Abstract
Leptin (OB), an adipocyte-secreted circulating hormone, and its receptor (OB-R) are key components of an endocrine loop that regulates mammalian body weight. In this report we have analyzed signal transduction activities of OB-R containing the fatty mutation [OB-R(fa)], a single amino acid substitution at position 269 (Gln → Pro) in the OB-R extracellular domain that results in the obese phenotype of the fatty rat. We find that this mutant receptor exhibits both ligand-independent transcriptional activation via interleukin 6 and hematopoietin receptor response elements and ligand-independent activation of signal transducer and activator of transcription (STAT) proteins 1 and 3. However, OB-R(fa) is unable to constitutively activate STAT5B and is highly impaired for ligand induced activation of STAT5B compared with OB-R(wt). Introduction of the fatty mutation into a OB-R/G-CSF-R chimera generates a receptor with constitutive character that is similar but distinct from that of OB-R(fa). Constitutive mutant OB-R(fa) receptor signaling is repressed by coexpression of OB-R(wt). The implications of an extracellular domain amino acid substitution generating a cytokine receptor with a partially constitutive phenotype are discussed both in terms of the mechanism of OB-R triggering and the biology of the fatty rat.
Keywords: obesity, constitutive leptin receptor signaling, signal transducer and activator of transcription
Leptin (OB) is an adipose tissue-secreted hormone that modulates food intake and energy expenditure (1–5). Leptin’s effects are predicted to be mediated by interaction with the leptin receptor (OB-R) in the hypothalamus, a region of the brain implicated in the control of body weight (6–8). OB-R is a single membrane-spanning protein with functional and structural homology to members of the class I cytokine receptor superfamily (9). Receptors of this class lack intrinsic tyrosine kinase activity, are activated by ligand-induced receptor homo-or heterodimerization, and utilize the Janus kinase and STAT families of signal transducing molecules to regulate gene transcription (10–12).
Multiple splice variants of OB-R mRNAs encoding proteins with identical extracellular domains but differing length intracellular domains have been detected (9, 13, 14). The predominant short OB-R isoform, highly expressed in the choroid plexus, is proposed to be involved in leptin transport from the blood into the cerebral spinal fluid (9, 15). The long OB-R transcript (OB-RL), expressed predominately in specific nuclei of the hypothalamus, encodes a protein with an extended intracellular domain containing motifs required for OB-R signal transducing activity (9, 16). The mouse mutant allele (db) of the OB-R gene encodes an OB-RL with a truncated cytoplasmic domain (13, 14) and more recent data demonstrate that this receptor is signaling inactive (17–19).
Obese Zucker (fatty) rats exhibit severe early onset obesity due to excessive food intake, hypometabolism, insulin resistance, and preferential energy storage in adipose tissue (20). These animals are homozygous for the fatty mutation that maps to the OB-R gene (21). A single missense mutation in the OB-R fatty allele at position 880 (A-C) introduces an amino acid substitution at position 269 (Gln → Pro) within the OB-R extracellular domain (22–24). Short OB-R isoform containing this change has an unaltered affinity for ligand, but exhibits a 6–8-fold decrease in cell surface expression levels (relative to the wild-type receptor), giving rise to speculation that these animals are obese due to diminished leptin transport into the central nervous system (24).
In this report we have analyzed cell surface expression levels and signal transducing activities of OB-RL containing the fatty mutation. The mutant receptor exhibits ligand-independent activation of STAT1 and -3 and constitutive activation of reporter gene transcription. In addition, we find that constitutive signaling by OB-RL(fa) is repressed by coexpression of wild-type OB-RL. These findings are discussed with regard to the mechanism of OB-R triggering as well as potential biochemical abnormalities that generate leptin resistance and loss of body weight homeostasis in the fatty rat.
MATERIALS AND METHODS
Cell Culture.
COS-1, COS-7, and GT1–7 cells were cultured as described (25, 26). 293 cells were grown in DMEM supplemented with 10% fetal bovine serum. Cells were mock stimulated in serum-free medium or treated in the same medium supplemented with 100 ng/ml of either mouse leptin (R & D Systems) or human granulocyte-colony stimulating factor (G-CSF) (Boehringer Mannheim).
Expression Vectors and Secreted Alkaline Phosphatase (SEAP) Reporter Gene Constructs.
Expression vectors for murine short OB-R (9), the OB-R/G-CSF-R, and G-CSF-R/OB-R chimeras (27) and rat STAT1, -3, and -5B have been described (28, 29). To construct mouse OB-RL, hypothalami were isolated, and total RNA was prepared using the RNeasy kit (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. cDNA was synthesized by the random priming method using Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s recommendations (Life Technologies, Grand Island, NY). Sequences encoding mouse OB-R were amplified by PCR using TAQ DNA polymerase (Hoffmann–LaRoche) and primers complementary to their respective OB-R cDNA sequences. Introduction of the fatty mutation into full-length mouse OB-R cDNA was performed by overlap extension PCR as has been described (30). For the generation of constructs for stable expression of wild-type and mutant mouse OB-R, each cDNA was subcloned into pIRES1neo (CLONTECH) and constructs were linearized prior to transfection. The reporter gene constructs pHRRE-SEAP and pILGRE-SEAP were generated by subcloning the hematopoietin receptor and interleukin 6 response elements (28, 31) into pSEAP-Promoter (CLONTECH).
