Abstract
The biogenesis of the Salmonella-containing vacuole within mammalian cells has been intensively studied over recent years. However, the ability of Salmonella to sense and adapt to the intracellular environment of different types of host cells has received much less attention. To address this issue, we report the transcriptome of Salmonella enterica serovar Typhimurium SL1344 within epithelial cells and show comparisons with Salmonella gene expression inside macrophages. We report that S. Typhimurium expresses a characteristic intracellular transcriptomic signature in response to the environments it encounters within different cell types. The signature involves the upregulation of the mgtBC, pstACS and iro genes for magnesium, phosphate and iron uptake, and Salmonella pathogenicity island 2 (SPI2). Surprisingly, in addition to SPI2, the invasion-associated SPI1 pathogenicity island and the genes involved in flagellar biosynthesis were expressed inside epithelial cells at later stages of the infection, while they were constantly downregulated in macrophage-like cells. To our knowledge, this is the first report of the simultaneous transcription of all three Type Three Secretion Systems (T3SS) within an intracellular Salmonella population. We discovered that S. Typhimurium strain SL1344 was strongly cytotoxic to epithelial cells after 6 h of infection and hypothesize that the time-dependent changes in Salmonella gene expression within epithelial cells reflects the bacterial response to host cells that have been injured by the infection process.
Introduction
In humans and many animals, Salmonella enterica causes a multistage systemic infection that involves invasion and crossing of the epithelial cell barrier and subsequent intracellular replication in monocytic cells. Regarding S. enterica, and the related enteric pathogens Shigella and Yersinia, much of the infection-associated cross-talk with host cells is directed by bacterial type III secretion systems (T3SSs) (Hansen-Wester and Hensel, 2001; Ghosh, 2004). T3SSs enable bacteria to translocate virulence proteins, also called effector proteins, from the bacteria into the host cell often to specifically interfere with host cell functions such as actin polymerization, signal transduction and apoptosis (Hansen-Wester and Hensel, 2001). S. enterica possesses two classical T3SSs encoded by Salmonella pathogenicity island 1 (SPI1) and Salmonella pathogenicity island 2 (SPI2) (Hansen-Wester and Hensel, 2001). In addition, Salmonella possesses a third T3SS responsible for the flagellar-based motility of the pathogen (Macnab, 2004; McCarter, 2006).
SPI1 plays a fundamental role in the early stages of mammalian infection through triggering Cdc42- and Rac1-mediated remodelling of the actin cytoskeleton of the host cell, and leading to internalization of the bacteria and subsequent penetration of the ileal mucosal lining (Galan and Curtiss, 1989; Hansen-Wester and Hensel, 2001). SPI1 also has a pro-inflammatory potential through its ability to activate JNK- and p38-dependent nuclear responses (Hobbie et al., 1997) and the release of IL-1β (Monack et al., 2001). Furthermore, selected SPI1-associated effector proteins cause a very strong pro-apoptotic effect in monocytic cells (Hersh et al., 1999). Concomitantly, SPI1-deficient mutants show a 10-fold reduction of LD50 in mice when given orally, whereas such mutants retain virulence via the intraperitoneal route, illustrating the importance of SPI1 in the early stages of infection, such as the invasion process (Bajaj et al., 1996).
Once intracellular in the endosomal compartment, S. Typhimurium takes control of the trafficking and evolution of the internalized vacuole, to form the Salmonella-containing vacuole (SCV). The SCV interacts with the early endocytic pathway but resists fusion with lysosomes in both non-phagocytic and phagocytic cells, enabling Salmonella to avoid host cell defences (Harrison et al., 2004; Marsman et al., 2004). Intracellular survival and systemic spread of Salmonella in the murine typhoid infection model relies on SPI2 and SPI2-associated effector proteins and their ability to interfere with vesicular trafficking of the host cell. SPI2 functions to protect the SCV from the effect of the phagocytic defence enzymes, such as phagocyte NADPH oxidase and inducible nitric oxide synthase (Mastroeni et al., 2000; Vazquez-Torres et al., 2000; Chakravortty et al., 2002). In addition, selected SPI2-associated effector proteins allow S. Typhimurium to polymerize actin in the vicinity of the SCV (Meresse et al., 2001; Poh et al., 2007). S. Typhimurium effectors also cause accumulation of microtubules around the SCV (Kuhle et al., 2004), acquisition of several endosomal markers (Steele-Mortimer et al., 1999) and recruitment of kinesin and dynein to regulate vacuolar membrane dynamics (Guignot et al., 2004; Marsman et al., 2004; Boucrot et al., 2005). Certain SPI2 effector functions are also associated with a slow-progressing induction of apoptosis in monocytic cells (Monack et al., 2001), and interference with the activity of the ubiquitination pathway in professional phagocytes (Rytkonen et al., 2007).
Whether or not the flagellar T3SS plays a distinct role in S. Typhimurium infection remains controversial. It is agreed that the flagella and motility system is generally associated with the extracellular life of the pathogen and that motility can assist invasion. The flagellar T3SS also possess a pro-inflammatory potential through its interaction with toll-like receptor (TLR) 5 (Reed et al., 2002; Choudhary et al., 2005) or with the cytosolic nod-like receptors (NLR), such Birc 1e and Ipaf (Franchi et al., 2006; Miao et al., 2006; Molofsky et al., 2006; Coers et al., 2007). The TLR-mediated recognition of flagellin triggers induction of a pro-inflammatory cascade, including IL-8 secretion that will recruit phagocytic cells such as neutrophiles to the site of infection (Ciacci-Woolwine et al., 1998; Eaves-Pyles et al., 2001; Gewirtz et al., 2001a,b; Hayashi et al., 2001; Zeng et al., 2006). The NLR-mediated cytosolic recognition of flagellin results in caspase 1-dependent IL-1β activation, leading to host cell death and a T cell-mediated immune response (Inohara et al., 2005).
While it is clear that internalization and subsequent intracellular replication of S. enterica are central T3SS-directed events of a systemic infection, these activities are not sufficient on their own to promote pathogenesis. The successful infection of a host by Salmonella requires a delicate interplay of several metabolic functions, including the ability to synthesize aromatic amino acids and nucleotides. Additionally, the capacity to express virulence functions must be integrated and controlled by the general gene regulatory programs that steers the responses and metabolic activities of the bacterial cell (Rhen and Dorman, 2005). For example, aroA or purE mutants retain their ability to invade mammalian cells but are unable to subsequently replicate in mice (Hoiseth and Stocker, 1981; Fields et al., 1986), whereas mutants defective in the conserved ompR/envZ or phoP/phoQ regulatory pathways remain attenuated in both cultured cells and in mice (Groisman, 2001). Although various nutritional and environmental signals necessary for the adaptation and survival of Salmonella have been investigated for decades, the environmental differences between host cell types and their impact upon Salmonella gene expression remain to be understood.
S. Typhimurium targets both epithelial cells and macrophages during infection of the gastrointestinal tract (Frost et al., 1997; Tsolis et al., 1999), but it is apparent that the two types of cells differ in their response to infection. For example, the timing of acquisition and exclusion of endosomal markers differs between macrophages and epithelial cells (Knodler and Steele-Mortimer, 2003); non-phagocytic cells also support different levels of S. Typhimurium intracellular replication, which begins at 3–4 h post infection (p.i.) in epithelial cells but is delayed until 4–8 h p.i. in macrophages (Gahring et al., 1990; Knodler and Steele-Mortimer, 2003; Eriksson-Ygberg et al., 2006). Initiation of the formation of Salmonella-induced filaments (Sifs) occurs at 5–6 h p.i. inside epithelial cells but not until 9 h p.i. inside macrophages (Garcia-del Portillo et al., 1993; Garcia-del Portillo and Finlay, 1995; Stein et al., 1996; Knodler et al., 2003). Finally, the maintenance of the SPI2-mediated condensation of actin appears to be essential for bacterial replication inside macrophage-like cells, but not for bacterial replication within epithelial cells (Meresse et al., 2001).
The recent ability to profile bacterial gene expression inside host cells (Eriksson et al., 2003; Hinton et al., 2004) has brought new insight into the infection biology of pathogens such as Listeria monocytogenes, S. enterica, Sh. flexneri and Mycobacterium tuberculosis (Eriksson et al., 2003; Schnappinger et al., 2003; Chatterjee et al., 2006; Lucchini et al., 2006). Furthermore, a combined approach using microarray and selective capture of combined sequences identified genes of S. enterica serovar Typhi that responded to the macrophage SCV environment (Faucher et al., 2006).
We describe here the transcriptomic analysis of S. Typhimurium SL1344 inside epithelial cells, and focus on similarities and differences to the bacterial gene expression profile inside macrophage-like cells reported by Eriksson et al. (2003). The HeLa epithelial cell line was chosen because it is a well-defined model for infection of mammalian cells with S. Typhimurium, and has been used to characterize the biogenesis and evolution of the SCV (Steele-Mortimer et al., 1999; Beuzon et al., 2000). Additionally, we identify novel time-dependent changes in Salmonella gene expression that occur inside epithelial cells, revealing simultaneous expression of the three Salmonella T3SS. We present the first evidence that S. Typhimurium not only alters its gene expression profile according to cell type, but also shows large transcriptomic changes that reflect alterations in host cell biology.
Results and discussion
HeLa cells have commonly been used to model infection of epithelial cells by S. Typhimurium and various other bacterial pathogens (Walz et al., 1985; Stibitz et al., 1988; Rosqvist et al., 1990; Garcia-del Portillo and Finlay, 1995; Wang et al., 1996; Boucrot et al., 2003; Jabado et al., 2003; Unsworth et al., 2004; Way et al., 2004; Birmingham et al., 2005; Lucchini et al., 2005). In our study, we have determined the gene expression profile of S. Typhimurium inside HeLa cells at 2, 4 and 6 h p.i., as these time points coincide with distinct stages of the intracellular infection process. At 2 h p.i., S. Typhimurium is inside a vacuolar compartment but has not yet begun to replicate intracellularly (Gorvel and Meresse, 2001; Steele-Mortimer et al., 1998). The 4 and 6 h time points reflect stages of infection when S. Typhimurium has begun to grow inside epithelial cells (Knodler and Steele-Mortimer, 2003).
To validate our experimental conditions, we first confirmed that S. Typhimurium SL1344 was replicating inside epithelial cells by measuring the segregation rate of a temperature-sensitive plasmid (pPir) (Posfai et al., 1997; Clements et al., 2002). We observed that four times fewer bacteria harboured the plasmid at 6 h p.i. compared with 2 h p.i. (Fig. 1A), indicating that at least two generations of replication had occurred intracellularly over this period of time. Additionally, we monitored the distribution of SL1344 inside epithelial cells at 2, 4 and 6 h p.i. This analysis showed that 75% of the infected epithelial cells harboured between one and 10 bacteria at 2 h p.i. (Fig. 1B), consistent with data obtained for another S. Typhimurium strain 12023 (also known as 14 038) in HeLa cell infection experiments (Steele-Mortimer et al., 1999; Beuzon et al., 2002). At 2 h p.i., less than 25% of the infected epithelial cells contained more than 10 bacteria. At 6 h p.i., this number rose to more than 50% (Fig. 1B), agreeing with the level of intracellular growth determined by plasmid segregation (Fig. 1A).
Fig. 1.
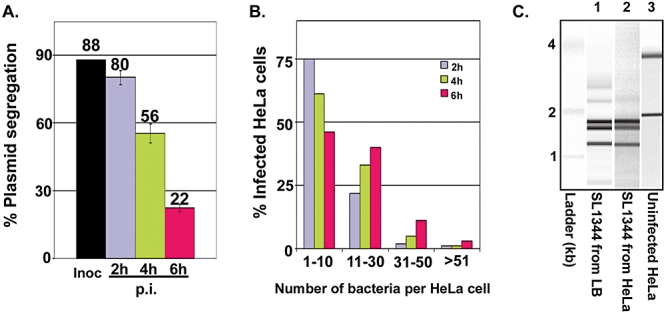
Intracellular replication and distribution of S. Typhimurium SL1344 Salmonella strain inside epithelial cells. A. Bacterial replication was measured by following the segregation of the pPIR plasmid, which only replicates at 30°C. During each bacterial division at 37°C, only one daughter cell inherits the pPIR plasmid. Plasmid segregation is expressed as the percentage of bacterial cells that retain pPIR. The difference in percentage of bacterial cells carrying the pPIR plasmid inside epithelial cells between 2 and 6 h p.i. reflects a fourfold increase in the population level. Inoculum (Black), Epithelial cells 2 h p.i. (Purple), epithelial cells 4 h (Green) and epithelial cells 6 h (Magenta). B. Distribution of S. Typhimurium cells inside epithelial cells at 2, 4 and 6 h p.i.; values shown are the percentage of epithelial cells containing different numbers of bacteria observed by light microscopy (Experimental procedures). Epithelial cells 2 h p.i. (Purple), epithelial cells 4 h p.i. (Green) and epithelial cells 6 h p.i. (Magenta). Over 150 infected HeLa cells were examined. C. Isolation of high quality bacterial RNA from infected HeLa cells and separation by size chromatography (Experimental procedures). Total RNA was extracted from S. Typhimurium grown in LB medium in vitro (lane 1), S. Typhimurium from infected epithelial cells (lane 2) and eukaryotic RNA isolated from uninfected HeLa cells (lane 3).
We optimized the maximal recovery of bacterial cells, and prepared stabilized bacterial RNA with minimal contamination by eukaryotic RNA as described previously (Eriksson et al., 2003; Hinton et al., 2004) (Fig. 1C). We determined the transcriptome of S. Typhimurium SL1344 inside HeLa epithelial cells, and compared it with transcriptomic data for SL1344 within J774-A.1 macrophages (Eriksson et al., 2003; Lucchini et al., 2005). We refer to these host cell types as epithelial and macrophage cells respectively. To determine the similarities and differences between the host cell types, we used a mid-logarithmic Luria–Bertani (LB) culture of SL1344 as a common comparator, and data were statistically filtered as described in Experimental procedures (Table S1). We note that transcriptomic and RT-PCR data represent the average bacterial gene expression occurring in a given environment as they reflect the entire population of intracellular Salmonella. We nevertheless intend that this novel transcriptomic data set will provide useful insights into the process of Salmonella infection.
