotting and nuclear p65 measurement
Immunoblotting was done as previously described.25 Primary antibodies used were anti-BCL6 (C-19), anti-STAT5 (C-17), anti-STAT3 (C-20), and antiactin (I-19) (all from Santa Cruz Biotechnology, Heidelberg, Germany), anti-PKC (Upstate, Huissen, The Netherlands), anti-IκBα, anti-pSTAT3(Tyr705), anti-pSTAT5(Tyr694), and antitubulin (11H10) (all from Cell Signaling Technology, Beverly, MA). Nuclear NF-κB p65 was measured using an enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's protocol (IMGENEX, San Diego, CA) with equal amounts of nuclear protein.
Immunohistochemistry
Immunohistochemistry for IL-21 was performed on paraffin-embedded tumor sections from HL patients using a rabbit polyclonal anti–IL-21 Ab (0.5 μg/mL; eBioscience, San Diego, CA). Functionality of the anti–IL-21 Ab was established on paraffin-embedded tonsil tissue as described.33 IL-21 staining was developed using the CSA-II kit (Dako Cytomation, Glostrup, Denmark) with DAB detection, and tissue was counterstained with hematoxylin.
Cell culture and reagents
B cells were cultured in complete medium at 37°C in humidified air containing 5% CO2. Irradiated CD40L-L cells were seeded at a density of 5 × 104 cells per well in 24-well tissue culture treated plates (Costar, Badhoevedorp, The Netherlands). Sorted B cells (5 × 105) were added together with IL-2 (20 U/mL) and IL-4 (10 ng/mL; R&D Systems, Minneapolis, MN). Phoenix, JY, Ramos, Ly-7, L428, and L1236 were maintained in IMDM supplemented with 10% FBS and antibiotics. The IL-21R-Fc chimeric protein was obtained from R&D Systems.
Luciferase reporter transactivation assay
The L1236 cells were transiently electroporated. Each electroporation was done in duplicate and luciferase activity was measured 3 days after transfection with the Dual-Luciferase Reporter Assay Kit (Promega Benelux, Leiden, The Netherlands) according to the manufacturer's protocol. Normalized values are reported as the means (± SD) from 3 independent transfections.
Flow cytometry
Monoclonal antibodies (all IgG1 unless otherwise indicated) to the human molecules CD3 (SK7), CD15 (80H5), CD19 (HIB19), CD20 (2H7, IgG2b), CD30 (Ber-H83), CD45 (2D1), CD56 (B159), CD70 (Ki24, IgG3), CD79b (CB3.1), D95 (DX2), CD123 (7G3), CD132 (AG184), and HLA-DR (L243; IgG2a; BD-Pharmingen, San Diego, CA) were directly labeled with FITC, PE, or APC. κ-Light chain (clone A8B5), λ-light chain (rabbit polyclonal), and CD138 (clone MI15), directly labeled with PE (Dako-Cytomation, Glostrup, Denmark), were used for flow cytometric analysis. Intracellular staining for pSTAT5 and pSTAT3 was done according to the manufacturer's instructions (BD-Pharmingen), using an Alexa-647–conjugated monoclonal antibody specific for pSTAT5 (Y694, clone 47, IgG1; BD), pSTAT3 (pY705, clone 4, IgG2a; BD-Pharmingen), or a control antibody. Stained cells were analyzed with a LSR II (BD-Immunocytometry Systems, San Jose, CA) and flow cytometric data were processed with CellQuest computer software (BD-Immunocytometry Systems).
Microdissection of HL cells, RNA isolation, and amplification
Microdissection of HL cells, RNA isolation, and amplification were done as previously described.34 The reliability of the amplification step was confirmed by comparing gene expression profile of nonamplified peripheral blood mononuclear cells (PBMCs) RNA and T7 promoter-based RNA amplification.34
RT-PCR
Reverse-transcription–polymerase chain reaction (RT-PCR) was performed on total cDNA using AmpliTaq gold polymerase (Roche, Almere, The Netherlands). The following primers were used: actin forward, 5′-GGATGCAGAAGGAGATCACTG-3′ and actin reverse, 5′-CGATCCACACGGAGTACTTG-3′ (60°C, 30 cycles); IL-21 forward, 5′-GAGGAAACCACCTTCCACAA-3′ and IL-21 reverse, 5′-CAGGAATCT-TCATTCCGTGT-3′ (60°C, 35 cycles); Gata3 forward, 5′-CTCATT-AAGCCCAAGCGAAG-3′ and Gata3 reverse, 5′-GCATTCCTCCTC-CAGAGTGT-3′ (60°C, 30 cycles); CD20 forward, 5′-AGCTAAT-CCCTCTGAGAAAAAC-3′ and CD20 reverse, 5′-CTGCTGACAGGA-GAACTATG-3′ (55°C, 40 cycles).
PCR primers for LMP-1, EBNA1/2, IPL, FER, PRAME, Rab13, and IL-13 have been described previously.25,35,36 Real-time RT-PCR analysis was done with an iCycler PCR (BioRad Laboratories, Veenendaal, The Netherlands). The iQ SYBR Green Supermix (BioRad) was used for amplification. After incubation at 95°C for 6 minutes, 40 cycles of amplification were done. Each cycle consisted of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. The primers used were as follows: actin forward, 5′-GGATGCAGAAGGAGATCACTG-3′ and actin reverse, 5′-CGATCCACACGGAGTACTTG-3′; E47 forward, 5′-GTCGGACAAAGCGCAGAC-3′ and E47 reverse, 5′-ACAGGCTGCTTTGGGATTCC-3′.
Results
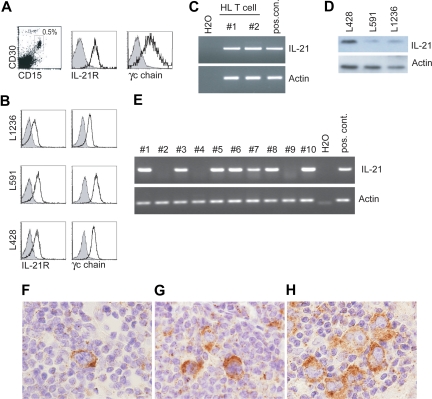
To determine whether the IL-21R was expressed on primary HL cells, we performed flow cytometric analysis using a single-cell suspension of primary HL. Figure 1A shows that cells coexpressing CD15 and CD30, which are highly specific markers for HL cells,13,14 expressed the IL-21R and the γc chain (CD132). Next we analyzed the expression of the IL-21R and the γc chain in the HL cell lines, L428, L591, and L1236. Similar to the ex vivo primary HL cells, both the IL-21R as well as the γc chain were expressed (Figure 1B).
Figure 1.