Cell Transfection and Analysis.
COS-1 cells were transfected by the DEAE-dextran method (32), COS-7, GT1–7, and 293 cells by the lipofectamine method (9). For the generation of pooled 293 cells expressing wild-type OB-RL [OB-RL(wt)] or OB-RL(fa), transfected cells were selected for growth in medium containing 0.9 mg/ml G418 (Life Technologies) for 3 weeks and >500 G418 resistant colonies were pooled to generate each population. For analysis of STAT protein activation, cells were maintained for 16 h in serum-free medium, followed by treatment with 100 ng/ml leptin for 15 min.
For SEAP assays, unless indicated otherwise, cells were transfected with the indicated reporter (1 μg) and receptor (3.0 μg) constructs. Forty-eight hours after transfection, cultures were washed twice with serum free medium and mock stimulated or treated with leptin for 24 h in nonsupplemented cell culture medium. SEAP reporter activities were measured by chemiluminescence using the Great EscApe alkaline phosphatase detection kit as described by the manufacturer (CLONTECH). Luminescence was measured in a Microbeta plus liquid scintillation counter (Wallac, Gaithersburg, MD) and expressed as arbitrary units of luminescence activity.
DNA binding by STAT proteins was analyzed by electrophoretic mobility shift assay (EMSA) using whole cell extracts as described (33). The radiolabeled double-stranded oligonucleotides SIEm67 (for STAT1 and -3) (32) and TB-2 (for STAT5B) (28) were used as substrates in the EMSA. OB-R expression in GT1–7 and 293 cells was quantified by cell surface binding of alkaline phosphatase (AP)-OB fusion protein as described previously (9, 34).
Western Blot Analysis Western blot analysis was done as described (18) and immunoreactive proteins were visualized by enhanced chemiluminescence detection as described by the manufacturer (Amersham).
Phosphospecific STAT3 antibody was from New England Biolabs, and STAT3 and -5B antisera were from Santa Cruz Biotechnology.
RESULTS
OB-R(fa) Is Constitutively Active for Reporter Gene Induction.
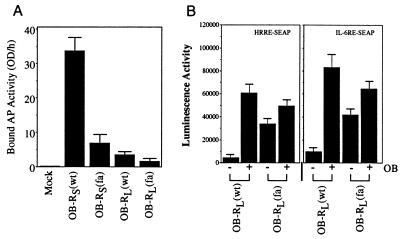
We compared the effect of the OB-R fatty mutation on cell surface expression of the murine short and long OB-R isoforms in the hypothalamic cell line GT1–7. Consistent with previous findings in COS-7 cells (24), the fatty mutation in short OB-R isoforms results in a 6–8-fold reduction in cell surface expression relative to its wild-type counterpart (Fig. 1A). In contrast, OB-RL containing the fatty mutation exhibits only a 2–3-fold reduction in cell surface expression compared with wild-type receptor (Fig. 1A). This reduction in cell surface expression of OB-RL(fa) relative to OB-RL(wt) was true at multiple cDNA input ratios (see below).
Figure 1.
Analysis of OB-R(fa) ligand binding and gene induction activities in GT1–7 cells. (A) GT1–7 cells were transfected with either control vector (column 1) or expression vectors for the short form (columns 2 and 3) or long form (columns 4 and 5) of murine OB-R(wt) (columns 2 and 4) or murine OB-R(fa) (columns 3 and 5). Two days after transfection, cells were incubated in 1 nM human AP-OB fusion protein and bound AP activity was determined (columns represent the average of two binding measurements and the bars reflect differences between the two). (B) GT1–7 cells were transfected with expression plasmids for the long form mouse OB-R(wt) or OB-R(fa) and the reporter constructs pHRRE-SEAP or pILGRE-SEAP. Two days after transfection, cells were mock stimulated (growth media alone) or treated with mouse leptin (100 ng/ml) for 24 h. The culture media was harvested and SEAP activity was determined (mean of two separate experiments).
To determine if reduced cell surface expression of OB-RL(fa) was correlated with decreased signal transducing activity, GT1–7 cells were cotransfected with either OB-RL(wt) or OB-RL(fa) cDNAs and reporter gene constructs containing regulatory elements responsive to OB-RL signals (18, 27). Cultures transfected with OB-RL(wt) exhibited minimal reporter gene activity in the absence of ligand stimulation and strong ligand-dependent induction of reporter gene activity on each of the response elements analyzed (Fig. 1B). In contrast, OB-RL(fa)-transfected cultures were found to have high levels of constitutive reporter gene activation. The constitutive reporter gene activation in OB-RL(fa)-transfected cultures was slightly induced (≈1.5-fold) upon treatment with ligand (Fig. 1B).