The intraepithelial transcriptome of S. Typhimurium shows that many genes required for Salmonella-mediated enteritis and growth within epithelial cells are highly expressed
We observed that 1377 S. Typhimurium genes were either upregulated or downregulated by at least twofold inside epithelial cells at 2 h p.i., and 1221 genes at 6 h p.i. compared with cultures grown in LB (Table 1). These included genes such as lpp, fadF (also known as yafH), rfbA, sitABCD and several SPI2-associated genes that have been shown to contribute to replication of S. Typhimurium in cultured epithelial cells (Watson et al., 1995; Spector et al., 1999; Maurer et al., 2000; Steele-Mortimer et al., 2002; Morgan et al., 2004) (Table S1).
Table 1.
Numbers of S. Typhimurium genes regulated during infection of epithelial cells and macrophages.
| Comparisons | Number of upregulated genes | Number of downregulated genes | Total number of differentially expressed genes |
|---|---|---|---|
| HeLa 2 h versus LB | 724 | 653 | 1377 |
| HeLa 6 h versus LB | 605 | 616 | 1221 |
| J774 4 h versus LB | 938 | 935 | 1873 |
| HeLa 6 h versus HeLa 2 h | 120 | 67 | 187 |
| J774 12 h versus J774 4 h | 49 | 4 | 53 |
| HeLa 2 h versus J774 4 h | 368 | 259 | 627 |
| HeLa 6 h versus J774 4 h | 529 | 503 | 1032 |
Numbers of S. Typhimurium genes that are differentially expressed between two conditions are shown after statistical analysis and data filtering (Experimental procedures); the values indicated are the numbers of genes that are upregulated or downregulated by at least than twofold.
Infection of epithelial cells is a prerequisite for the induction of enteritis in the calf infection model, which has been proposed as a tool to simulate human Salmonella-mediated gastroenteritis (Frost et al., 1997). Morgan et al. used signature-tagged mutagenesis to identify 98 genes that were essential for calf enteritis (Frost et al., 1997; Tsolis et al., 1999; Morgan et al., 2004). To functionally validate our transcriptomic data, we established the relationship between our Salmonella gene expression data in HeLa cells and the genes previously shown to be essential for calf enteritis. This comparison (Table 2) lists the 50 S. Typhimurium genes that are required for bovine enteritis and that are most highly expressed in HeLa cells (Table 2 and Table S17). Our data showed that the adaptation of Salmonella to cultivated epithelial cells involved the expression of genes with proven roles in Salmonella virulence. It was apparent that genes listed that showed high levels of expression inside epithelial cells were expressed at lower levels inside macrophages (Table 2).
Table 2.
Genes required for Salmonella-associated enteritis in the bovine infection model and for survival and replication in cultivated epithelial cells are expressed within HeLa cells.
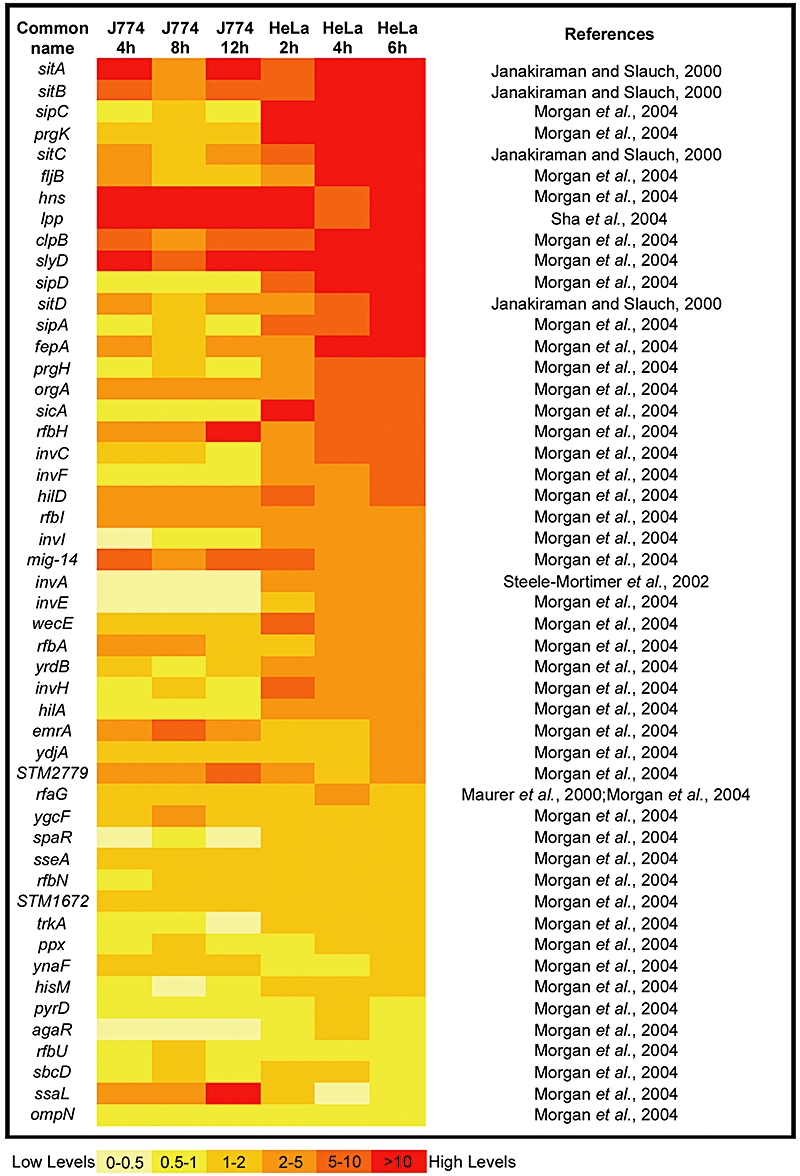 |
The table shows 50 S. Typhimurium genes that are required for replication in epithelial cells (Janakiraman and Slauch, 2000; Maurer et al., 2000; Steele-Mortimer et al., 2002) or enteritis in the bovine infection model (Morgan et al., 2004) and are highly expressed in HeLa cells. Each gene is colour-coded according to its level of gene expression (i.e. signal ratio of cDNA versus genomic DNA). Highly expressed genes are red,and weakly expressed genes are pale yellow. The gene list was sorted by values for S. Typhimurium gene expression in HeLa at 6 h p.i., and the data are taken from Table S1.
Our transcriptomic data also confirmed previous reporter gene-based analyses of S. Typhimurium in MDCK epithelial cells that showed upregulation of genes involved in magnesium (mgtBC) and phosphate transport (pstACS) (Garcia-del Portillo et al., 1992), and the virulence plasmid pSLT-associated intracellular replication gene spvB (Fierer et al., 1993). The expression of the genes involved in de novo biosynthesis of purines was induced up to sevenfold in epithelial cells, and the pnuC gene encoding a nucleoside/purine/pyrimidine transporter was twofold upregulated in epithelial cells at 6 h p.i. compared with LB (Table S1).
The transcriptomic data reported here are supported by both mutant and gene fusion-based studies that involve epithelial cells, indicating that our approach is valid for the investigation of early events of Salmonella infection.
Similarities between the S. Typhimurium transcriptome within epithelial and macrophage cells
As the intracellular infection of epithelial cells and macrophages by S. Typhimurium differ in terms of kinetics and maturation of the SCV, we compared the S. Typhimurium SL1344 transcriptomes within HeLa epithelial cells with the data we previously obtained for S. Typhimurium SL1344 in macrophage-like J774-A.1 cells (Eriksson et al., 2003).
Inside macrophages, most changes in the S. Typhimurium transcriptome were observed at 4 h p.i. and the expression of very few genes changed at later stages of infection (Eriksson et al., 2003; Lucchini et al., 2005). We therefore used the 4 h time point in macrophages to compare with the intraepithelial transcriptome of Salmonella at 2 h and 6 h p.i. (Fig. 2). We classified the genes that were differentially regulated in both cell types into functional categories (Fig. 3). The analysis revealed that a substantial pool of S. Typhimurium genes were upregulated or downregulated in both epithelial cells and macrophages. This represents the qualitative intracellular transcriptomic signature of S. Typhimurium, which is apparent from the intersecting areas of Fig. 2A and B and is detailed in Tables S6 and S13. This revealed that certain functional categories of genes showed the same expression profiles in both cell types (Figs 2A, B, 3A and B, Tables S6 and S13), suggests that many common environmental cues exist within both cell types, and have similar impacts upon S. Typhimurium gene expression. We note that, although the same categories of genes are changing in both cell types compared with LB (Fig. 3A and B), the level of expression is often different in each cell type. For example, the mgtBC and pstACS magnesium and phosphate transport systems showed upregulation in both cell types, indicative of a common lack of magnesium and phosphate in both SCVs. Interestingly, while the levels of expression of the mgtBC genes remained high in macrophages at all three time points, both genes were substantially downregulated inside epithelial cells between the 2 h and 6 h time points (Fig. 4A, Table S1). This could imply that S. Typhimurium utilizes alternative ways of acquiring magnesium, or that the requirement for Salmonella to actively transport magnesium changes later in infection. Alternatively, the availability of magnesium to Salmonella could change at later stages of infection.
Fig. 2.
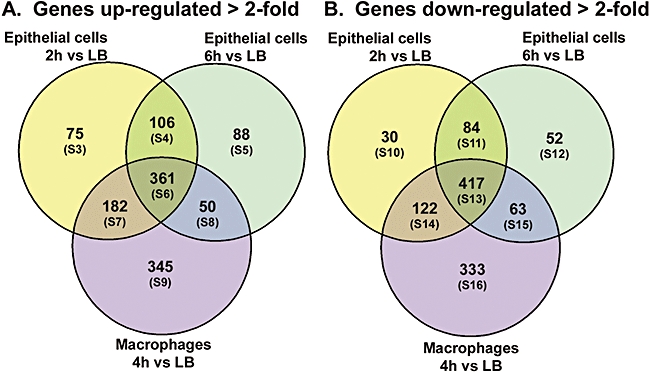
Common intracellular transcriptomic signature and cell type specific responses of S. Typhimurium in epithelial cells and macrophages. Non-proportional Venn diagrams are shown. A. The S. Typhimurium genes significantly upregulated by at least twofold from epithelial (2 h p.i.), epithelial (6 h p.i.) and macrophage cells (4 h p.i.), all in comparison with LB. B. Genes significantly downregulated by at least twofold in the same samples. The common intracellular signature of S. Typhimurium is represented by the central area of each Venn diagram which contains genes that show similar patterns of regulation inside both types of mammalian cells (Tables S6 and S13). The Venn diagrams were also used to define lists of genes that were only upregulated or downregulated inside macrophages (Tables S9 and S16) or only inside epithelial cells (Tables S3–S5 and S10–S12).
Fig. 3.
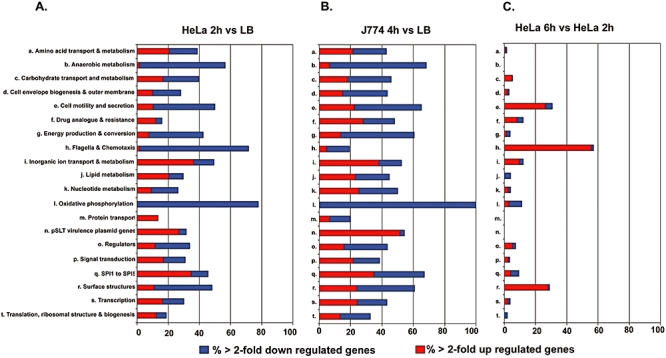
Functional categories of S. Typhimurium genes changing in expression in epithelial cells and macrophages. The Red and Blue bars indicate the percentage of genes of each functional category upregulated or downregulated, respectively, inside epithelial cells (2 h p.i.) versus LB (A), or inside macrophages (4 h p.i.) versus LB (B). The classes of genes differentially regulated in epithelial cells at 6 h p.i. compared with the 2 h time point are shown in (C). The numbers of genes involved in each analysis are shown in Table 1. The list of genes included in each functional category was obtained from the Kyoto Encyclopedia of Genes and Genomes, KEGG (http://www.genome.jp/kegg/).
Fig. 4.
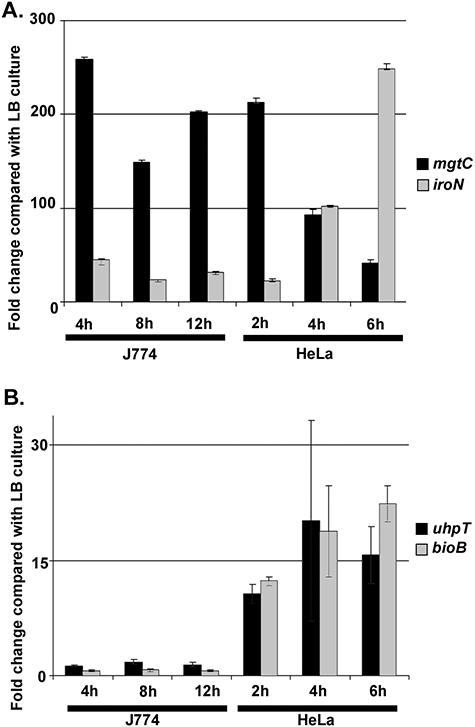
Selected S. Typhimurium genes that show differential expression between epithelial cells and macrophages. Transcriptomic data are shown for the S. Typhimurium mgtC and iroN genes (A), uhpT and bioB genes (B) inside both macrophage and epithelial cells. Data for the expression of specific genes are taken from Table S1. Error bars indicate the standard error of the mean.
Within both cell types, we noted a strong induction of the ent (Fig. 7A), fep, fhu, iro (Fig. 4A), sit and suf genes compared with LB (Table S1). Many of these metal acquisition systems import iron, which forms an essential component of respiratory enzymes which contain iron-sulfur clusters (Beinert et al., 1997; Tokumoto et al., 2002) and regulatory proteins such as FUR (Tsolis et al., 1995). Late infection of epithelial cells (6 h p.i.) showed a 100-fold greater level of expression of these iron uptake systems than the early time point (2 h p.i.), whereas their expression profiles remained similar at all time points inside macrophages. Induction of these bacterial iron-uptake systems within both macrophages and epithelial cells was also observed for Shigella flexneri, which has a cytosolic lifestyle inside mammalian cells (Runyen-Janecky and Payne, 2002; Lucchini et al., 2005), suggesting that uptake of iron is triggered in both the macrophage cytosol for Shigella and within the SCV for Salmonella. Additionally, we observed that the sfbAB genes, encoding the periplasmic iron-binding lipoprotein SfbA and the nucleotide-binding ATPase SfbB, are up to sixfold more highly expressed in epithelial cells than in macrophages. The sfbABC operon has been identified as a pathogenicity islet in S. enterica sv. Enteritidis, important for ferric iron acquisition in BALB/c mice (Pattery et al., 1999). Overall, our observation of the upregulation of both ferrous and ferric iron uptake systems suggests that Salmonella is actively acquiring iron inside epithelial cells.