IL-21R, CD132, and IL-21 expression on HL cells. Flow cytometric analysis for IL-21R and γc chain (CD132) expression on (A) primary HL tissue and (B) HL cell lines, L1236, L591, and L428. For IL-21R expression on primary HL cells we gated on the CD30+CD15+ cells and γc chain expression is gated on CD15+ cells. Gray histogram is isotype control. Representative of 5 different donors. (C) RT-PCR for IL-21 and actin expression on HL-infiltrating sorted T cells (n = 2). (D) Immunoblot analysis for IL-21 and actin as a loading control was performed on 3 HL cell lines (n = 3). (E) RT-PCR for IL-21 and actin on cDNA generated from laser capture-isolated purified HL cells from 10 different donors (nos. 1-10). (F-H) Multinucleated HL cells stained positive for IL-21 protein by immunohistochemistry (brown staining). Stainings from 3 different representative patient tumor samples (of 6 patients tested) are shown. Immunohistochemistry for IL-21 was performed on paraffin-embedded tumor sections from HL patients using a rabbit polyclonal anti–IL-21 Ab (0.5 μg/ml; eBioscience). Functionality of the anti–IL-21 Ab was established on paraffin-embedded tonsil tissue as described.33 IL-21 staining was developed using the CSA-II kit (Dako Cytomation) with DAB detection; tissue was counterstained with hematoxylin. Slides were visualized on an Olympus BX51 light microscope (Olympus, Zoeterwoude, The Netherlands) using a UPlan/Apo 40×/0.85 objective and Olympus DP70 camera. Images were captured with Olympus DP Controller software version 1.2.1.108 and were processed with Adobe Photoshop 7.0. (Adobe Systems, San Jose, CA).
To determine whether HL-infiltrating T cells expressed IL-21, we sorted CD3+CD4+ T cells from a primary HL tumor by flow cytometry. Both patient samples tested expressed IL-21 mRNA as assessed by RT-PCR (Figure 1C). Upon analyzing the expression of IL-21 protein by immunoblot analysis in tumor cell lines, we unexpectedly observed that this cytokine is expressed in HL cell lines (Figure 1D). The presence of IL-21 protein in HL cell lines prompted us to examine the expression in fresh HL tumor samples. We first determined IL-21 mRNA expression in primary HL cells isolated by laser capture microdissection. Approximately 1000 HL cells from different HL cases were isolated, and subsequently the isolated RNA was amplified, as previously described.34 Seven of the 10 HL cases tested were positive for IL-21 mRNA (Figure 1E). Immunohistochemistry on 6 patients showed that HL cells are positive for IL-21 protein (Figure 1F-H). We could also detect IL-21 in the smaller cells, most likely T cells (Figure 1F-H). We confirmed that this antibody also detected IL-21 produced by CD4+ T cells activated by beads coated with anti-CD3 and anti-CD28 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
IL-21 activates both STAT3 and STAT5 in HL cell lines
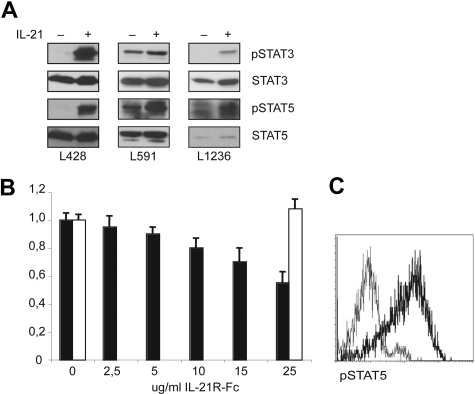
To determine whether IL-21 signaling was active in HL cell lines, we determined the effects of exogenous IL-21 on STAT3 and STAT5. The HL cells were first starved for 6 hours in serum-free medium and then incubated with exogenous IL-21 for 15 minutes. Two of the 3 cell lines, L428 and L1236, did not express tyrosine-phosphorylated STAT3 (pSTAT3) after serum starvation, and IL-21 induced pSTAT3 in these lines. Serum-starved L428 cells expressed pSTAT5 only after activation with IL-21. In contrast, L591 cells showed constitutive expression of pSTAT3 and pSTAT5, which were increased after addition of IL-21. Phosphorylation of STAT5 in starved L1236 cells was increased after incubation with IL-21 (Figure 2A). These data show that IL-21 either induced or increased the phosphorylation status of STAT3 and STAT5, which indicates that the IL-21R is functional in 3 analyzed HL cell lines. We next investigated the effects of endogenously produced IL-21 on the proliferation of HL cells. We cultured the HL cell lines L428 and L591 for 3 days with an IL-21R-Fc fusion protein, which blocks IL-21 signaling. Proliferation was measured by determining [3H] thymidine incorporation at 72 hours after treatment. The proliferation of L591 cells was suppressed to 40% of that of untreated control cells (Figure 2B). Treatment of the cell lines L428 and L1236 with the IL-21R-Fc fusion protein had no effect on the proliferation (Figure 2B and data not shown). These data raise the possibility that the relatively high level of expression of pSTAT3 and pSTAT5 in L591 cells may be due to triggering of the IL-21 receptor by endogenously produced IL-21. The finding that IL-21 activated both STAT3 and STAT5 in HL cell lines is in line with previous observations that (nonstarved) HL cell lines expressed phosphorylated STAT3 and STAT5.21 We confirmed and extended these findings by showing that 5 samples of primary HL cells defined by the coexpression of CD15 and CD30 expressed high levels of pSTAT5 by flow cytometric analysis (Figure 2C). In contrast, 2 of the 4 primary HL samples tested shown negative for pSTAT3 (data not shown).
Figure 2.
Signaling through IL-21R activates STAT3 and STAT5. (A) Tyrosine phosphorylation of STAT3 and STAT5 upon IL-21 treatment. Immunoblot analysis for tyrosine-phosphorylated STAT3 (pSTAT3) and pSTAT5 was performed on the HL cell lines L1236, L591, and L428. As loading controls, blotting for total STAT3 and STAT5 was performed (n = 3). (B) Proliferation assay was performed on L591 (□) and L1236 (■) with different concentrations of the IL-21R-Fc protein to block IL-21 signaling. Proliferation was measured at 72 hours by [3H]-thymidine incorporation. The y-axis shows the proliferation of the treated culture as a percentage of the untreated control cells, calculated as the mean of triplicate samples (n = 3). (C) Flow cytometric analysis of pSTAT5 expression in cells from primary HL tissue. The pSTAT5 (bold line) histogram was obtained after gating on CD30+CD15+ cells, and the expression level was compared with an isotype control antibody gated on the CD30+CD15+ cells (thin line). Isotype control histogram gave similar results as when gated on the CD30−CD15− cells. Representative example of 5 different patients is shown.