Fig. 7.
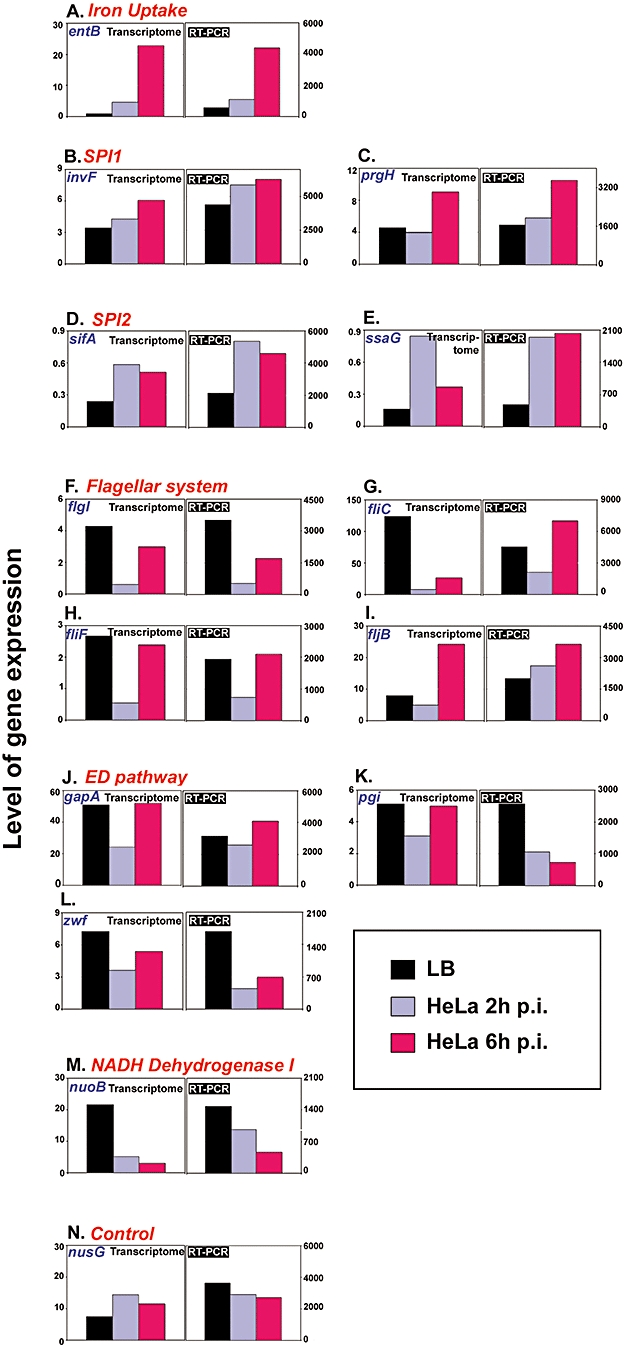
RT-PCR confirmation of transcriptomic data. RNA was extracted from Salmonella cells released from infected epithelial cells at 2 and 6 h p.i. and from mid-exponential LB cultures, reverse transcribed to cDNA and used as template for RT-PCR amplification of entB (A), invF (B), prgH (C), sifA (D), ssaG (E), flgI (F), fliC (G), fliF (H), fljB (I), gapA (J), pgi (K), zwf (L), nuoB (M) and nusG (N) cDNAs using specific primers pairs (Table 5 and Experimental procedures). Each panel shows the expression levels observed from the transcriptomic data (graph on the left) and the RT-PCR analyses (graph on the right). Black bars show expression levels determined from LB culture. Purple and magenta bars show expression levels obtained inside epithelial cells at 2 and 6 h p.i. respectively.
The SCVs of both macrophage and epithelial cells have been reported to be limiting for purines, pyrimidines and aromatic amino acids (Finlay et al., 1991; Leung and Finlay, 1991). Consistent with these data, we observed that expression of the genes involved in de novo biosynthesis of purines was induced up to sevenfold in both macrophages and epithelial cells (Table S1).
Differences in Salmonella Typhimurium gene expression within the SCV of epithelial and macrophage cells
A direct comparison between the S. Typhimurium transcriptomes in epithelial cells (2 and 6 h p.i.) and macrophages (4 h p.i.) identified 627 and 1032 genes, respectively, that were up- or downregulated by more than twofold (Table 1). This suggests that S. Typhimurium gene expression within the macrophage SCV is most similar to the early epithelial SCV (2 h p.i.) (Fig. 2). We also identified clusters of genes that showed epithelial cell-specific or macrophage-specific patterns of expression (Fig. 2); 269 and 166 genes are upregulated or downregulated in epithelial cells compared with LB but not in macrophages, respectively (Fig. 2A and B, Tables S3–S5 and S10–S12). In contrast, 345 and 333 genes were up- or downregulated in macrophages but not in epithelial cells (Fig. 2A and B, Tables S9 and S16).
Subsequent time-dependent changes in the S. Typhimurium transcriptome occurred at 6 h p.i. in epithelial cells, with expression of 187 genes changing at 6 h compared with 2 h p.i. (Table 1). Among these differences, genes encoding the superoxide dismutase (sodBC), alkyl hydroxyperoxidase (ahpCF), catalase (katE) and flavohemoglobin (hmpA) showed low levels of expression, reflecting the lack of oxidative or nitrosative stress in HeLa cells (Table S1; Fig. S1). The main differences are described and discussed in more detail below.
Biotin synthesis
We noted that biotin synthesis genes were highly induced in epithelial cells, showing up to 10-fold higher expression than in macrophages (Fig. 4B). Biotin enzymes function as carboxyl group transfer enzymes, and are necessary for fatty acid biosynthesis (Keseler et al., 2005). Induction of Sh. flexneri biotin biosynthesis genes were also observed during infection of epithelial cells but not macrophages (Lucchini et al., 2005), suggesting differences in the availability or importance of biotin during infection of these mammalian cells. We investigated the role of the bio genes by infecting epithelial cells with a bioABFCD deletion mutant of S. Typhimurium. Although, this gene deletion prevented growth of the mutant on minimal medium that lacked biotin, the strain was not attenuated in its ability to invade or replicate within epithelial cells compared with its parental strain SL1344 (Table 3). This suggests that biotin synthesis is not crucial for the survival and replication of S. Typhimurium inside cultured epithelial cells. Differences in the induction of biotin biosynthesis genes by Salmonella during infection of epithelial cells and macrophages could reflect fluctuations in the intracellular availability of biotin, or that other signals trigger bio gene expression inside epithelial cells.
Table 3.
Genes involved in biotin utilization of hexose phosphate utilization are not required for intracellular replication of S. Typhimurium.
| Strains | Genotype | Invasiona(% inoculum) | Change in intracellular population from 2 to 6 h p.i. compared with wtb |
|---|---|---|---|
| SL1344 | wt | 18 ± 1.15 | 1 |
| JH3214 | SL1344Δbio | 25 ± 1.76 | 0.93 ± 0.09 |
| JH3216 | SL1344ΔuhpT | 34 ± 1.20 | 0.88 ± 0.04 |
Invasion ability is the ratio of the intracellular bacterial cell number at 2 h p.i. over that of the inoculum (± standard error of the mean).
Ratio of change in intracellular bacterial numbers at 6 h compared with 2 h p.i. for each mutant over that of wt SL1344 strain.
Intra-vacuolar pH
Upregulation of acid-inducible S. Typhimurium genes, such as pagC (Alpuche Aranda et al., 1992), cysB, adiY, marAB inside the macrophage vacuole is consistent with the acidic nature of the SCV (Rathman et al., 1996; Eriksson et al., 2003). Inside epithelial cells, expression of four of these five acid-inducible genes was either downregulated or did not change compared with LB. Only the cysB transcriptional activator showed a fourfold to sevenfold induction in this cell type (Table S1). These data suggest that the macrophage SCV is more acidic than the epithelial SCV.
Cell type-specific expression of FUN genes
S. Typhimurium SL1344 carries over 900 genes of unknown function (FUN genes (Hinton, 1997)). Expression of about 30% of these genes was altered inside epithelial cells compared with LB. These genes are designated as ‘putative’ or ‘hypothetical’ in our Supplementary Tables. Thirty-two of these genes were only upregulated in epithelial cells at 2 h p.i. and not in macrophages, whereas 96 genes were only induced in macrophages after 4 h infection and not in epithelial cells. One hundred and twenty-eight FUN genes were upregulated in both cell types (Table S1). The FUN genes that show cell type-specific expression may reveal new virulence determinants, and are currently under investigation.
All three T3SS are upregulated inside epithelial cells
Focusing on the T3SS of Salmonella Typhimurium such as SPI1, SPI2 and the flagellar system, we observed major differences in expression profiles between the two host cell types. The role of the three T3SS of Salmonella is often perceived as follows: flagella genes are seen as being important for the extracellular life of Salmonella, and for reaching the epithelial barrier, while SPI1 mediates the invasion process and SPI2 ensures intracellular survival. It might have been thought that sequential expression of the T3SS would facilitate successful infection but we have now shown that all three T3SS are upregulated simultaneously inside epithelial cells.
SPI1 genes are expressed inside epithelial cells
SPI1 and SPI1-associated genes are necessary for the invasion of mammalian cells, for full virulence in orally infected mice or infected murine ileal loops, and for causing enteritis in bovine and porcine infection models (Penheiter et al., 1997; Barthel et al., 2003; Morgan et al., 2004; Stecher et al., 2005; Carnell et al., 2007). We have previously shown that SPI1 structural genes are strongly downregulated inside macrophages at 4 h p.i. (Eriksson et al., 2003), and under in vitro conditions that mimicked the environment of the macrophage SCV (Eriksson-Ygberg et al., 2006). Surprisingly, SPI1 genes remained expressed not only once S. Typhimurium was inside the epithelial SCV (at 2 h p.i.), but some of these genes were actually upregulated at 4 and 6 h p.i. compared with 2 h p.i. (Figs 5, 7B and C, Table S2). This suggests that the intracellular environment encountered by Salmonella at late infection inside epithelial cells contains more SPI1-inducing cues than at earlier stages. As observed in macrophages (Eriksson et al., 2003), SPI4 and some SPI5 genes such as pipC (Fig. 5), sopB, orfX, STM1089 and STM1093 (Table S1) were coexpressed with SPI1 genes in epithelial cells. Intracellular expression of SPI1 would be consistent with the additional roles of SPI1 and SPI1-related proteins in interfering with the host response during the later stages of infection (Murray and Lee, 2000; Drecktrah et al., 2005). Furthermore, SPI1 genes are known to contribute to the induction of the host pro-inflammatory response involving the recruitment of phagocytes into the gut mucosa (Galyov et al., 1997; Jones et al., 1998; Gewirtz et al., 1999).
Fig. 5.
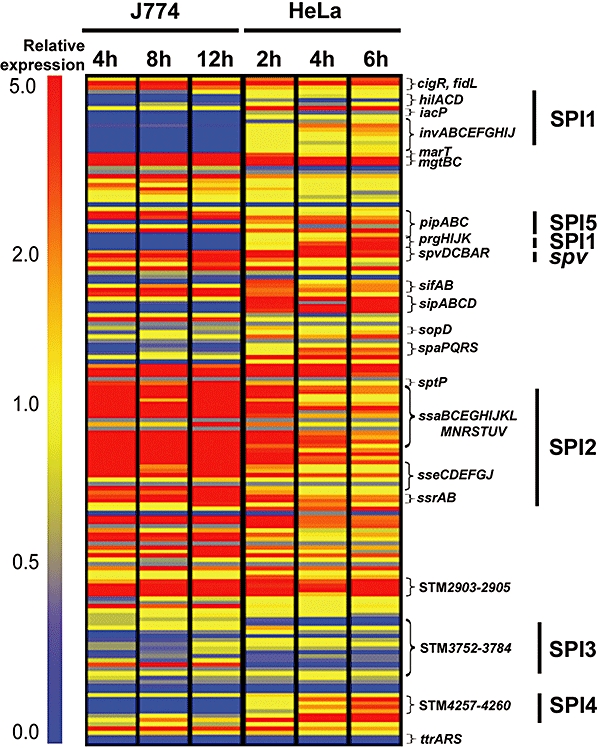
SPI1 and SPI2 genes are upregulated inside epithelial cells. The expression profiles of SPI1 to SPI5 genes, their known effectors (Abrahams and Hensel, 2006; McClelland et al., 2001), and the spvABCDR virulence plasmid genes inside macrophage and epithelial cells are shown relative to the LB comparator. Each horizontal bar represents the expression level of a single gene. The post-infection sampling time is indicated. Blue shows that the gene is downregulated, yellow that it is expressed at similar levels and red that it is upregulated, compared with LB. Data are taken from Table S1, and are summarized in Table S2.
SPI2 genes are expressed in both macrophages and epithelial cells
The SPI2 island is crucial for the intracellular survival and replication of S. Typhimurium in phagocytic and non-phagocytic cells (Hensel, 2000). SPI2 effectors and structural proteins are necessary for the trafficking of the SCV (Salcedo and Holden, 2003; Guignot et al., 2004), for the recruitment of the appropriate endosomal markers and for the maintenance of integrity of the SCV membrane (Beuzon et al., 2000; Beuzon et al., 2002). In macrophages, SPI2 gene expression was strongly induced and remained induced at all time points (Eriksson et al., 2003). As expected, SPI2 genes and the SsrA regulon (Rytkonen et al., 2007) were also upregulated within epithelial cells. We noticed that the levels of expression of SPI2 and SPI2-related genes were similar to those in macrophages early during epithelial infection (2 h p.i.), but were slightly reduced at subsequent time points (4 and 6 h p.i.) (Figs 5, 7D and E; Table S2). As observed in macrophages (Eriksson et al., 2003), some SPI5 genes such as pipABD showed similar expression patterns to SPI2 genes inside epithelial cells, consistent with the important role of SPI5 in intracellular survival (Ehrbar et al., 2002). We hypothesize that SPI2 expression inside epithelial cells results from the low level of iron or phosphate present in the host cells, because SPI2 genes have been reported to be upregulated under low iron- and low-phosphate conditions (Zaharik et al., 2002; Lober et al., 2006).
S. Typhimurium SL1344 produces flagellin inside epithelial cells
The importance of flagella in pathogenesis has long been established for various bacteria (Achtman et al., 1986; Akerley et al., 1992; Legnani-Fajardo et al., 1996; Feldman et al., 1998; Nuijten et al., 2000) but flagella have become increasingly relevant to the study of intravacuolar pathogens, such as Legionella and Salmonella (Amer et al., 2006; Franchi et al., 2006; Miao et al., 2006; Miao et al., 2007). Although it was initially believed that flagella merely accelerated the invasion process of Salmonella (van Asten et al., 2004), studies have shown that lack of flagella caused attenuation at several different levels during infection (Schmitt et al., 2001). Furthermore, recent studies propose that, in some circumstances, Salmonella secretes flagella from the SCV into the cytosol of macrophages, in a SPI1 T3SS-dependent manner (Miao et al., 2007; Sun et al., 2007). It has been suggested that intracellular flagellin secretion has at least two purposes: first, cytosolic recognition of flagellin through the NLR-mediated signalling pathway would contribute to trigger a pro-inflammatory response (Sun et al., 2007); second, production of flagellin inside mammalian cells could lead to the escape of Salmonella from dying macrophages (Sano et al., 2007). These observations suggest that the flagellin protein plays a direct role in Salmonella pathogenesis, which is independent of the role of flagella in motility. However, we note that this phenomenon was only observed when macrophages were infected with Salmonella grown in SPI1-inducing conditions. When Salmonella were grown in non-SPI1 inducing conditions, we did not observe de novo transcription of fliC and fljB flagellin-encoding genes inside macrophages (Eriksson et al., 2003).
Our protocol involved the infection of epithelial cells with bacteria which had been grown in SPI1-inducting conditions; the transcriptomic analysis revealed time-dependent upregulation of S. Typhimurium flagella genes. Over 50 genes are required for a functional flagellar system (Chilcott and Hughes, 2000; Macnab, 2007). In macrophages, most flagellar genes were downregulated and remained downregulated at all time points (see Fig. 6A) (Eriksson et al., 2003). A similar pattern of expression was observed inside epithelial cells after 2 h infection when most flagella genes were also downregulated (Fig. 6A). Strikingly, the expression of 33 flagella genes were subsequently upregulated after 4 and 6 h infection of epithelial cells, reaching similar levels of expression to those seen in LB. Immunogold and immuno-fluorescent labelling against the FliC and FljB flagellins showed that fliC- and fljB-encoded proteins are present at detectable levels in infected epithelial cells at 6 h p.i. (Fig. 6C and D). The fluorescent micrograph on Fig. 6B shows discrete polymerized Salmonella flagella within infected cells. This was confirmed by confocal microscopy (data not shown).
Fig. 6.
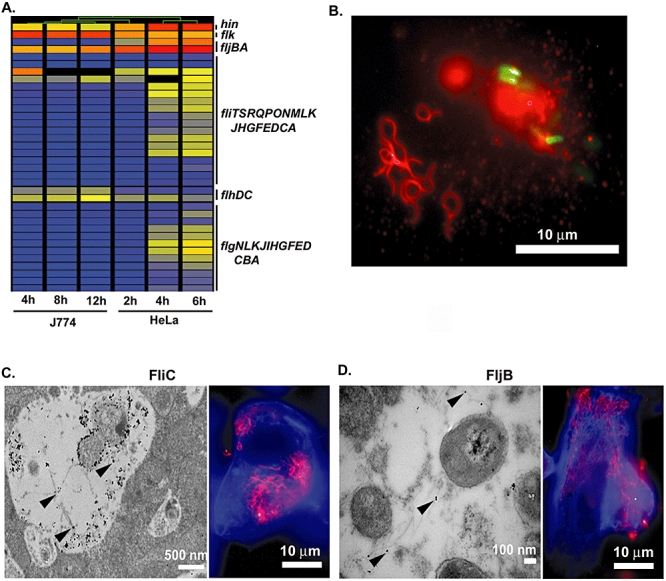
Expression of flagella by S. Typhimurium SL1344 inside epithelial cells. Cluster diagram of the expression profile of 36 flagella genes inside macrophage and epithelial cells, relative to the LB comparator (A). The colours are as Fig. 5A. Fluorescence micrograph shows polymerized flagella (red) separated from ssaG::gfp+ expressing JH3009 Salmonella cells (green) within epithelial cells at 6 h p.i. (B). Fluorescence and Transmission immunogold electron micrographs showing presence of FliC and FljB inside epithelial cells at 6 h p.i. (C, D). Flagella proteins were visualized with Anti-FliC (C) and Anti-FljB (D) antibodies. The arrow heads show immunogold labelling of FliC (C) and FljB (D) on TEM micrographs. The fluorescence micrographs in panels C and D show actin (blue) and large amounts of intracellular flagellin (magenta).
Verification of de novo production of flagellin and SPI2 protein within epithelial cells
In order to support our observation concerning fliC and fljB expression, we tested transcription of flagellar genes by RT-PCR, and observed increased flagellar gene expression at 6 h p.i. inside epithelial cells (Fig. 7F–I; Table S1).
As the detection of flagella proteins inside epithelial cells could have resulted from flagellin transcytosis (Lyons et al., 2004), we tested de novo synthesis of flagellin by assessing the levels of flagellin protein throughout infection in the presence or absence of chloramphenicol to inhibit protein synthesis (Experimental procedures). We first confirmed that bacterial protein synthesis had been inhibited intracellularly by quantifying MopA (GroEL) at 2 h and 6 h p.i. MopA was selected because transcription levels of the mopA gene remain fairly constant at all time points inside epithelial cells and in LB broth culture (Table S1). When protein synthesis was active, twice as much MopA was detected at 6 h p.i. compared with the 2 h time point. When bacterial protein synthesis was inhibited, the amount of MopA was similar at both time points (Fig. 8A), confirming that bacterial protein synthesis was effectively inhibited by chloramphenicol inside mammalian cells. Similar results were observed for the SPI2 protein SseC, a Salmonella protein that is synthesized inside mammalian cells (Fig. 8B). A greater than threefold increase was observed in SseC between 2 h and 6 h p.i. inside epithelial cells, and this increase was dependent upon protein synthesis. This confirmed that the SPI2 apparatus was synthesized de novo inside epithelial cells. We note that the increase of the total amount of SseC and MopA reflects the level of intracellular bacterial replication.
Fig. 8.
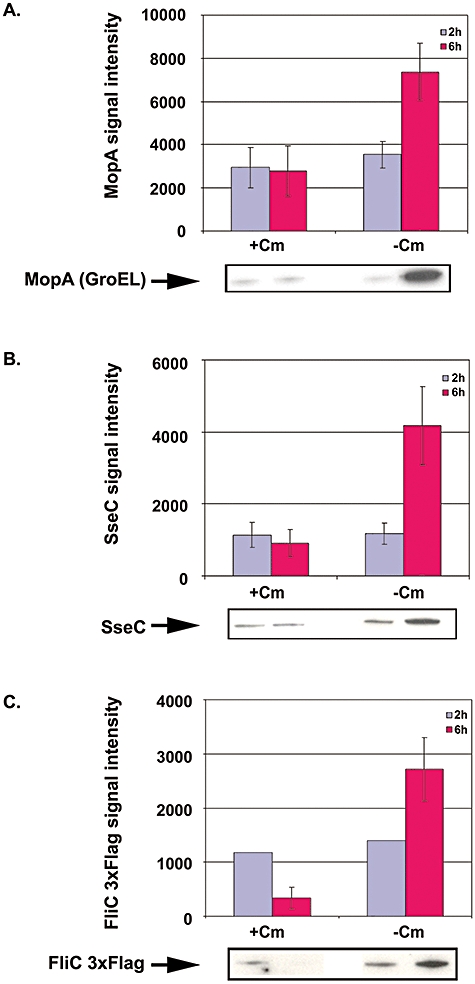
Confirmation of the de novo production of MopA, SseC and FliC 3xFlag by S. Typhimurium SL1344 inside epithelial cells. The levels of MopA (GroEL) (A), SseC (B) and 3xflag FliC (C) proteins detected at 2 h (purple) and 6 h (magenta) post infection in lysates of infected epithelial cells. De novo protein synthesis was assessed by treating intracellular bacterial cells with chloramphenicol and comparing protein levels with that of untreated intracellular bacteria. Experiments were performed in triplicate. The error bars indicate the SEM. For each protein, a representative example of the Western blot is given below the bar chart.
This system was used to show that flagellin was produced de novo inside epithelial cells (Fig. 8C). A twofold increase of FliC 3xFlag was detected inside epithelial cells at 6 h p.i. compared with the 2 h time point, and this was dependent upon bacterial protein synthesis. When bacterial protein synthesis was inhibited, around three times less FliC 3xFlag was detected at 6 h than at 2 h p.i. (Fig. 8C), whereas the level of the MopA control remained stable at the two time points (Fig. 8A). The reduced amount of intracellular flagellin at 6 h could result from degradation of FliC in the intraepithelial environment where substantial proteolysis may occur; MopA would be less exposed to the host proteolytic action of the host cell as it remains inside the bacterial cells. The twofold increase of FliC 3xFlag is probably an underestimation of the intracellular production of FliC, as it represents both proteolytic degradation of FliC and de novo synthesis. Clearly, this increase reflects bacterial intracellular replication, and shows for the first time that flagellin is produced within epithelial cells at this stage of infection.
The induction of expression of a fliC::gfp transcriptional gene fusion has been reported during murine infection (Cummings et al., 2006). The authors noted that fliC::gfp transcription was restricted to an unknown cell type located in Peyer's patches, and was not observed either in the mesenteric lymph nodes or in the spleen of infected mice. Our study builds on the work of Cummings et al. and provides the first description of de novo production of flagella by S. Typhimurium inside epithelial cells. It is not clear why Salmonella should do this, but it is conceivable that the flagellar T3SS could secrete other proteins including virulence effectors, as has been shown for other pathogens such as Yersinia enterocolitica (Young et al., 1999) and postulated to occur in Salmonella (Komoriya et al., 1999). Alternatively, production of flagellin could reflect bacterial adaptation to the fate of the host cell (see below).
S. Typhimurium SL1344 is highly cytotoxic to epithelial cells
The intracellular production of three different T3SSs was unexpected and may reflect a difference in the morphology of the epithelial cells between early and late infection stages. The epithelial cells responded promptly to SL1344 infection by inducing nearly twice as much IL-6 and IL-8 secretion at 2 h p.i. compared with uninfected cells. By 6 h p.i., the level of secretion of both pro-inflammatory cytokines was over 10-fold higher than in non-infected cells (Fig. 9A). Inhibition of bacterial protein synthesis did not change the level of release of IL-6 and IL-8 upon infection (data not shown), suggesting that cytokine secretion is induced at an early stage of Salmonella infection. We performed microscopic analysis of Salmonella-infected epithelial cells, which revealed striking cytotoxic effects. At 6 h p.i., TEM and light microscopic observations showed that 70% of the epithelial cells showed signs of cell death, such as chromatin condensation or vacuolization of the cell cytoplasm (Zychlinsky and Sansonetti, 1997; Fink and Cookson, 2005) (Fig. 9B). This alteration in cell morphology coincided with an increased release of the cytoplasmic lactate dehydrogenase (LDH) into the culture medium at 4 and 6 h p.i. (Fig. 9C). Such cytotoxicity by S. Typhimurium SL1344 was not restricted to HeLa epithelial cells, but also observed upon infection of two other epithelial cell lines, MDCK and Caco-2 (Fig. 9D). At similar time points, infected J774 A.1 macrophage cells retained their integrity (data not shown).
Fig. 9.
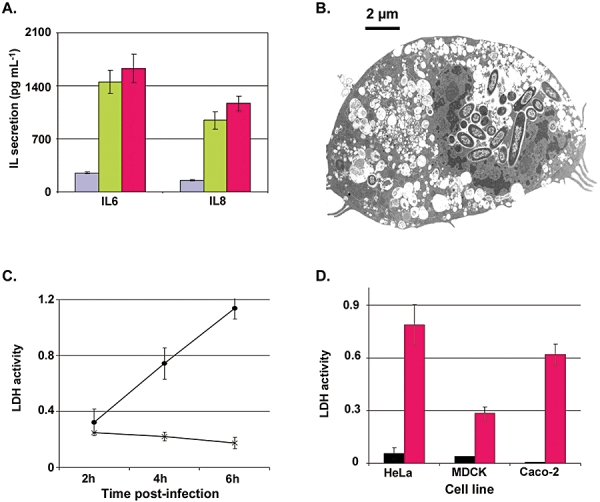
S. Typhimurium is cytotoxic to epithelial cells. Levels of IL-6 and IL-8 secreted by HeLa cells infected with SL1344 were measured by flow cytometry (Experimental procedures). Levels of secreted IL from epithelial cells at 2 h (purple), 4 h (green) and 6 h (magenta) post infection (A). Error bars in panels A, C and D show SEM. Transmission electron micrograph showing a dying epithelial cell at 6 h post infection which was representative of 70% of the infected epithelial cell sections that were observed (B). Cytotoxic effect of SL1344 strain (closed circle) on epithelial cells after 2, 4 and 6 h infection compared to non-infected cells (cross) (C). All cytotoxicity data were obtained from three independent replicates. Cytotoxic effect of SL1344 strain on HeLa, MDCK and Caco-2 epithelial cell lines. Black bars show LDH levels obtained on uninfected epithelial cells. Magenta bars show the LDH levels obtained on infected epithelial cells after 6 h infection (D).
The substantial and rapid cytotoxicity caused by S. Typhimurium SL1344 in epithelial cells leads us to suggest a two-step hypothesis to explain the temporal change in Salmonella gene expression patterns that were observed. During the early adaptation of Salmonella to the epithelial SCV a component of the transcriptomic changes could lead to cytotoxicity. This cell death might serve to moderate epithelial cell pro-inflammatory cytokine responses. Second, the time-dependent changes in the intraepithelial transcriptome of S. Typhimurium SL1344, such as the fluctuations in expression of mgtBC, iron uptake systems and flagella genes (Figs 4 and 6), could reflect the response of Salmonella to the increasing levels of epithelial cell death.