Immortalization of primary B cells with CA-STAT5B
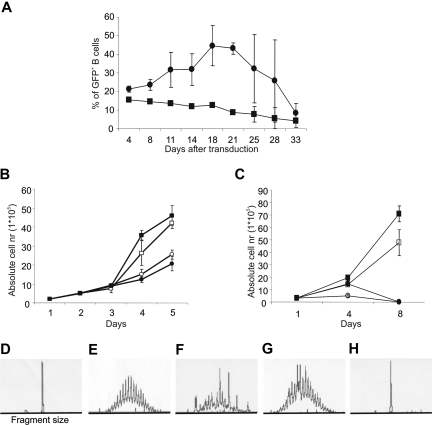
Next we investigated whether the downstream targets of IL-21, STAT3, and STAT5 might be involved in generation of HL. We previously documented that constitutive active STAT5a (CA-STAT5a) or CA-STAT5b mutants and inducible STAT5bER mutant in human primary B cells resulted in a significant extension of their replicative lifespan.25 Here we addressed whether constitutive activation of STAT3 would have similar effects on the proliferation of primary B cells. We cocultured purified tonsil CD19+ B cells with CD40L-expressing mouse fibroblasts (CD40L-L cells) in the presence of IL-2 and IL-4. At day 7, the B cells were transduced with retrovirus expressing STAT3ER upstream of an internal ribosomal entry site–GFP (IRES-GFP) cassette and cultured these cells in the presence of CD40L-L cells, with IL-2 and IL-4, and in the presence or absence of tamoxifen for 5 weeks. The activation of STAT3ER upon addition of tamoxifen to STAT3ER–transduced B cells led to a brief increase in frequency (Figure 3A) and numbers (data not shown) of the transduced cells, but did not lead to long-term proliferation of the transduced B cells. The failure of tamoxifen to induce proliferation of the STAT3ER-transduced B cells was not due to functional inactivity of the construct since addition of tamoxifen to STAT3ER–transduced B cells resulted in induction of Blimp-1 and strong differentiation into antibody-producing cells.27 Following our observation that CA-STAT5a or CA-STAT5b mutants and inducible STAT5bER mutant in human primary B cells resulted in a significant extension of their replicative lifespan in the presence of cytokines,25 we asked whether proliferation of CA-STAT5b+ B cells required cytokines and CD40L-L cells. Therefore we cultured CA-STAT5b+ cells obtained from tonsil or peripheral blood (PB) with or without cytokines and with or without CD40L-L cells. Both tonsil and PB CA-STAT5b+ B cells proliferated independently of cytokines (Figure 3B), indicating that the constitutive activation of STAT5 was sufficient to replace the signal induced by IL-2 and IL-4. Unexpectedly, we observed that CA-STAT5b+ tonsil B cells could proliferate independently of CD40 ligation, while PB B cells proliferated without cytokines, but remained CD40L dependent (Figure 3C). Both the CA-STAT5b+ PB B cells cultured with CD40L-L cells in the absence of cytokines and tonsil B cells cultured without cytokines or CD40L-L cells could be maintained for periods of more than 6 months. All cytokine and CD40L independently proliferating CA-STAT5b+ B-cell tonsil lines were Epstein-Barr virus (EBV) negative as determined by the absence of LMP-1, EBNA1, and EBNA2 (Figure 7). PB and tonsil B cells transduced with CA-STAT5b remained polyclonal when cultured on CD40L-L cells as determined with PCR-based genescan analysis of IgVH gene segments37 (Figure 3D-H). However the cells became monoclonal following culture in the absence of CD40L-L cells (Figure 3D-H). The monoclonal outgrowth of CA-STAT5b+ B cells from tonsil suggests that only B cells with additional mutations in genes important for survival and proliferation are able to proliferate in a CD40L-L cell–independent manner.
Figure 3.
STAT5, but not STAT3, induces proliferation in human B cells. (A) Total tonsil CD19+ B cells were transduced with LZRS-control-IRES-GFP (■) or LZRS-STAT3ER-IRES-GFP (●) and cultured on CD40L-L cells with IL-2 and IL-4 in the presence of tamoxifen (1 μM). The percentage of GFP-positive cells was determined continuously throughout the culture period. Data represent means (± SD) of 2 independent experiments. (B) Absolute numbers in time of CA-STAT5b+ tonsil B cells cultured in the presence or absence of CD40L-expressing L cells with or without IL-2 and IL-4. ■ represent plus CD40L plus cytokines; □, plus CD40L minus cytokines; ●, minus CD40L plus cytokines; and ○, minus CD40L minus cytokines. Data represent means (± SD) of 3 independent experiments. Identical results were obtained with 8 different donors. (C) Absolute numbers in time of CA-STAT5b+ PB B cells cultured in the presence or absence of CD40L-expressing L cells and with or without IL-2 and IL-4 as indicated in panel B. Data represent means (± SD) of 4 independent experiments. Genescan analysis for VH FR3-JH region. (D) L428: positive control for monoclonality. (E) Tonsil B cells: positive control for polyclonality. (F) CA-STAT5b+ tonsil B cells cultured on CD40L-expressing L cells. (G) CA-STAT5b+ PB B cells cultured on CD40L-expressing L cells. (H) CA-STAT5b+ tonsil B cells cultured without CD40L-expressing L cells. Analysis of the VH FR1-JH and VH FR2-JH region gave identical results. Similar results were obtained with 4 other tonsil CA-STAT5b+ B-cell lines and 2 other PB CA-STAT5b+ B-cell lines.
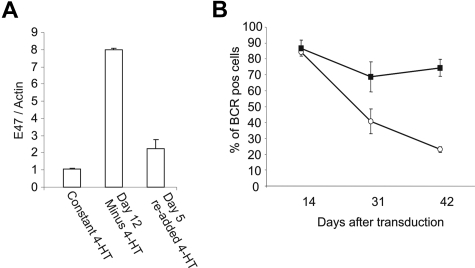
Figure 7.
E47 can rescue the downmodulation of the BCR induced by CA-STAT5b. (A) E47 expression is repressed upon STAT5bER activation as assessed by real-time RT-PCR analysis (n = 3). (B) Flow cytometric analysis of the BCR determined by the surface expression of kappa and lambda light chains. PB B cells double transduced with CA-STAT5b plus control GFP (■) or CA-STAT5b plus E47 (○) cultured on CD40L-L cells plus IL-2 and IL-4. Data represent means (± SD) of 3 independent experiments.
Ectopic expression of CA-STAT5 leads to down-regulation of the BCR
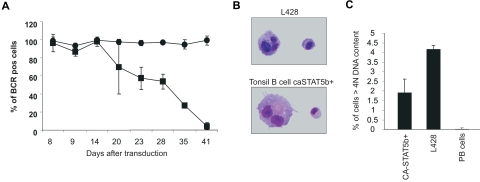
One of the hallmarks of HL is the lack of BCR expression. We therefore determined whether ectopic expression of CA-STAT5b had an effect on the BCR. The data in Figure 4A clearly show that the BCR was gradually down-regulated in CA-STAT5b–transduced B cells during the first 6 weeks of culture. The reduction in surface BCR expression was not a result of differentiation into plasma cells as the CA-STAT5b+ B cells that became BCR negative were unable to secrete Ig (data not shown) and failed to express the plasma cell marker CD138 (Figure 5).