Expression of T3SS is associated with increased carbohydrate metabolism and energy requirement by Salmonella inside epithelial cells
The simultaneous expression of the three T3SS inside epithelial cells is a remarkable event which raises several questions that are currently under investigation. We also hypothesize that such a drain on cellular energy levels would be reflected in the expression patterns of the main metabolic pathways. In E. coli, the demand for ATP controls over 75% of the glycolytic flux (Koebmann et al., 2002). Our transcriptomic data showed that genes involved in ATP synthesis were transcribed at higher levels in epithelial cells than in macrophages (Table S1). These observations suggest that more glucose is catabolized by Salmonella inside epithelial cells than in macrophages. Elevated glucose metabolism of intraepithelial Salmonella also correlates with the upregulation of ptsG and uhpT genes (Fig. 4B), encoding a high affinity glucose transporter (Melton et al., 1976) and a hexose phosphate transporter, respectively (Table S1), suggesting that both sugars and sugar phosphates might be available to intraepithelial Salmonella. A similar observation was made for Listeria monocytogenes and Shigella flexneri (Chico-Calero et al., 2002; Lucchini et al., 2005). When Salmonella lacked the uhpT gene, it showed reduced growth on glucose 6-phosphate as sole carbon source in vitro (data not shown); however, no reduction in growth was observed in epithelial cells compared with the parental strain (Table 3), as observed for Shigella (Runyen-Janecky and Payne, 2002), suggesting either that glucose 6-phosphate is not the only available intracellular carbon source or that it is transported by other sugar transporters that are induced intracellularly but not in vitro (Table S1).
Many Salmonella genes encoding enzymes of the Entner–Doudoroff (ED) pathway were expressed in both epithelial cells (Fig. 7J–L) and macrophages (Table S1), with genes such as zwf, pgi and gapA showing up to twofold higher expression in epithelial cells than in macrophages (Fig. 10A and B). The ED pathway facilitates growth on sugar acids such as galacturonate that are found in the gastrointestinal tract (Salyers and Leedle, 1983; Allen, 1984). Together these data suggest that S. Typhimurium may use host-sugar acids as intracellular carbon sources, in both macrophages and epithelial cells, in addition to host-derived glucose in epithelial cells.
Fig. 10.
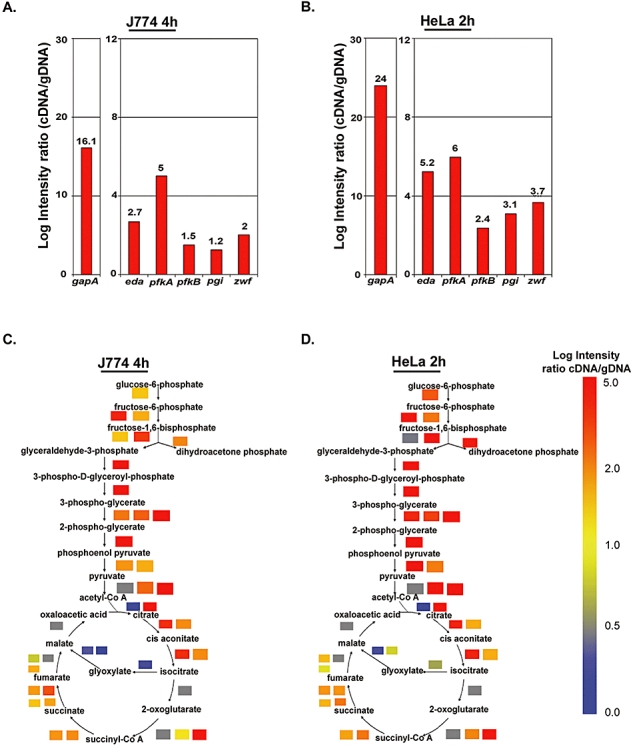
The S. Typhimurium TCA cycle, Entner–Doudoroff and Glycolytic pathways are more active inside epithelial cells than macrophage cells. Panels A and B show expression levels of S. Typhimurium genes involved in the Entner–Doudoroff pathway inside macrophages at 4 h p.i. (A) and inside epithelial cells at 2 h p.i. (B). These bar charts show the approximate level of each mRNA from the un-normalized transcriptomic data set. C and D show colour-coded expression levels for genes involved in each step of the S. Typhimurium glycolysis and TCA pathways. This figure does not involve normalization to a comparator but uses transcriptomic data to show the approximate level of each mRNA from the non-normalized Log intensity ratios between cDNA and genomic reference DNA. Each coloured block refers to a specific gene that encodes a relevant enzyme involved in particular metabolic pathways. Red indicates that the gene is highly expressed, yellow that the gene is expressed at intermediate levels, blue that the gene is poorly expressed and grey that no data were available for that gene. Panel C shows the gene expression data from Salmonella isolated from macrophages (4 h p.i.) and (D) data obtained from Salmonella isolated from epithelial cells (2 h p.i.). Key differences can be seen by comparing particular coloured blocks across C and D.
TCA cycle and glycolysis genes were expressed in Salmonella isolated from both infected macrophages and epithelial cells (Fig. 10C and D; Table S1), implying that both pathways are still active inside these host cells. Our results are consistent with a recent study which detected Salmonella enzymes involved in the TCA cycle inside infected spleenic macrophages (Becker et al., 2006). We observed that the central metabolic pathway of glycolysis, a major route for glucose catabolism, was expressed at higher levels in infected epithelial cells than in macrophages (Fig. 10C and D, Table S1).
The elevated activity of the Salmonella glycolytic and TCA pathways in epithelial cells fits with the higher expression levels of several Salmonella genes involved in the aerobic electron transport chain and ATP generation, such as the nuo (Fig. 7M), cyd, hem and atp genes (Table S1). The demand for energy production could therefore be expected to be reflected in the metabolic fluxes of intraepithelial Salmonella, involving the production of ATP through glycolysis, TCA cycle and ED pathways. In summary, our data suggest that glucose, phosphate sugars and sugar acids may be important carbon and energy sources inside epithelial cells. Energy from the generated ATP could be utilized for iron, phosphate and other transport systems (Kadner, 1996), and for biosynthetic activities. As the Salmonella SL1344 strain replicates only twice between 2 and 6 h p.i. inside epithelial cells, the apparently larger demand for ATP synthesis in epithelial cells than in macrophages could also be linked to the higher levels of expression of T3SS and flagellar machineries within the epithelial cells.
Concluding remarks
By comparing global gene expression of strain SL1344 inside epithelial cells and macrophages (Eriksson et al., 2003), we identified the intracellular transcriptomic signature of S. Typhimurium. We have also gained insights into the strategies used by this pathogen to adapt to the two main mammalian cell types in which S. Typhimurium can replicate. The key Salmonella gene expression profiles inside epithelial cells were validated with RT-PCR on additional biological replicates.
Our transcriptomic data are consistent with previous studies that showed that the SCV of both macrophages and epithelial cells contain limiting levels of magnesium, phosphate and iron, and are summarized in Fig. 11. We were interested to see that the genes required for bovine enteritis are highly expressed in HeLa cells (Table S17; Morgan et al., 2004). Recently, 95 S. Typhimurium genes necessary for porcine intestinal infection were identified (Carnell et al., 2007) and these closely parallel the genes implicated in the calf enteritis infection model. Perhaps HeLa cells will be a useful model for the intracellular aspect of epithelial cell-mediated infection, despite the fact that HeLa cells are not differentiated.
Fig. 11.
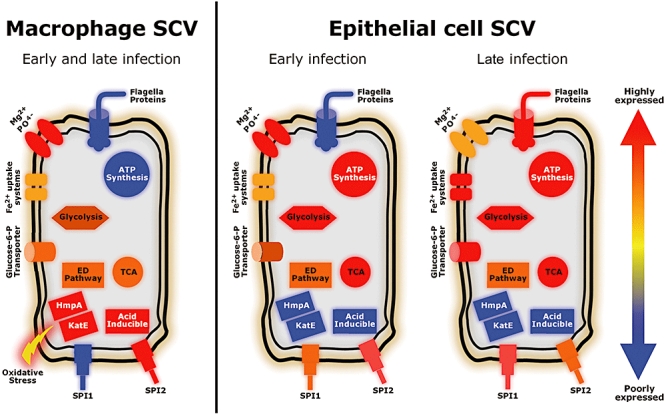
Model proposed for the responses of Salmonella Typhimurium to the intracellular environment of macrophages and epithelial cells. The response of S. Typhimurium to the macrophage SCV is relatively stable by 4 h p.i. and hardly varies at later time points. Conversely, the epithelial SCV changes through time, probably because Salmonella responds to alterations in the epithelial cells from the early to the late stages of infection. Each symbol represents groups of functionally related proteins and is coloured according to expression levels of the appropriate genes under each condition.
Clear differences in virulence gene expression patterns were observed between the macrophages and epithelial cells; for example, oxidative and nitrosative stresses caused higher expression of many detoxification genes inside macrophages than within epithelial cells. This fits with the dogma that epithelial cells are not designed to kill pathogens, and are permissive for bacterial replication.
Genes belonging to the SPI2 and the SsrA regulon were upregulated in both cell types, but their expression levels were higher in macrophages than in epithelial cells (Fig. 11). This expression of SPI2 genes is consistent with the requirement of SPI2 effectors for the biogenesis and evolution of both macrophage and epithelial SCVs, interfering with basic intracellular transport processes such as the host-actin cytoskeleton and microtubule network, as well as the innate and adaptive immune system (Abrahams and Hensel, 2006). The spvABCDR genes were also upregulated in both cell types, reflecting the importance of these plasmid-encoded genes for intracellular survival and replication of S. Typhimurium (Rhen et al., 1993; Gulig et al., 1998; Eriksson-Ygberg et al., 2006).
Strikingly, SPI1 genes were expressed inside epithelial cells at all time points post invasion, whereas they were strongly downregulated in macrophages (Fig. 11). SPI1 gene products are required for the invasion of mammalian cells but also contribute to the induction of pro-inflammatory responses (Murray and Lee, 2000) and the SPI1 effector protein SopB can stimulate nitric oxide production by macrophages (Drecktrah et al., 2005).The SPI1 T3SS machinery can also translocate certain SPI2-dependent effectors, such as SspH1, SlrP or SseK1 (Abrahams and Hensel, 2006). Such cross-talk between SPI1 and SPI2 could occur intracellularly and is consistent with the reported ability of the SPI1 regulator HilA to impact upon transcription of the SPI2 saaH gene (Thijs et al., 2007).
The second unexpected expression profile involved the flagella biosynthetic genes; these were strongly downregulated inside macrophages at all time points and inside epithelial cells at 2 h p.i., but were induced in a time-dependent manner inside epithelial cells at later stages of infection (Fig. 11). Monomeric flagellin is the ligand of the TLR5 receptor and triggers secretion of the neutrophil chemoattractant IL-8 by epithelial cells (Gewirtz et al., 2001b). S. Typhimurium flagellin is also transcytosed in a SPI2-dependent manner to the basolateral side of epithelial cells, in a separate compartment to the SCV (Lyons et al., 2004). Additionally, flagellin present in the cytosol of infected macrophages can be recognized by the Birc- 1e/Naip5 and Ipaf cytosolic NOD receptor molecules (Inohara et al., 2005; Franchi et al., 2006; Miao et al., 2007), triggering the pro-inflammatory signalling cascade as well as pyroptosis. Recently, monomeric FliC has been shown to be produced and secreted by extracellular Salmonella Typhi and Typhimurium upon contact with epithelial cell-derived lysophospholipids (Subramanian and Qadri, 2006). Our results show for the first time that S. Typhimurium can produce flagellin de novo inside epithelial cells, and the consequent biological implications are currently under investigation. Additionally, it is possible that expression of flagellar genes impacts upon intracellular expression of SPI1 genes, as observed previously in vitro (Lucas et al., 2000).
The intracellular production of flagellin coincided with a strong cytotoxic effect of S. Typhimurium strain SL1344 upon epithelial cells at the late stage of infection. It is possible that the pathogen senses the imminent death of the host epithelial cells and/or the pro-inflammatory signalling mediators, resulting in IL-6 and IL-8 secretion. Upregulation of the genes encoding flagella and the SPI1 T3SS could facilitate the invasion of neighbouring uninfected cells, and might build on the recent report that intracellular flagellin production mediated the escape of Salmonella from macrophages undergoing oncosis (Sano et al., 2007). It is possible that the SPI1 T3SS could secrete flagellin inside epithelial cells as has been reported in macrophages (Sun et al., 2007). This secretion process would be energy demanding and would be consistent with the elevation of S. Typhimurium carbon metabolism in epithelial cells compared with macrophages (Fig. 11).
In summary, we have identified likely similarities and differences between the SCVs of epithelial and macrophage cells. Our discovery that all three of the Salmonella T3SSs were transcribed simultaneously inside epithelial cells lead us to hypothesize that Salmonella is undergoing radical adaptation to epithelial cells that are close to death.
Experimental procedures
Bacterial strains and growth conditions
The S. Typhimurium SL1344 used in this study, and in our previous transcriptomic analyses (see http://www.ifr.ac.uk/Safety/MolMicro/pubs.html for references), was kindly provided by Catherine Lee (Hoiseth and Stocker, 1981; Shea et al., 1999). All strains are listed in Table 4. Strains were grown in 25 ml of LB broth (Sambrook and Russell, 2001) at 37°C, unless stated otherwise. Cultures were shaken in 250 ml flasks at 250 r.p.m. For invasion assays, a 1:10 dilution of a 5 ml overnight bacterial culture was grown in 5 ml of LBS (LB containing a total of 0.3 M NaCl) in 30 ml universals until an optical density at 600 nm (A600) of 1.2. Antibiotics were added as required at the following final concentrations (ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; chloramphenicol, 10 μg ml−1).
Table 4.
Bacterial strains used in this study.
| Name | Description | Source orReference |
|---|---|---|
| SL1344 | 4/74 hisG rpsL | Hoiseth and Stocker (1981) |
| JH3009 | SL1344 Φ(ssaG′-gfp+), Cmra | Hautefort et al. (2003) |
| JH3214 | SL1344 ΔbioABFCD (Cmr) | This study |
| JH3216 | SL1344 ΔuhpT, Cmr | This study |
| JH3053 | SL1344 (fliC-3xflag), Kmrb | This study |
| JH3227 | SL1344 (pPIR), Ampr | This study |
The gfp gene fusions are inserted in the putPA locus at positions 1 210 040–1211 657 on the LT2 genome (Hautefort et al., 2003). For these strains, ‘Φ’ indicates transcriptional gene fusion.