Figure 4.
Ectopic expression of CA-STAT5b leads to BCR multinucleated cells. (A) Surface BCR expression on CA-STAT5b–transduced (■) or control-transduced (●) PB B cells was determined in time by flow cytometric analysis of surface of kappa and lambda light chain expression. Cells were cultured on CD40L plus IL-2 and IL-4. Data represent means (± SD) of 3 independent experiments. Identical results were obtained with 12 different donors. Similar results were obtained with tonsil (n = 6). (B) Cytospins of CA-STAT5b+ tonsil B cells and L428 were prepared, and images in Figure 4B were visualized and captured on an Olympus BX51 light microscope with a 100×/1.3 oil objective. Giemsa staining was performed to visualize the nuclei of the cells at a total magnification of 1000×. The presence of mononucleated and multinucleated cells strongly resembled the Hodgkin and Reed-Sternberg cells, respectively (n = 6). (C) To enumerate the percentage of cells with multiple nuclei, DNA content was measured using propidium iodide staining and flow cytometric analysis. Single cells were gated and cells with more than 4N DNA were considered to be cells with multiple nuclei. Data represent means (± SD) of 3 CA-STAT5b+ tonsil B-cell lines in comparison with L428 and freshly isolated PB B cells.
Figure 5.
Comparison of CA-STAT5b+ B cells and HL cell lines. (A) Flow cytometric analysis of L428 and CA-STAT5b+ tonsil B cells. Thin lines are matched isotype controls. Data are from 1 representative experiment of 5 originated from different donors. (B,C) Comparison of expression of a number of genes in CA-STAT5b+ B cells and HL cell lines. The EBV-transformed B-cell line JY, the GC-type diffuse large B-cell lymphoma cell line LY-7, and the Burkitt lymphoma cell line Ramos served as negative controls, and 2 HL cell lines L428 and L1236 served as positive controls. (B) Four different CA-STAT5b+ tonsil B-cell lines cultured with CD40L-expressing L cells were analyzed. (C) PB and tonsillar B cells transduced with CA-STAT5b were cultured with or without CD40L-expressing L cells and analyzed for EBV- and HL-specific gene expression.
Presence of large multinucleated cells in CA-STAT5+ B cells
We examined the CA-STAT5+ B cells for the presence of multinucleated cells, another crucial hallmark of HL. Giemsa staining on cytospin preparations of CA-STAT5b+ B cells revealed the presence of multinucleated cells both in CA-STAT5b+ and as a positive control, the HL cell line L428 (Figure 4B). The presence of such multinucleated cells was not unique for tonsil CA-STAT5b+ B cells since similar results were obtained with PB B cells transduced with CA-STAT5b (Figure S2). Flow cytometric analysis of DNA content using propidium iodide (PI) staining demonstrated that the proportion of multinucleated (> 4n) cells was comparable with what we found in the HL cell lines tested (Figure 4C).
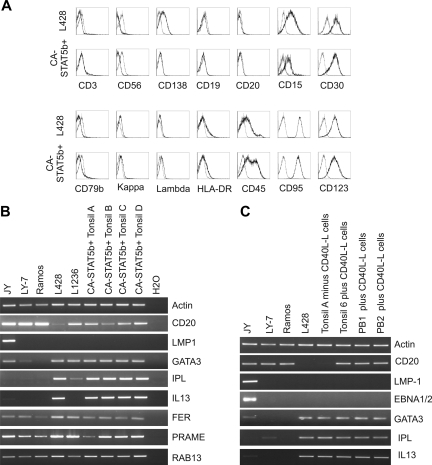
Cell surface phenotypes of CA-STAT5b+ B cells and HL cells are similar
We next compared the phenotype of CA-STAT5b+ tonsil B cells with that of well-characterized HL cell lines (Figure 5A). None of CA-STAT5b–generated cell lines expressed the T and natural killer (NK) lineage markers cells CD3 or CD56. Whereas B cell–specific markers CD20, CD79b, and Ig kappa and lambda light chains were expressed on the B cells before transduction with CA-STAT5b (data not shown), these antigens were absent on the CA-STAT5b+ B cells (Figure 5A), in agreement with previous studies.8,38 Furthermore, the expression of HLA-DR, CD45, FAS (CD95), IL-3Rα (CD123), and γ common chain (CD132) on CA-STAT5b–transduced B cells and HL cells was comparable. Importantly, like HL cells, CA-STAT5b+ B cells coexpressed CD30 and the myeloid marker CD15, which is a conspicuous characteristic for HL.13,14 A similar phenotype was observed in CA-STAT5b+ PB B cells cultured in the presence of CD40L-L cells with one exception. CA-STAT5b+ PB B cells and CA-STAT5b+ tonsil B cells expanded with CD40L-expressed CD19, whereas this antigen was absent on CA-STAT5b+ tonsil B cells proliferating in the absence of CD40L and on HL cell lines (Figure 5A and data not shown).
Limited gene expression profile of CA-STAT5b+ B cells is similar to that of HL cells
Genetic profiling of HL cell lines and other B-cell tumors revealed a number of genes specifically expressed in HL: IPL, FER, PRAME, Rab13, T cell–associated gene GATA3,36 and IL13.35 Using RT-PCR, we determined the expression of these genes in CA-STAT5b+ tonsil B-cell lines and 2 HL cell lines (L428 and L1236); the EBV-transformed B-cell line (JY); and the GC-type diffuse large B-cell lymphoma line (LY-7); the Burkitt lymphoma line Ramos served as control (Figure 5B). Both HL and CA-STAT5b+ tonsil B-cell lines expressed CD20 transcripts consistent with their B-cell nature. The observation that CD20 protein was undetectable by flow cytometric analysis may imply that the expression of the protein was too low to detect or that the translation of CD20 is inhibited in these cells. GATA3, IPL, and IL-13 mRNAs were specifically expressed in the CA-STAT5b+ tonsil B cells and in the HL cell lines, although IL13 was not expressed by one HL cell line L1236. FER, PRAME, and Rab13 were present in both HL cell lines and CA-STAT5b+ B cells but also expressed in JY, LY-7, and Ramos, albeit at lower levels. A similar gene expression profile was found in CA-STAT5b+ tonsil and PB B cells cultured in the presence of CD40L-L cells (Figure 5C). Since BCL6 is a direct target of STAT5 in B cells,25 we determined the BCL6 protein levels. Both the CA-STAT5b–transduced B-cell lines as well as the HL cells lines expressed similar levels of BCL6 protein (Figure S3). Taken together, our data indicate that CA-STAT5b+ B cells strongly resemble the HL cell lines.
Proliferation of CA-STAT5b+ and HL cell lines depends on active NF-κB, which is partially controlled by STAT5
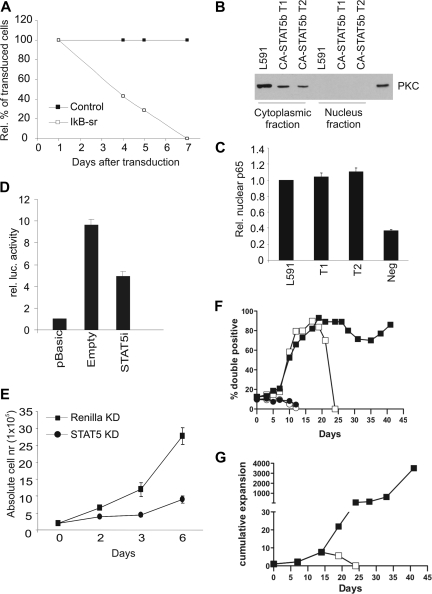
It is well documented that CD40 signaling results in NF-κB activation.39 The fact that the CA-STAT5b+ tonsil B cells could be cultured without CD40L-L cells suggested that proliferation of these cells could be driven by constitutive activation of the NF-κB pathway. To test the role of NF-κB in proliferation of CA-STAT5b+ tonsil B cells, we introduced the NF-κB superrepressor IκBSR40 in an IRES-ΔNGFR cassette into these cells. We observed that the percentage of cultured IκBSR-transduced cells decreased in time, whereas the percentage of control ΔNGFR-transduced B cells remained stable (Figure 6A). Ectopic expression of IκBSR indeed led to higher IκB protein levels as assessed in IκBSR-transduced L591 cells (Figure S4). To extend the findings of the IκBSR overexpression, we determined whether p65, which is part of the NF-κB complex, was present in the nucleus. The HL cell line and the 2 CA-STAT5b+ tonsil B cells showed similar p65 levels in the nuclear fractions as determined by a p65 ELISA (Figure 6C). The isolated nuclear fractions were not contaminated with the cytosolic fraction since no PKC protein, known to be present only in the cytosol, could be detected by immunoblot (Figure 6B). Together these data indicate that CA-STAT5b+ tonsil B cells exhibit constitutively active NF-κB, thereby allowing for proliferation of tonsil B cells in the absence of CD40L stimulation. These data suggest a link between STAT5b and NF-κB in HL. Indeed, it has been reported that activated STAT5 can control NF-κB activity in the pro-B-cell line Ba/F3.41 To determine whether NF-κB–dependent transcriptional activity could be affected by modulation of STAT5 we cotransfected HL cells with a NF-κB-luciferase reporter construct together with a vector containing STAT5 siRNA or with a control siRNA vector. We observed a 47% (± 7%, n = 3) reduction of NF-κB activity in HL cells when STAT5 was knocked down compared with the control, suggesting that STAT5 contributes to the activation of NF-κB in HL (Figure 6D). As a control, we determined whether STAT5 RNAi affected a signal transduction pathway different from the NF-κB pathway. We used the retinoic acid receptor signal transduction pathway using a retinoic acid receptor element (RARE) luciferase reporter vector as a readout. In Ramos cells, STAT5 RNAi, which resulted in reduced STAT5 protein levels, did not influence the retinoic acid signal transduction pathway (Figure S7). These data support the notion that STAT5 does not mediate nonspecific inhibitory effects and hence validates the effect that STAT5 positively regulates NF-κB activity. Next we determined the effects of decreased STAT5 levels on the proliferation. Figure 6E shows that HL cells stably expressing siRNA targeting STAT5, which efficiently knocked down STAT5 expression (Figure S5), proliferated much less than control-transduced cells.
Figure 6.
STAT5 inhibits NF-κB signaling and proliferation in HL cells. (A) Percentage of IkBSR-IRES-ΔNGFR (□) or control ΔNGFR (■) retrovirally transduced CA-STAT5b+ tonsil B cells over time (n = 3). (B) Purity of the nuclear protein fraction of L591 and 2 CD40L-independent CA-STAT5b tonsil B-cell lines was tested by PKC expression. (C) p65 ELISA was performed on the nuclear fractions using equal protein loading. The values obtained from the nuclear lysate from L591 were set as one (n = 2). Negative control is not incubated with protein lysate. (D) L1236 cells were electroporated with control luciferase vector (pBasic) or a luciferase vector with a NF-κB–responsive element in combination with a pSuper vector that knocks down STAT5 using siRNA or an empty control. (n = 3). At 72 hours, firefly luciferase activity was measured, normalized to the cotransfected Renilla luciferase activity. L428 gave similar results. (E) L428 was retrovirally transduced with a GFP-marked vector expressing siRNAs targeting either STAT5 (●) or Renilla (■) as a control. The absolute numbers of GFP-purified cells over time are depicted. Data represent means (± SD) of 3 independent experiments. Similar results were obtained using another HL cell line L1236. (F) Percentage of double-transduced CA-STAT5bER-IRES-ΔNGFR plus CA-IKK2-IRES-GFP cells (□ ■) or CA-STAT5bER-IRES-ΔNGFR plus control GFP cells (○ ●) in time, cultured in the absence of CD40L and in the presence (● ■) or absence (○ □) of tamoxifen. (G) Cumulative expansion of double-transduced CA-STAT5bER/CA-IKK2 cells cultured in the presence (●) or absence (■) of tamoxifen. For panels F and G, 1 representative of 2 experiments is shown.
To determine whether activation of NF-κB is required for the survival and proliferation of the monoclonal CA-STAT5+ tonsil B cells lines, we made use of a constitutively active IKK2 (CA-IKK2) mutant that leads to constitutive NF-κB signaling.42 As shown in Figure 3C, CA-STAT5b+ PB B cells required CD40L-L cells to survive and proliferate. For this reason, we transduced PB B cells with CA-STAT5bER-IRES-ΔNGFR and CA-IKK2-IRES-GFP or with control GFP. In addition, we introduced CA-IKK2 into B cells that were already transduced with CA-STAT5bER-IRES-GFP and cultured already for 2 months in the presence of tamoxifen. After transduction, the cells were cultured in the absence of CD40L-L cells in the presence or absence of tamoxifen. In both samples of cells, the percentage and the absolute numbers of double-transduced CA-STAT5bER plus control GFP cells cultured with or without tamoxifen did not increase (Figure 6F and data not shown). This was expected, as CD40 was not triggered. The percentage of CA-STAT5bER plus CA-IKK2 double-transduced cells cultured without tamoxifen increased in time, indicating some survival-inducing effect of CA-IKK2 in the absence of STAT5 activation, however this effect was transient and these cells eventually died. Most importantly, only the cells cultured with tamoxifen, thus with activation of both STAT5b and NF-κB, increased in absolute cell number (Figure 6G). Both samples of cells could be cultured for at least 2 months without CD40 ligation. To assess whether all CA-IKK2– and CA-STAT5bER–transduced cells expand under these conditions or whether clonal selection occurred, we determined the clonality of the double-transduced B cells in time by genescan analysis (Figure S6). The B cells that were cotransduced with CA-STAT5bER and CA-IKK2 and cultured with tamoxifen for 49 days were multiclonal, while the cells that were first transduced with CA-STAT5ER, cultured for 48 days, then transduced with CA-IKK2 and cultured for another 49 days (97 days in culture) had a much more restricted clonality. Together, these data indicate that expression of CA-IKK2 overcame the requirement for CD40 ligation for maintaining expansion of CA-STAT5bER–transduced B cells. The observation that long-term cultured CA-IKK2 and CA-STAT5bER double transduced cells have a restricted clonality suggests that continued activation of both STAT5 and CA-IKK2 is required for survival and expansion of primary B cells, but that additional mutations occurring in certain clones are required for full transformation.