Carries a protein fusion of the 3xFlag tag to the C-terminal part of FliC.
Construction of deletion mutants and epitope-tagged chromosomal genes
The entire bioABFCD operon and uhpT structural genes were replaced by PCR-generated antibiotic resistance cassettes on the S. Typhimurium chromosome. Briefly, either a chloramphenicol or kanamycin resistance cassette was amplified by PCR from pKD4 and pKD3 plasmid templates using primers listed in Table 5. Each of these primers includes at its 5′ ends a 40–50 base-long extension showing homology with the flanking regions the target gene. The PCR products were used to replace the coding sequence of the target genes on S. Typhimurium chromosome using the Lambda Red recombination system (Datsenko and Wanner, 2000); recombinant strains JH3214 and JH3216 were selected for antibiotic resistance and verified by analytical PCR. Strain JH5053 (Table 3), carrying fliC gene tagged with the 3xFlag epitope sequence, was constructed as described previously (Uzzau et al., 2001). Recombinant transfer of the 3xFlag sequence into the fliC gene was achieved according to the Lambda Red recombination method from Datsenko and Wanner (2000). All deletions or recombinant fliC gene were subsequently transduced with P22-phage into a clean S. Typhimurium SL1344 background to avoid possible non-intended recombination events (Gemski and Stocker, 1967). Strain JH5053 remained motile, showing that the 3xFlag-tagged flagellin proteins were functional.
Table 5.
Oligonucleotides used in this study.
| Name | Sequence | Use in this study |
|---|---|---|
| bio_F | CTTGCCCGACGAAGAAGCGTTGAAGCGCCGCCATATTGACCGGATGCGTG gtgtaggctggagctgcttca | Construction of deletion mutant |
| bio_R | AAAAACAGCAGGATCGTCTGATCCTGCAAGAAGGCAGTTTTTAAGCGCAGcatatgaatatcctccttaga | Construction of deletion mutant |
| uhpT_F | TTTACAATGCCTGCCATTCGCAGGTATAAAAATTAGCTCAGGAGTAATCCgtgtaggctggagctgcttca | Construction of deletion mutant |
| uhpT_R | GGCGTCCAGCGCGGCGAACGTGCCCGCCCAGCCGGTCAGACCAAAAACGGcatatgaatatcctccttaga | Construction of deletion mutant |
| fliC-3xFlag_F | GGCGAACCAGGTTCCGCAAAACGTCCTCTCTTTACTGCGTgactacaaagaccatgacgga | Construction of FliC-3xFlag protein |
| fliC-3xFlag_R | CCTTGATTGTGTACCACGTGTCGGTGAATCAATCGCCGGAcatatgaatatcctccttaga | Construction of FliC-3xFlag protein |
| entB RT_F2 | gtttatgcgcgacattaagc | RT-PCR confirmatory experiments |
| entB RT_R2 | ccatcatgcgtactgaatcc | RT-PCR confirmatory experiments |
| flgI RT_F2 | ggcggacattcaaaatatgg | RT-PCR confirmatory experiments |
| flgI RT_R2 | acatcagatccatcggcgtc | RT-PCR confirmatory experiments |
| fliC RT_F2 | tgacagcagcaggtgttacc | RT-PCR confirmatory experiments |
| fliC RT_R2 | gttgtgaccttcggctttac | RT-PCR confirmatory experiments |
| fliF RT_F2 | gaatgtgggcgatattgagc | RT-PCR confirmatory experiments |
| fliF RT_R2 | tgttgccagaagggcagttc | RT-PCR confirmatory experiments |
| fljB RT_F2 | ggtaagacaattgaaggcgg | RT-PCR confirmatory experiments |
| fljB RT_R2 | ccagctctggttgtgctttg | RT-PCR confirmatory experiments |
| gapA RT_F2 | tactccgaacgtatccgttg | RT-PCR confirmatory experiments |
| gapA RT_R2 | tcgaacacggaagtgcatac | RT-PCR confirmatory experiments |
| invF RT_F2 | aagatcgtaaacgctgcgag | RT-PCR confirmatory experiments |
| invF RT_R2 | attgggtgatgttctcgtgg | RT-PCR confirmatory experiments |
| nuoB RT_F2 | tatgaccaaatgctggagcc | RT-PCR confirmatory experiments |
| nuoB RT_R2 | tgccaatcgattcttgcagc | RT-PCR confirmatory experiments |
| nusG RT_F2 | tggtccagatggtcatgaac | RT-PCR confirmatory experiments |
| nusG RT_R2 | ggcgagacttctcataatcg | RT-PCR confirmatory experiments |
| pgi RT_F | gggtaacatggaatccaacg | RT-PCR confirmatory experiments |
| pgi RT_R | taccctgatcgcgatattcc | RT-PCR confirmatory experiments |
| prgH RT_F | gagatacgttgtgggctcgt | RT-PCR confirmatory experiments |
| prgH RT_R | gcgctctcagcttttgactt | RT-PCR confirmatory experiments |
| sifA RT_F | ccatgcgaatatatccgaaa | RT-PCR confirmatory experiments |
| sifA RT_R | aaaatggcgtgaaaaacctg | RT-PCR confirmatory experiments |
| ssaG RT_F2 | aggccaggccattaatgaca | RT-PCR confirmatory experiments |
| ssaG RT_R2 | tagcaatgattccactaagc | RT-PCR confirmatory experiments |
| zwf RT_F2 | tcaagacgcctgaactgaac | RT-PCR confirmatory experiments |
| zwf RT_R2 | cttcggtaatggagtccacc | RT-PCR confirmatory experiments |
Uppercase sequences indicate homology with the flanking regions of the target genes or operon.
Epithelial cell infection by Salmonella strains
Human HeLa and Caco-2 epithelial cells (ECACC, No. 93021013, 86010202, respectively) and MDCK canine epithelial cells (ECACC, No. 85011435) were grown in medium containing DMEM (Cat. D-5796, Sigma) supplemented with 10% Fetal Bovine Serum (FBS; Cat. F-7524, Sigma), 2 mM L-Glutamine (Cat. G-7513, Sigma) and 10 mM HEPES buffer (Cat. H-0887, Sigma). This was subsequently called complete medium.
For the transcriptome analysis of S. Typhimurium inside epithelial cells, a total of 108 epithelial cells were seeded in 75 cm2 tissue culture flasks (12 flasks in total). For microscopic observations, between 5 × 105 and 106 epithelial cells were seeded in 6-well plates. All epithelial cells were incubated statically at 37°C, 10% CO2. S. Typhimurium bacteria were grown overnight in LB broth at 37°C, 250 r.p.m. The strains were then subcultured in LBS as described above. The late logarithmic cultures were diluted in complete medium without serum and used to infect the epithelial cell monolayer using a multiplicity of infection of 100 bacteria per epithelial cell. Contact with the epithelial cell monolayer was maximized by 5 min centrifugation at 1500 r.p.m. at room temperature. Infected epithelial cells were immediately incubated for 30 min at 37°C, 10% CO2. The time zero of an experiment was set at the beginning of this incubation. Remaining extracellular bacteria were then killed by replacement of the medium with complete medium containing 30 μg ml−1 gentamicin. After a 30 min incubation at 37°C, 10% CO2, the gentamicin concentration was reduced to 5 μg ml−1 of gentamicin until the end of the assay (i.e. for a further 1, 3 or 5 h).
To assess intracellular bacterial replication in epithelial cells, the SL1344 strain carrying the pPir plasmid (JH3227) was used (Posfai et al., 1997; Clements et al., 2002). Intracellular replication was assessed as described previously (Eriksson et al., 2000). During growth above 30°C, each round of replication results in a proportion of bacterial cells that do not inherit the plasmid, reflecting the level of bacterial replication.
De novo bacterial protein synthesis was assessed during epithelial cell infection for strains SL1344wt and JH3053 (Table 3). To inhibit bacterial protein synthesis, chloramphenicol was added to 10 μg ml−1 final concentration at the stage where extracellular bacteria are killed in gentamicin-containing medium, 30 min after the start of the assay, until the end of the assay.
RNA extraction
At each time point, infected epithelial cells were lysed on ice for 30 min in 0.1% SDS, 1% acidic phenol and 19% ethanol in water as described previously (Eriksson et al., 2003; Hinton et al., 2004). S. Typhimurium cells were pelleted after centrifugation of the pooled epithelial cell lysates and RNA was prepared using the Promega SV total RNA purification kit. Bacterial RNA was further purified by an extraction in a 50% acidic phenol-50% chloroform mix. Approximately, 5 × 108 bacteria were isolated from each time point, yielding between 0.5 and 10 μg of total RNA. The transcriptome of SL1344 grown in tissue culture media was considered to be an inappropriate comparator because many infection-relevant and environmentally responsive genes are upregulated in both DMEM and RPMI media, including mgtC, ent, fep, fhu, iro, bio, pur, some SPI2 genes and many genes of unknown function (data not shown). Of these genes, the iron-regulated iroN genes, the mgt magnesium transporter and SPI2 genes were already known to be upregulated inside epithelial and macrophage cells (Garcia-del Portillo et al., 1992; Smith et al., 1998). Furthermore, a study of Legionella pneumophila demonstrated that intracellular nutrition of Legionella was independent of the monocytic cell growth conditions and truly reflected the intracellular environment (Wieland et al., 2005). We assumed that the best comparator available would be a completely unrelated environment, such as LB broth. Control RNA from in vitro grown bacteria was obtained by growing Salmonella in LB as described previously (Lucchini et al., 2005). Size chromatography of RNA was carried out with an Agilent 2100 Bioanalyser.
Microarray procedures
Microarray techniques were performed essentially as described before (Lucchini et al., 2005; Nagy et al., 2006). All our transcriptome analyses were defined using microarrays covering 92% of the genes common between S. Typhimurium LT2 and SL1344 strains (Kelly et al., 2004).
Probe preparation and scanning
Because the amount of bacterial RNA extracted from infected epithelial cells were small, RNA were first reverse transcribed into cDNA and subsequently labelled by random priming with Klenow Fragment according to the ‘Labelling protocol for reduced amounts of RNA’ described at http://www.ifr.ac.uk/safety/microarrays/protocols.html
Fluorescently labelled genomic DNA was used as a reference channel for each experiment. Slides were scanned on an Axon 4000A scanner (Axon Instruments) using GenePix version 1.4 software (Axon Instruments). Each hybridization was performed twice. Two biological replicates were performed for the 2 and 6 h time points, and one for the 4 h time point.
Transcriptomic data analysis
Microarray data analysis was performed as described (Eriksson-Ygberg et al., 2006). Spots showing a reference signal lower than background plus two standard deviations or obvious blemishes were excluded from subsequent analyses. Local background was subtracted from spot signals, and fluorescence ratios were calculated using the Genepix version 1.4 software (Agilent). In a few cases, when comparing results from different hybridizations, we observed slight deviations which were dependent on gene expression levels. These were corrected using the Loess function in R (Limma package). To compensate for unequal dye incorporation or any effect of the amount of template, data centring was performed by bringing the median natural logarithm of the ratios for each group of spots printed by the same pin to zero. The complete data set is available as supplementary material. Data that passed the quality controls were analysed using Genespring version GX7.3 software (Agilent). Significance of the centred data at P = 0.05 was determined using a parametric-based statistical test adjusting the individual P-value with the Benjamini and Hochberg false discovery rate multiple test correction (Benjamini and Hochberg, 1995). We report reproducible changes in gene expression of more than twofold during infection. The LB and HeLa complete data sets have been submitted to the ArrayExpress repository with the accession number E-MEXP-1368.
Confirmation of transcriptomic data
RT-PCR analyses were performed on transcripts using gene-specific primer pairs (Table 4). Primers were designed in silico (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to minimize primer–primer complementarity and to yield predicted amplicons in the 100–300 bp range, corresponding to the terminal part of each gene coding sequence. Total Salmonella RNA was isolated from bacteria released from infected epithelial cells using the RiboPure-Bacteria RNA purification kit (Ambion), and converted to cDNA by using Stratascript (Stratagene) as described by the manufacturer. The cDNA was used as a template for PCR (PCR Master Mix, Promega). An appropriate number of cycles was chosen for each gene to ensure that the RT-PCR amplification was in the linear range (25–35). RT-PCR products were analysed by agarose gel electrophoresis. The product generated on the nusG (also known as rfaH) cDNA was used as a reference that shows a low level of variation in expression level in our experiments. PCR product intensities on agarose gels were measured using the AlphaDigidoc AD-1200 densitometry software (Alpha Innotech).
Fixation of infected mammalian cells for fluorescence microscopy
For fluorescence microscopic observations of infected mammalian cells, samples were immediately fixed for 15 min, at room temperature in 3% paraformaldehyde (w/v) (Sigma) freshly prepared in Phosphate Buffer Saline (PBS) pH 7.2. Fixed samples were subsequently washed in PBS and quenched in PBS containing 10 mM NH4Cl overnight at 4°C.
Fluorescence Immunohistochemistry
The rabbit anti-Salmonella FliC and FljB antibodies (Cat. 228 241 and 224 741 respectively, Biosciences Pharmingen) were used at dilution 1:400. Donkey Texas Red-conjugated Anti-Rabbit secondary IgG antibody (Cat. 715 075 150, Jackson Immunoresearch) was used at a final dilution of 1:100. All antibodies were diluted in PBS containing 10% horse serum and 0.1% Saponin. Briefly, samples were washed twice in PBS containing 0.1% Saponin (PBS-0.1% Saponin) and incubated for 1 h at room temperature in the primary antibody solution. The samples were subsequently washed once in PBS-0.1% Saponin and incubated for 1 h at room temperature in the secondary Antibody solution. The coverslips were then washed twice in PBS-0.1% Saponin, once in PBS and finally in H2O. Samples were mounted in Aqua Poly Mount (Cat. 18 606, Polysciences). Samples were observed on a Zeiss LSM510 META laser-scanning confocal microscope or an upright BX51 Olympus Optical Company fluorescence microscope.
Preparation of samples for Light and Transmission electron microscopy
For immunogold antiflagellin antibody labelling and light microscope observations, Salmonella-infected epithelial cells were fixed for 1 h in freshly prepared 4% paraformaldehyde (pH 7). The cells were then harvested and mixed with a drop of 2% low-melting-point agarose (Cat. A-4018, Sigma) near its setting point. The sample was cooled at 5°C for 10 min, chopped into small pieces and then dehydrated in an ethanol series from 10% to 90% with stepwise increases of 10%. After three changes in 100% ethanol, the sample was transferred to 50% LR White resin (London Resin) in ethanol, infiltrated with 100% LR White resin and polymerized at 60°C.