Forced expression of E47 rescues the down-regulation of BCR
Recently it has been suggested that decreased E47 transcriptional activity is involved in the loss of the B cell–specific expression pattern of HL cells, including the lack of BCR expression.11 For this reason, we examined whether STAT5 influenced expression of E47. We assessed the expression of E47 by real-time RT-PCR in STAT5bER+ B cells cultured with CD40L-L cells plus IL-2 and IL-4 with tamoxifen and cells cultured for 12 days without tamoxifen to ensure inactivation of the ectopically expressed STAT5bER (Figure 7A). The expression level of E47 was decreased when STAT5bER+ B cells were cultured with tamoxifen compared with STAT5bER+ B cells that were deprived of tamoxifen for 12 days. Importantly, when tamoxifen was added back to the culture for 2 days, E47 expression decreased again. mRNA levels of other factors involved in E-protein signaling, Id2, Id3, and Abf-1, did not change upon STAT5bER activation (data not shown). Next, we determined whether the BCR down-regulation could be rescued by ectopic expression of E47. We double transduced primary B cells with CA-STAT5b-IRES-ΔNGFR plus control GFP or CA-STAT5b-IRES-ΔNGFR plus E47-IRES-GFP and cultured the cells with CD40L-L cells plus IL-2 and IL-4. As expected from the data in Figure 4A, B cells double transduced with CA-STAT5b plus control GFP lost the expression of the BCR (Figure 7B). In contrast, B cells double transduced with CA-STAT5b plus E47 remained BCR positive (Figure 7B). Similar to what has been described in HL cell lines, ectopic expression of E47 in CA-STAT5b–transduced B cells did not have an effect on proliferation. Furthermore overexpression of E47 did not influence the percentage of multinucleated cells and the expression of CD15 and CD30 (data not shown). Together these data demonstrate that STAT5 negatively influenced E47 at the transcriptional level and that the decreased BCR expression in CA-STAT5b+ B cells could be rescued by ectopic expression of E47.
Discussion
IL-21 is a major proliferation and differentiation factor for normal human B cells and might therefore be involved in survival and proliferation of malignant B cells as well. Here we show that ex vivo isolated HL cells express high levels of the IL-21R. It has been well documented that IL-21 is produced by T cells,29 and our observation that IL-21 was expressed by HL cells and HL tumor–infiltrating T cells suggested that IL-21 may promote HL proliferation and possibly survival in an IL-21–dependent manner. Consistent with this notion, we demonstrated that IL-21 can increase the number of phosphorylated STAT3 and STAT5 molecules in HL cell lines and moreover observed that blocking endogenous IL-21 signaling resulted in decreased proliferation of the HL cell line L591. We observed that primary HL cells express pSTAT5 consistent with earlier observations that STAT5 localizes in the nucleus of primary HL cells.21 On the basis of these data, we hypothesize that IL-21 is involved in proliferation and survival of HL cells, probably in early stages of the oncogenic process, via paracrine and autocrine loops. Besides IL-21, other factors such as IL-13 might be involved in provoking continued STAT activation.35,43 Continued activation by cytokines may prime the cells for additional mutations and make HL cells independent of cytokines.
These findings prompted us next to examine the roles of the STATs that are stimulated by IL-21 in proliferation of human B cells. We found that ectopic expression of constitutively active mutants of both STAT5a (data not shown) and STAT5b in primary human B cells results in in vitro immortalization of primary B cells, while ectopic expression of a constitutively active STAT3 mutant did not induce proliferation but induced plasma cell differentiation.27 Thus although STAT3 can be activated in some HL cells lines as published elsewhere20 and confirmed here, STAT3 does not seem to have a role in the in vitro immortalization of human primary B cells. Whether STAT3 is involved in the oncogenic process of HL remains to be determined.
Besides expressing a number of HL-specific genes and a highly similar set of cell surface antigens, CA-STAT5b+ B cells are like HL cells also with respect to molecular requirements for proliferation. CA-STAT5b+ B cells proliferated in a cytokine-independent manner. Pim-1, which is a direct target of STAT5, is also described to endow cytokine-independent proliferation to hematopoietic cells44,45 and is expressed in HL cell lines.46 Previously it has been documented that proliferation of HL cell lines is dependent upon constitutively active NF-κB.16,17 This turned out also to be the case for CA-STAT5b+ tonsil B cells since ectopic expression of IκBSR resulted in inhibition of proliferation of these cells. The high expression of a number of NF-κB target genes such as CD25, cFLIP, IL-15Rα, and CD44 (data not shown) in CA-STAT5b+ B cells is consistent with the notion that these cells have activated NF-κB. STAT5 knockdown resulted in a significant reduction of NF-κB activity as determined with a reporter assay. STAT5 knockdown did not alter the expression of p65 (results not shown), thus the mechanism underlying the STAT5 regulation of NF-κB activation remains to be elucidated. Although these data suggest that continued activation of STAT5 results in increased NF-κB activity, this was clearly not sufficient to maintain proliferation of CA-STAT5b–transduced PB B cells since CD40 ligation was still needed. Introduction of CA-IKK2 overcame the requirement of CD40L for proliferation of CA-STAT5bER–transduced PB B cells, indicating that CD40 mediates its effect on proliferation of CA-STAT5b–transduced PB B cells through activating NF-κB. We furthermore observed that expression of IκBSR in CA-STAT5b–immortalized tonsil B cells resulted in inhibition of proliferation. Together with the demonstration that STAT5 is also needed for continued expansion, our data suggest that the STAT5 and NF-κB pathways collaborate in inducing proliferation of human B cells and that constitutive activation of both these pathways may result in phenotypic changes characteristic for HL. Upon introduction of CA-IKK2 into PB CA-STAT5b+ B cells, monoclonal cells eventually emerge in long-term cultures. It is tempting to hypothesize that in those cells additional mutations occurred, resulting in full transformation and immortalization.
One hallmark of HL is the absence of BCR expression. Ectopic expression of E47 rescues the block in BCR expression in CA-STAT5–transduced B cells, clearly showing that continued activation of STAT5 plays a crucial role in the loss of the B-cell identity via downmodulation of E47. However other typical features of HL cells were not affected by down-regulation of E47, suggesting that continued activation of STAT5 mediates appearance of these features, such as induction of CD15 in a manner independent of E47 down-regulation. The mechanism of E47 down-regulation upon CA-STAT5b activation is unknown. As STAT5b is an activator of transcription, it is, therefore, expected that the down-regulation of E47 is an indirect process. It is possible that STAT5b may contribute to HL by regulating genes that induce epigenetic events, resulting in methylated Ig promoter9 and possibly also the E47 promoter.
Based on our data, we hypothesize that molecular alterations leading to continued activation of NF-κB and STAT5 are causally linked to HL genesis. Although no mutations were found in the STAT5 gene,47 it is conceivable that mutations upstream of STAT5 that lead to constitutive activation of the STAT5 pathway may eventually lead to transformation. It is possible that a negative regulator of STAT5 is altered, resulting in increased tyrosine-phosphorylated STAT5 levels. Recently, mutations in the suppressor of cytokine signaling 1 (SOCS1) have been shown to occur frequently in primary HL and HL cell lines.48 SOCS1 normally down-regulates the kinase activity of Jaks, leading to decreased levels of phosphorylated STATs.49,50 For this reason, mutation of the SOCS1 gene is thought to result in higher levels of phosphorylated STATs. In addition, molecular changes in the NF-κB pathway have been associated with HL.5,18 For instance, mutations in the gene encoding IκBα were found in 30% of primary Hodgkin lymphomas. Genetically affected STAT5 and NF-κB pathways most likely have both overlapping and distinct roles in HL genesis. The availability of an in vitro system to alter primary B cells into HL-like cells offers unique opportunities to study the molecular events leading to HL genesis and may provide new targets for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Berend Hooijbrink for his help with fluorescence-activated cell sorting (FACS) and maintenance of the FACS facility; Ae-Ri van Noort, Maho Nagasawa, Etsuko Yasuda, and Erwin Wijnands for their expert technical assistance; A. R. Musler and Dr C. M. van der Loos for help with the IL-21 immunohistochemistry; Remko Schotte for helpful discussions; the Department of Otolaryngology, Academic Medical Center, Amsterdam, The Netherlands (Prof W. Fokkens) for providing tonsil tissue; Dr Matthew Poynter (University of Vermont, Burlington, VT) for providing IκBαSR; and J. J. Schuringa (University of Groningen, Groningen, The Netherlands) for providing the CA-IKK2 construct.
This work was supported by Netherlands Organization for Scientific Research (NWO) grant 901-08-093 (H.S.) and by National Institute of Allergy and Infectious Diseases–National Institutes of Health (NIAID-NIH) grant F32-AI063846 (S.A.D.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: F.A.S. performed, designed, and analyzed research, and wrote the paper; L.A.S., M.N., R.J.B., and B.B. performed research; S.A.D. and T.B. performed, designed, and analyzed research; K.K. and K.O. provided critical reagents; C.J.M.N. designed and analyzed research; H.S. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hergen Spits, Department of Immunology, Genentech, South San Francisco, CA 94080; e-mail: spits.hergen@gene.com.
References
- 1.Kuppers R, Rajewsky K. The origin of Hodgkin and Reed/Sternberg cells in Hodgkin's disease. Annu Rev Immunol. 1998;16:471–493. doi: 10.1146/annurev.immunol.16.1.471. [DOI] [PubMed] [Google Scholar]
- 2.Thomas RK, Re D, Wolf J, Diehl V. Part I: Hodgkin's lymphoma: molecular biology of Hodgkin and Reed-Sternberg cells. Lancet Oncol. 2004;5:11–18. doi: 10.1016/s1470-2045(03)01319-6. [DOI] [PubMed] [Google Scholar]
- 3.Kuppers R, Rajewsky K, Zhao M, et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci U S A. 1994;91:10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwering I, Brauninger A, Klein U, et al. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;101:1505–1512. doi: 10.1182/blood-2002-03-0839. [DOI] [PubMed] [Google Scholar]
- 5.Kanzler H, Kuppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertel CB, Zhou XG, Hamilton-Dutoit SJ, Junker S. Loss of B cell identity correlates with loss of B cell-specific transcription factors in Hodgkin/Reed-Sternberg cells of classical Hodgkin lymphoma. Oncogene. 2002;21:4908–4920. doi: 10.1038/sj.onc.1205629. [DOI] [PubMed] [Google Scholar]
- 7.Jundt F, Kley K, Anagnostopoulos I, et al. Loss of PU. 1 expression is associated with defective immunoglobulin transcription in Hodgkin and Reed-Sternberg cells of classical Hodgkin disease. Blood. 2002;99:3060–3062. doi: 10.1182/blood.v99.8.3060. [DOI] [PubMed] [Google Scholar]
- 8.Stein H, Marafioti T, Foss HD, et al. Down-regulation of BOB. 1/OBF. 1 and Oct2 in classical Hodgkin disease but not in lymphocyte predominant Hodgkin disease correlates with immunoglobulin transcription. Blood. 2001;97:496–501. doi: 10.1182/blood.v97.2.496. [DOI] [PubMed] [Google Scholar]
- 9.Ushmorov A, Ritz O, Hummel M, et al. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood. 2004;104:3326–3334. doi: 10.1182/blood-2003-04-1197. [DOI] [PubMed] [Google Scholar]
- 10.Bain G, Gruenwald S, Murre C. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol Cell Biol. 1993;13:3522–3529. doi: 10.1128/mcb.13.6.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathas S, Janz M, Hummel F, et al. Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat Immunol. 2006;7:207–215. doi: 10.1038/ni1285. [DOI] [PubMed] [Google Scholar]
- 12.Murre C. Regulation and function of the E2A proteins in B cell development. Adv Exp Med Biol. 2007;596:1–7. doi: 10.1007/0-387-46530-8_1. [DOI] [PubMed] [Google Scholar]
- 13.Hsu SM, Jaffe ES. Leu M1 and peanut agglutinin stain the neoplastic cells of Hodgkin's disease. Am J Clin Pathol. 1984;82:29–32. doi: 10.1093/ajcp/82.1.29. [DOI] [PubMed] [Google Scholar]
- 14.Gruss HJ, DaSilva N, Hu ZB, Uphoff CC, Goodwin RG, Drexler HG. Expression and regulation of CD30 ligand and CD30 in human leukemia-lymphoma cell lines. Leukemia. 1994;8:2083–2094. [PubMed] [Google Scholar]