Light microscopic observations were performed on over 100 1 μm thick sections, mounted on glass slides and stained with 1% toluidine blue in 1% borax (pH 11) using a BX60 microscope (Olympus) and Acquis software (Syncroscopy).
For observations of epithelial cell structure, Salmonella-infected epithelial cells were briefly fixed in 3% glutaraldehyde (Agar Scientific) in 0.1 M cacodylate buffer (pH 7.2). The cells were subsequently scraped from the plates, pelleted and re-suspended in fresh fixative for 2 h, then washed and stored overnight in buffer alone. Cells were then pelleted and mixed with a drop of 2% low-melting-point agarose near its setting point. Samples were cooled at 5°C for 10 min, chopped into small pieces, then post-fixed in 2% aqueous osmium tetroxide (Agar Scientific) for 2 h and dehydrated in an ethanol series from 10% to 90% with stepwise increases of 10%. After three changes in 100% ethanol, the pieces were transferred to acetone, then infiltrated and embedded in Spurr epoxy resin (Agar Scientific) which was polymerized overnight at 75°C (Spurr, 1969). Sections approximately 75 nm thick were cut with a diamond knife, collected on copper grids, and stained sequentially with uranyl acetate (saturated in 50% ethanol) and Reynold's lead citrate (Reynolds, 1963). Sections were examined and photographed in a JEOL 1200 EX/B transmission electron microscope (TEM) at 80 kV.
Immunogold labelling
The anti-Salmonella FliC and FljB antibodies were used as for fluorescence immunohistochemistry (see above), and were diluted at 1:400. The secondary antibody, goat anti-rabbit conjugated to ultra-small gold (Cat. 800.011, Aurion) was diluted 1:75. Briefly, the free aldehydes in both LR White resin and Spurr-resin sections (previously etched for 10 s in sodium ethoxide, then rinsed in 100% ethanol) and LRW embedded sections, were blocked in 50 mM glycine solution in PBS for 15 min. Sections were subsequently blocked in goat serum blocking solution (Cat. 905.002, Aurion) for 30 min, washed three times for 5 min in acetylated-BSA (BSA-c) buffer (PBS containing 0.1% BSA-c, pH 7.4) and incubated overnight in primary antibody solutions in BSA-c buffer (Cat. 900.099, Aurion). Control sections were incubated in buffer alone. Sections were washed six times for 5 min in BSA-c buffer, and incubated in secondary goat anti-rabbit antibody for 2 h. Sections were washed six times for 5 min in BSA-c buffer, and three times for 5 min in PBS only. Sections were then post-fixed in 2% glutaraldehyde in PBS for 5 min, washed in PBS for 5 min and five times for 2 min in distilled water. Gold labelling was subsequently silver enhanced with Silver R-Gent SE-EM (Cat. 500.033, Aurion), for 25 min. Samples were then washed five times for 2 min in distilled water, stained in uranyl acetate, washed and air dried, before analysis by TEM as above.
Pro-Inflammatory IL-6 and IL-8 cytokine detection
The quantification of interleukin IL-6 and IL-8 in the supernatants of cultured epithelial cells was determined by using the cytometric bead array for Human IL-6 and IL-8 flex sets (Cat. 558 276 and 558 277 respectively, BD Bioscience, San Jose, CA, USA). The operations were performed according to the manufacturer's instructions. The intensity of the fluorescence signal was acquired on a FACScalibur fluorescence activated cell sorter flow cytometer (BD Becton Dickinson), and analysed using CellQuest software.
Protein methods
To visualize bacterial proteins by Western blotting, infected epithelial cells were lysed at each time point in 2 ml per well of 0.1% SDS in PBS pH 7.3. Samples were kept at −80°C until further processing. Ten microlitres of each sample was first heated at 100°C for 5 min run on a 12% SDS-PAGE (Cat. NP0342, Invitrogen). Transfer to PVDF membranes was performed as described by the manufacturer (Invitrogen). The membranes were then blocked for 45 min in 10% Marvel Milk prepared in 0.05% PBS-Tween 20. Immunodetection of proteins was performed using a chemiluminescence-based Super signal West Pico Rabbit detection kit (Cat. 34 083, Pierce). When monoclonal primary antibodies were used the goat anti-mouse IgG secondary antibody was used as described by the manufacturer (Cat. 31 430, Pierce). Signal intensity obtained for all proteins detected was compared with that obtained for MopA (GroEL). To increase sensitivity in our Western blot detection method, the C-terminal end of FliC was tagged with 3xFlag. The membrane was subsequently incubated for 1 h at room temperature in a 1:2000 dilution of an Anti-flag mouse monoclonal antibody (Cat. F-3165, Sigma), or a 1:15 000 dilution of an Anti-SseC polyclonal antibody (Kindly provided by Michael Hensel) or a 1:10 000 dilution of an Anti-GroEL polyclonal antibody (Cat. G-6532, Sigma), in 0.05% PBS-Tween 20 containing 5% Marvel milk. Fifteen minutes washes were repeated three times in 0.05% PBS-Tween 20 at room temperature, completed with two additional washes in PBS only after the secondary antibody incubation step. Detection was performed according to the manufacturer's instructions. Protein band signal intensities were measured using the AlphaDigidoc AD-1200 densitometry software (Alpha Innotech).
Cytotoxicity assay
Epithelial cells were infected as mentioned above except that DMEM without Phenol Red was used to prevent interference with this assay. Furthermore, the FBS content of the complete tissue culture medium was reduced to 5% because of the associated Lactate De-Hydrogenase activity it contains in order to reduce the background level. Lactate De-Hydrogenase enzyme activity released from the mammalian cells was measured in the tissue culture medium using the Cytotox 96 non-radioactive cytotoxicity assay kit (Cat. G1780, Promega) and was considered to reflect cytotoxicity of S. Typhimurium.
Acknowledgments
We are very grateful to David Holden for technical advice and valuable discussions, and to Francisco Garcia-del Portillo for reviewing our data on the cytotoxicity effect of SL1344 on epithelial cells. We thank Muna Anjum and Michael Hensel for providing us with monoclonal Anti-FliC and polyclonal Anti-SseC antibodies and Kathryn Gotts for her assistance with light microscopy. We appreciate initial discussions of microarray data with Nicholas Walton and Matthew Rolfe for contributing to the GeneSpring microarray data analysis. We thank Regis Stentz for his help and advice in our protein work, Gary Rowley for important suggestions throughout the whole project and Corinne Appia-Ayme for her comments on the manuscript. We are grateful to Maria José Proença for technical help during preliminary large scale epithelial cell infection experiments. The help of Paul Pople with Fig. 11 was much appreciated. We acknowledge funding from the Core Strategic Grant of BBSRC to Jay Hinton, from VR, Cancerfonden and STINT to Mikael Rhen and from the Defra VTRI VT104 to Vittoria Danino.
Supplementary material
The following supplementary material is available for this article online:
Salmonella is not exposed to oxidative or nitrosative stressors within epithelial cells. Transcriptomic data are shown for the S. Typhimurium hmpA and katE genes inside both macrophage and epithelial cells. Data for the expression of these genes are taken from Table S1. Error bars indicate the standard error of the mean.
The in vivo gene expression profile of S. Typhimurium SL1344 strain (4747 gene data set in MS Excel format).
Data taken from Table S1, including Salmonella Typhimurium known virulence genes, as depicted on Fig. 5.
Data taken from Table S1 showing the genes specifically upregulated in HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in HeLa cells at 2 and 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in both macrophages and HeLa cells, as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in macrophages and only HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in macrophages and only HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in macrophages at 4 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in HeLa cells at 2 and 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in both macrophages and HeLa cells, as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in macrophages and only HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in macrophages and only HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in macrophages at 4 h p.i., as depicted on Fig. 1.
Genes required for Salmonella-associated enteritis in the bovine infection model and for survival and replication in cultivated epithelial cells are expressed within HeLa cells.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1462-5822.2007.01099.x
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abrahams GL, Hensel M. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 2006;8:728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- Achtman M, Heuzenroeder M, Kusecek B, Ochman H, Caugant D, Selander RK, et al. Clonal analysis of Escherichia coli O2: K1 isolated from diseased humans and animals. Infect Immun. 1986;51:268–276. doi: 10.1128/iai.51.1.268-276.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Monack DM, Falkow S, Miller JF. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol. 1992;174:980–990. doi: 10.1128/jb.174.3.980-990.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A. The Structure and Function of Gastrointestinal Mucous. Boca Raton, FL: CRC Press; 1984. [Google Scholar]
- Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer A, Franchi L, Kanneganti T-D, Body-Malapel M, Ozoren N, Brady G, et al. Regulation of Legionella Phagosome Maturation and Infection through Flagellin and Host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- van Asten FJ, Hendriks HG, Koninkx JF, van Dijk JE. Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int J Med Microbiol. 2004;294:395–399. doi: 10.1016/j.ijmm.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Selbach M, Rollenhagen C, Ballmaier M, Meyer TF, Mann M, Bumann D. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- Beinert H, Holm RH, Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzon CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Jiang X, Ohlson MB, Miller SI, Brumell JH. Salmonella-induced filament formation is a dynamic phenotype induced by rapidly replicating Salmonella enterica serovar Typhimurium in epithelial cells. Infect Immun. 2005;73:1204–1208. doi: 10.1128/IAI.73.2.1204-1208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Beuzon CR, Holden DW, Gorvel JP, Meresse S. Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J Biol Chem. 2003;278:14196–14202. doi: 10.1074/jbc.M207901200. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Henry T, Borg JP, Gorvel JP, Meresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- Carnell SC, Bowen A, Morgan E, Maskell DJ, Wallis TS, Stevens MP. Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium secreted components identified by signature-tagged mutagenesis. Microbiology. 2007;153:1940–1952. doi: 10.1099/mic.0.2006/006726-0. [DOI] [PubMed] [Google Scholar]
- Chakravortty D, Hansen-Wester I, Hensel M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med. 2002;195:1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, et al. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, et al. Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. PNAS. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary A, Tiwari RP, Koul A, Chanana V, Gupta S, Rishi P. Role of Salmonella surface components in immunomodulation of inflammatory mediators. Mol Cell Biochem. 2005;270:167–175. doi: 10.1007/s11010-005-4506-x. [DOI] [PubMed] [Google Scholar]
- Ciacci-Woolwine F, Blomfield IC, Richardson SH, Mizel SB. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect Immun. 1998;66:1127–1134. doi: 10.1128/iai.66.3.1127-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements MO, Eriksson S, Thompson A, Lucchini S, Hinton JC, Normark S, Rhen M. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc Natl Acad Sci USA. 2002;99:8784–8789. doi: 10.1073/pnas.132047099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Knodler LA, Galbraith K, Steele-Mortimer O. The Salmonella SPI1 effector SopB stimulates nitric oxide production long after invasion. Cell Microbiol. 2005;7:105–113. doi: 10.1111/j.1462-5822.2004.00436.x. [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles T, Murthy K, Liaudet L, Virag L, Ross G, Soriano FG, et al. Flagellin, a novel mediator of Salmonella-induced epithelial activation and systemic inflammation: I kappa B alpha degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J Immunol. 2001;166:1248–1260. doi: 10.4049/jimmunol.166.2.1248. [DOI] [PubMed] [Google Scholar]
- Ehrbar K, Mirold S, Friebel A, Stender S, Hardt WD. Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium. Int J Med Microbiol. 2002;291:479–485. doi: 10.1078/1438-4221-00156. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Bjorkman J, Borg S, Syk A, Pettersson S, Andersson DI, Rhen M. Salmonella typhimurium mutants that downregulate phagocyte nitric oxide production. Cell Microbiol. 2000;2:239–250. doi: 10.1046/j.1462-5822.2000.00051.x. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Eriksson-Ygberg SE, Clements MO, Rytkonen A, Thompson A, Holden DW, Hinton JCD, Rhen M. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect Immun. 2006;74:1243–1254. doi: 10.1128/IAI.74.2.1243-1254.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. PNAS. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary Infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. PNAS. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer J, Eckmann L, Fang F, Pfeifer C, Finlay BB, Guiney D. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect Immun. 1993;61:5231–5236. doi: 10.1128/iai.61.12.5231-5236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Chatfield S, Leung KY, Dougan G, Falkow S. Characterization of a Salmonella choleraesuis mutant that cannot multiply within epithelial cells. Can J Microbiol. 1991;37:568–572. doi: 10.1139/m91-095. [DOI] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat Immunol. 2006;30:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- Frost AJ, Bland AP, Wallis TS. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet Pathol. 1997;34:369–386. doi: 10.1177/030098589703400501. [DOI] [PubMed] [Google Scholar]
- Gahring LC, Heffron F, Finlay BB, Falkow S. Invasion and replication of Salmonella typhimurium in animal cells. Infect Immun. 1990;58:443–448. doi: 10.1128/iai.58.2.443-448.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Curtiss R. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. 3rd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Wood MW, Rosqvist R, Mullan PB, Watson PR, Hedges S, Wallis TS. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- Garcia-del Portillo F, Finlay BB. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J Cell Biol. 1995;129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F, Foster JW, Maguire ME, Finlay BB. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol Microbiol. 1992;6:3289–3297. doi: 10.1111/j.1365-2958.1992.tb02197.x. [DOI] [PubMed] [Google Scholar]
- Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc Natl Acad Sci USA. 1993;90:10544–10548. doi: 10.1073/pnas.90.22.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemski P, Jr, Stocker BA. Transduction by bacteriophage P22 in nonsmooth mutants of Salmonella typhimurium. J Bacteriol. 1967;93:1588–1597. doi: 10.1128/jb.93.5.1588-1597.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Siber AM, Madara JL, McCormick BA. Orchestration of neutrophil movement by intestinal epithelial cells in response to Salmonella typhimurium can be uncoupled from bacterial internalization. Infect Immun. 1999;67:608–617. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001a;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- Gewirtz AT, Simon PO, Schmitt CK, Taylor LJ, Hagedorn CH, O'Brien AD, et al. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J Clin Invest. 2001b;107:99–109. doi: 10.1172/JCI10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvel JP, Meresse S. Maturation steps of the Salmonella-containing vacuole. Microbes Infect. 2001;3:1299–1303. doi: 10.1016/s1286-4579(01)01490-3. [DOI] [PubMed] [Google Scholar]
- Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignot J, Caron E, Beuzon C, Bucci C, Kagan J, Roy C, Holden DW. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J Cell Sci. 2004;117:1033–1045. doi: 10.1242/jcs.00949. [DOI] [PubMed] [Google Scholar]