- 15.Atayar C, Poppema S, Blokzijl T, Harms G, Boot M, van den Berg A. Expression of the T-cell transcription factors, GATA-3 and T-bet, in the neoplastic cells of Hodgkin lymphomas. Am J Pathol. 2005;166:127–134. doi: 10.1016/S0002-9440(10)62238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bargou RC, Leng C, Krappmann D, et al. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood. 1996;87:4340–4347. [PubMed] [Google Scholar]
- 17.Bargou RC, Emmerich F, Krappmann D, et al. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin's disease tumor cells. J Clin Invest. 1997;100:2961–2969. doi: 10.1172/JCI119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuppers R. Molecular biology of Hodgkin's lymphoma. Adv Cancer Res. 2002;84:277–312. doi: 10.1016/s0065-230x(02)84009-x. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki Y, Schmidt-Supprian M, Derudder E, Rajewsky K. Role of NFkappaB signaling in normal and malignant B cell development. Adv Exp Med Biol. 2007;596:149–154. doi: 10.1007/0-387-46530-8_13. [DOI] [PubMed] [Google Scholar]
- 20.Kube D, Holtick U, Vockerodt M, et al. STAT3 is constitutively activated in Hodgkin cell lines. Blood. 2001;98:762–770. doi: 10.1182/blood.v98.3.762. [DOI] [PubMed] [Google Scholar]
- 21.Hinz M, Lemke P, Anagnostopoulos I, et al. Nuclear factor kappaB-dependent gene expression profiling of Hodgkin's disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity. J Exp Med. 2002;196:605–617. doi: 10.1084/jem.20020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skinnider BF, Kapp U, Mak TW. The role of interleukin 13 in classical Hodgkin lymphoma. Leuk Lymphoma. 2002;43:1203–1210. doi: 10.1080/10428190290026259. [DOI] [PubMed] [Google Scholar]
- 23.Shimoda K, van Deursen J, Sangster MY, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 24.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med. 2000;192:1841–1848. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheeren FA, Naspetti M, Diehl S, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 26.Fornek JL, Tygrett LT, Waldschmidt TJ, Poli V, Rickert RC, Kansas GS. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood. 2006;107:1085–1091. doi: 10.1182/blood-2005-07-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diehl SA, Schmidlin H, Nagasawa M, et al. STAT3-mediated upregulation of BLIMP1 is coordinated with BCL6 downregulation to control human plasma cell differentiation. J Immunol. 2008;180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozaki K, Kikly K, Michalovich D, Young PR, Leonard WJ. Cloning of a type I cytokine receptor most related to the IL-2 receptor beta chain. Proc Natl Acad Sci U S A. 2000;97:11439–11444. doi: 10.1073/pnas.200360997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish-Novak J, Dillon SR, Nelson A, et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 2000;408:57–63. doi: 10.1038/35040504. [DOI] [PubMed] [Google Scholar]
- 30.Asao H, Okuyama C, Kumaki S, et al. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J Immunol. 2001;167:1–5. doi: 10.4049/jimmunol.167.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Jaffa ESVJ. Lyon, France: IARC Press; 2001. World Health Organization classification of tumours. [Google Scholar]
- 32.Onishi M, Nosaka T, Misawa K, et al. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Loos C. A focus on fixation. Biotechnic Histochem. 2007;82:141–154. doi: 10.1080/10520290701375302. [DOI] [PubMed] [Google Scholar]
- 34.Karube K, Ohshima K, Suzumiya J, Kawano R, Kikuchi M, Harada M. Gene expression profile of cytokines and chemokines in microdissected primary Hodgkin and Reed-Sternberg (HRS) cells: high expression of interleukin-11 receptor alpha. Ann Oncol. 2006;17:110–116. doi: 10.1093/annonc/mdj064. [DOI] [PubMed] [Google Scholar]
- 35.Kapp U, Yeh WC, Patterson B, et al. Interleukin 13 is secreted by and stimulates the growth of Hodgkin and Reed-Sternberg cells. J Exp Med. 1999;189:1939–1946. doi: 10.1084/jem.189.12.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuppers R, Klein U, Schwering I, et al. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J Clin Invest. 2003;111:529–537. doi: 10.1172/JCI16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 38.Drexler HG. Recent results on the biology of Hodgkin and Reed-Sternberg cells, I: biopsy material. Leuk Lymphoma. 1992;8:283–313. doi: 10.3109/10428199209051008. [DOI] [PubMed] [Google Scholar]
- 39.Berberich I, Shu GL, Clark EA. Cross-linking CD40 on B cells rapidly activates nuclear factor-kappa B. J Immunol. 1994;153:4357–4366. [PubMed] [Google Scholar]
- 40.Haskill S, Beg AA, Tompkins SM, et al. Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura T, Ouchida R, Kodama T, et al. Cytokine receptor common beta subunit-mediated STAT5 activation confers NF-kappa B activation in murine proB cell line Ba/F3 cells. J Biol Chem. 2002;277:6254–6265. doi: 10.1074/jbc.M109878200. [DOI] [PubMed] [Google Scholar]
- 42.Schepers H, Eggen BJ, Schuringa JJ, Vellenga E. Constitutive activation of NF-kappa B is not sufficient to disturb normal steady-state hematopoiesis. Haematologica. 2006;91:1710–1711. [PubMed] [Google Scholar]
- 43.Rolling C, Treton D, Pellegrini S, Galanaud P, Richard Y. IL4 and IL13 receptors share the gamma c chain and activate STAT6, STAT3 and STAT5 proteins in normal human B cells. FEBS Lett. 1996;393:53–56. doi: 10.1016/0014-5793(96)00835-6. [DOI] [PubMed] [Google Scholar]
- 44.Nosaka T, Kitamura T. Pim-1 expression is sufficient to induce cytokine independence in murine hematopoietic cells, but is dispensable for BCR-ABL-mediated transformation. Exp Hematol. 2002;30:697–702. doi: 10.1016/s0301-472x(02)00808-1. [DOI] [PubMed] [Google Scholar]
- 45.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. Embo J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jucker M, Schaadt M, Diehl V, Poppema S, Jones D, Tesch H. Heterogeneous expression of proto-oncogenes in Hodgkin's disease derived cell lines. Hematol Oncol. 1990;8:191–204. doi: 10.1002/hon.2900080404. [DOI] [PubMed] [Google Scholar]
- 47.Yamada K, Ariyoshi K, Onishi M, et al. Constitutively active STAT5A and STAT5B in vitro and in vivo: mutation of STAT5 is not a frequent cause of leukemogenesis. Int J Hematol. 2000;71:46–54. [PubMed] [Google Scholar]
- 48.Weniger MA, Melzner I, Menz CK, et al. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25:2679–2684. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- 49.Endo TA, Masuhara M, Yokouchi M, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 50.Krebs DL, Hilton DJ. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.