- Gulig PA, Doyle TJ, Hughes JA, Matsui H. Analysis of host cells associated with the Spv-mediated increased intracellular growth rate of Salmonella typhimurium in mice. Infect Immun. 1998;66:2471–2485. doi: 10.1128/iai.66.6.2471-2485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- Harrison RE, Brumell JH, Khandani A, Bucci C, Scott CC, Jiang X, et al. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell. 2004;15:3146–3154. doi: 10.1091/mbc.E04-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautefort I, Proença MJ, Hinton JCD. Single copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Env Microbiol. 2003;69:7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Hensel M. Salmonella pathogenicity island 2. Mol Microbiol. 2000;36:1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton JC. The Escherichia coli genome sequence: the end of an era or the start of the FUN? Mol Microbiol. 1997;26:417–422. doi: 10.1046/j.1365-2958.1997.6371988.x. [DOI] [PubMed] [Google Scholar]
- Hinton JC, Hautefort I, Eriksson S, Thompson A, Rhen M. Benefits and pitfalls of using microarrays to monitor bacterial gene expression during infection. Curr Opin Microbiol. 2004;7:277–282. doi: 10.1016/j.mib.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Hobbie S, Chen LM, Davis RJ, Galan JE. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Inohara N, Chamaillard M, McDonald C, Nunez G. NOD-LRR PROTEINS: role in host–microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Jabado N, Cuellar-Mata P, Grinstein S, Gros P. Iron chelators modulate the fusogenic properties of Salmonella-containing phagosomes. Proc Natl Acad Sci USA. 2003;100:6127–6132. doi: 10.1073/pnas.0937287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman A, Slauch JM. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol Microbiol. 2000;35:1146–1155. doi: 10.1046/j.1365-2958.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- Jones MA, Wood MW, Mullan PB, Watson PR, Wallis TS, Galyov EE. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadner RJ. Cytoplasmic membrane. In: Neidhardt FC, editor. Escherichia Coli and Salmonella. Washington, DC: ASM Press; 1996. pp. 58–87. [Google Scholar]
- Kelly A, Goldberg MD, Carroll RK, Danino V, Hinton JCD, Dorman CJ. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Steele-Mortimer O. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic. 2003;4:587–599. doi: 10.1034/j.1600-0854.2003.00118.x. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Vallance BA, Hensel M, Jackel D, Finlay BB, Steele-Mortimer O. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol Microbiol. 2003;49:685–704. doi: 10.1046/j.1365-2958.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. The glycolytic flux in Escherichia coli Is controlled by the demand for ATP. J Bacteriol. 2002;184:3909–3916. doi: 10.1128/JB.184.14.3909-3916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa S-I. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol Microbiol. 1999;34:767–779. doi: 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- Kuhle V, Jackel D, Hensel M. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic. 2004;5:356–370. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- Legnani-Fajardo C, Zunino P, Piccini C, Allen A, Maskell D. Defined mutants of Proteus mirabilis lacking flagella cause ascending urinary tract infection in mice. Microb Pathog. 1996;21:395–405. doi: 10.1006/mpat.1996.0070. [DOI] [PubMed] [Google Scholar]
- Leung KY, Finlay BB. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lober S, Jackel D, Kaiser N, Hensel M. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int J Med Microbiol. 2006;296:435–447. doi: 10.1016/j.ijmm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Liu H, Jin Q, Hinton JCD, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun. 2005;73:88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons S, Wang L, Casanova JE, Sitaraman SV, Merlin D, Gewirtz AT. Salmonella typhimurium transcytoses flagellin via an SPI2-mediated vesicular transport pathway. J Cell Sci. 2004;117:5771–5780. doi: 10.1242/jcs.01500. [DOI] [PubMed] [Google Scholar]
- McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Flagella and motility. In: Neidhardt FC, editor. Escherichia Coli and Salmonella. Washington, DC: ASM Press; 2007. pp. 1–34. [Google Scholar]
- Marsman M, Jordens I, Kuijl C, Janssen L, Neefjes J. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell. 2004;15:2954–2964. doi: 10.1091/mbc.E03-08-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni P, Vazquez-Torres A, Fang FC, Xu Y, Khan S, Hormaeche CE, Dougan G. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med. 2000;192:237–248. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer JJ, Doggett TA, Burns-Keliher L, Curtiss R. Expression of the rfa, LPS biosynthesis promoter in Salmonella typhimurium during invasion of intestinal epithelial cells. Curr Microbiol. 2000;41:172–176. doi: 10.1007/s002840010113. 3rd. [DOI] [PubMed] [Google Scholar]
- Melton T, Kundig W, Hartman PE, Meadow N. 3-deoxy-3-fluoro-d-glucose-resistant Salmonella typhimurium mutants defective in the phosphoenolpyruvate: glycose phosphotransferase system. J Bacteriol. 1976;128:794–800. doi: 10.1128/jb.128.3.794-800.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meresse S, Unsworth KE, Habermann A, Griffiths G, Fang F, Martinez-Lorenzo MJ, et al. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell Microbiol. 2001;3:567–577. doi: 10.1046/j.1462-5822.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;30:549–551. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Miao EA, Andersen-Nissen E, Warren SE, Aderem A. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol. 2007;10:10. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Detweiler CS, Falkow S. Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell Microbiol. 2001;3:825–837. doi: 10.1046/j.1462-5822.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, et al. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2004;54:994–1010. doi: 10.1111/j.1365-2958.2004.04323.x. [DOI] [PubMed] [Google Scholar]
- Murray RA, Lee CA. Invasion genes are not required for Salmonella enterica serovar Typhimurium To breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect Immun. 2000;68:5050–5055. doi: 10.1128/iai.68.9.5050-5055.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emody L, et al. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun. 2006;74:5914–5925. doi: 10.1128/IAI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuijten PJM, van den Berg AJG, Formentini I, van der Zeijst BAM, Jacobs AAC. DNA rearrangements in the flagellin locus of an flaA mutant of Campylobacter jejuni during colonization of chicken Ceca. Infect Immun. 2000;68:7137–7140. doi: 10.1128/iai.68.12.7137-7140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattery T, Hernalsteens J-P, De Greve H. Identification and molecular characterization of a novel Salmonella enteritidis pathogenicity islet encoding an ABC transporter. Mol Microbiol. 1999;33:791–805. doi: 10.1046/j.1365-2958.1999.01526.x. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Mathur N, Giles D, Fahlen T, Jones BD. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- Poh J, Odendall C, Spanos A, Boyle C, Liu M, Freemont P, Holden DW. SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell Microbiol. 2007;10:20–30. doi: 10.1111/j.1462-5822.2007.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai G, Koob MD, Kirkpatrick HA, Blattner FR. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157 : H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathman M, Sjaastad MD, Falkow S. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun. 1996;64:2765–2773. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KA, Hobert ME, Kolenda CE, Sands KA, Rathman M, O'Connor M, et al. The Salmonella typhimurium flagellar basal body protein FliE is required for flagellin production and to induce a proinflammatory response in epithelial cells. J Biol Chem. 2002;277:13346–13353. doi: 10.1074/jbc.M200149200. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead cirtate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen M, Dorman CJ. Hierarchical gene regulators adapt Salmonella enterica to its host milieus. Int J Med Microbiol. 2005;294:487–502. doi: 10.1016/j.ijmm.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Rhen M, Riikonen P, Taira S. Transcriptional regulation of Salmonella enterica virulence plasmid genes in cultured macrophages. Mol Microbiol. 1993;10:45–56. doi: 10.1111/j.1365-2958.1993.tb00902.x. [DOI] [PubMed] [Google Scholar]
- Rosqvist R, Forsberg A, Rimpilainen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- Runyen-Janecky LJ, Payne SM. Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect Immun. 2002;70:4379–4388. doi: 10.1128/IAI.70.8.4379-4388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. PNAS. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Leedle JAZ. Carbohydrate Metabolism in the Human Colon. New York: Academic Press; 1983. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sano G-i, Takada Y, Goto S, Maruyama K, Shindo Y, Oka K, et al. Flagella facilitate escape of Salmonella from oncotic macrophages. J Bacteriol. 2007;189:8224–8232. doi: 10.1128/JB.00898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CK, Ikeda JS, Darnell SC, Watson PR, Bispham J, Wallis TS, et al. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect Immun. 2001;69:5619–5625. doi: 10.1128/IAI.69.9.5619-5625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Fadl AA, Klimpel GR, Niesel DW, Popov VL, Chopra AK. The two murein lipoproteins of Salmonella enterica Serovar Typhimurium contribute to the virulence of the organism. Infect Immun. 2004;72:3987–4003. doi: 10.1128/IAI.72.7.3987-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Beuzon CR, Gleeson C, Mundy R, Holden DW. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RL, Kaczmarek MT, Kucharski LM, Maguire ME. Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtCB during invasion of epithelial and macrophage cells. Microbiology. 1998;144:1835–1843. doi: 10.1099/00221287-144-7-1835. [DOI] [PubMed] [Google Scholar]
- Spector MP, DiRusso CC, Pallen MJ, Garcia del Portillo F, Dougan G, Finlay BB. The medium-/long-chain fatty acyl-CoA dehydrogenase (fadF) gene of Salmonella typhimurium is a phase 1 starvation-stress response (SSR) locus. Microbiology. 1999;145:15–31. doi: 10.1099/13500872-145-1-15. [DOI] [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Stecher B, Macpherson AJ, Hapfelmeier S, Kremer M, Stallmach T, Hardt W-D. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect Immun. 2005;73:3228–3241. doi: 10.1128/IAI.73.6.3228-3241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O, Meresse S, Gorvel JP, Finlay BB. Early events in the biogenesis of the Salmonella typhimurium-containing vacuole in cultured epithelial cells. Mol Biology Cell. 1998;9:2241. [Google Scholar]
- Steele-Mortimer O, Meresse S, Gorvel J-P, Toh B-H, Finley BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- Stein MA, Leung KY, Zwick M, Garcia-del Portillo F, Finlay BB. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol Microbiol. 1996;20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- Stibitz S, Weiss AA, Falkow S. Genetic analysis of a region of the Bordetella pertussis chromosome encoding filamentous hemagglutinin and the pleiotropic regulatory locus. Vir J Bacteriol. 1988;170:2904–2913. doi: 10.1128/jb.170.7.2904-2913.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian N, Qadri A. Lysophospholipid sensing triggers secretion of flagellin from pathogenic Salmonella. Nat Immunol. 2006;30:30. doi: 10.1038/ni1336. [DOI] [PubMed] [Google Scholar]
- Sun Y-H, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- Thijs IMV, De Keersmaecker SCJ, Fadda A, Engelen K, Zhao H, McClelland M, et al. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J Bacteriol. 2007;189:4587–4596. doi: 10.1128/JB.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto U, Nomura S, Minami Y, Mihara H, Kato S, Kurihara T, et al. Network of protein–protein interactions among iron-sulfur cluster assembly proteins in Escherichia coli. J Biochem (Tokyo) 2002;131:713–719. doi: 10.1093/oxfordjournals.jbchem.a003156. [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Baumler AJ, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Adams LG, Ficht TA, Baumler AJ. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth KE, Way M, McNiven M, Machesky L, Holden DW. Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cell Microbiol. 2004;6:1041–1055. doi: 10.1111/j.1462-5822.2004.00417.x. [DOI] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz W, Schmidt MA, Labigne-Roussel AF, Falkow S, Schoolnik G. AFA-I, a cloned afimbrial X-type adhesin from a human pyelonephritic Escherichia coli strain. Purification and chemical, functional and serologic characterization. Eur J Biochem. 1985;152:315–321. doi: 10.1111/j.1432-1033.1985.tb09200.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- Watson PR, Paulin SM, Bland AP, Jones PW, Wallis TS. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect Immun. 1995;63:2743–2754. doi: 10.1128/iai.63.7.2743-2754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way SS, Thompson LJ, Lopes JE, Hajjar AM, Kollmann TR, Freitag NE, Wilson CB. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol. 2004;6:235–242. doi: 10.1046/j.1462-5822.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- Wieland H, Ullrich S, Lang F, Neumeister B. Intracellular multiplication of Legionella pneumophila depends on host cell amino acid transporter SLC1A5. Mol Microbiol. 2005;55:1528–1537. doi: 10.1111/j.1365-2958.2005.04490.x. [DOI] [PubMed] [Google Scholar]
- Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. PNAS. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharik ML, Vallance BA, Puente JL, Gros P, Finlay BB. Host–pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. PNAS. 2002;99:15705–15710. doi: 10.1073/pnas.252415599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Wu H, Sloane V, Jones R, Yu Y, Lin P, et al. Flagellin/TLR5 responses in epithelia reveal intertwined activation of inflammatory and apoptotic pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G96–G108. doi: 10.1152/ajpgi.00273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinsky A, Sansonetti PJ. Apoptosis as a proinflammatory event: what can we learn from bacteria-induced cell death? Trends Microbiol. 1997;5:201–204. doi: 10.1016/S0966-842X(97)01044-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Salmonella is not exposed to oxidative or nitrosative stressors within epithelial cells. Transcriptomic data are shown for the S. Typhimurium hmpA and katE genes inside both macrophage and epithelial cells. Data for the expression of these genes are taken from Table S1. Error bars indicate the standard error of the mean.
The in vivo gene expression profile of S. Typhimurium SL1344 strain (4747 gene data set in MS Excel format).
Data taken from Table S1, including Salmonella Typhimurium known virulence genes, as depicted on Fig. 5.
Data taken from Table S1 showing the genes specifically upregulated in HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in HeLa cells at 2 and 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in both macrophages and HeLa cells, as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in macrophages and only HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in macrophages and only HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically upregulated in macrophages at 4 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in HeLa cells at 2 and 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in both macrophages and HeLa cells, as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in macrophages and only HeLa cells at 2 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in macrophages and only HeLa cells at 6 h p.i., as depicted on Fig. 1.
Data taken from Table S1 showing the genes specifically downregulated in macrophages at 4 h p.i., as depicted on Fig. 1.
Genes required for Salmonella-associated enteritis in the bovine infection model and for survival and replication in cultivated epithelial cells are expressed within HeLa cells.