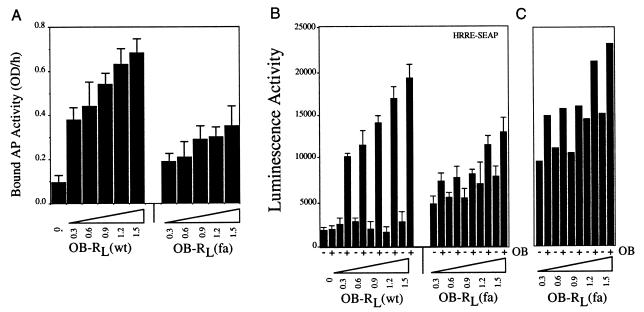
To estimate the effect of reduced cell surface expression of OB-RL(fa) on the magnitude of signal transduction, cells were transfected with the reporter gene construct HRRE-SEAP and increasing amounts of cDNA’s encoding OB-RL(wt) or OB-RL(fa). Transfected cells were then analyzed for both cell surface ligand binding and reporter gene activation. For both OB-RL(wt) and OB-RL(fa)-transfected cultures, strong correlations were found among quantities of transfected cDNA, cell surface binding activity and ligand inducible reporter gene activation (Fig. 2 A and B). However, only in OB-RL(fa)-transfected cell cultures was cDNA input-dependent constitutive reporter gene activation detected. Interestingly, when OB-RL(fa) and OB-RL(wt) signaling data are normalized to reflect equivalent cell surface expression levels, we find that the fatty mutation does not appreciably alter maximal-ligand stimulated receptor signaling activity (Fig. 2C).
Figure 2.
Comparative ligand binding and signal transducing activities of OB-RL(wt) and OB-RL(fa). GT1–7 cells were cotransfected with the reporter gene construct HRRE-SEAP (1 μg/ml) and the indicated amounts of cDNAs encoding either murine OB-RL(wt) or OB-RL(fa). 24 h after transfection, cultures were sub-divided and analyzed for (A) AP-OB binding activity or (B) treated with mouse leptin (100 ng/ml) for 24 h and assayed for reporter gene induction as described in Fig. 1 (columns represent the averages of two measurements and the bars reflect differences between the two). (C) Hypothetical constitutive and ligand induced signaling activity for OB-RL(fa) when normalized to OB-RL(wt) cell surface binding activity (as measured in A).
OB-R(fa) Constitutively Activates STAT1 and -3 But Is Impaired for Constitutive and Inducible STAT5B Activation.
Ligand-induced OB-R activation stimulates the DNA binding activity of members of the STAT family (17–19, 35). Therefore, we analyzed the ability of OB-RL(fa) to activate members of the STAT family under both ligand-independent and ligand-dependent conditions. COS-1 cells were cotransfected with expression vectors for either OB-RL(wt) or OB-RL(fa) and a subset of STAT proteins. Transfected cultures were then analyzed for STAT activation by EMSA analysis. OB-RL(wt) activates endogenous COS STAT1 and -3 or coexpressed STAT1, -3, or -5B under conditions of ligand-induced receptor activation (Fig. 3A). In contrast, OB-RL(fa) constitutively activates both endogenous COS STAT1 and -3 and coexpressed STAT1 and -3, and activation of these STATs was further increased by the addition of ligand. Dose response indicates that the reduced level of STAT1 and -3 activation in OB-RL(fa) cultures is due to both the reduced cell surface expression of this receptor compared with OB-RL(wt) (Fig. 2A) and a reduced intrinsic ability of OB-RL(fa) to activate STAT1 and -3 compared with OB-RL(wt) (data not shown). In contrast to our findings for STAT1 and -3, we find that OB-RL(fa) is unable to constitutively activate cotransfected STAT5B and is greatly impaired for ligand-induced activation of STAT5B compared with OB-RL(wt) (Fig. 3A). Immunoblot analysis indicates this effect is not due to reduced expression of this transcription factor in these cultures (Fig. 3B).
Figure 3.
OB-RL(fa) constitutively activates STAT1 and -3. COS-1 cells were transfected with expression vectors for murine OB-RL(wt) or OB-RL(fa) and the indicated STAT proteins. Controls received empty expression vector. Cells were mock stimulated or treated with mouse leptin (100 ng/ml) for 15 min, extracts were prepared and analyzed. (A) EMSA for STAT protein DNA binding activity was performed using the indicated radiolabeled substrate oligonucleotides. Autoradiographs illustrating shift patterns are for 16 and 72 h exposures (Upper and Lower, respectively) (B) Protein extracts prepared for quantitation of DNA binding activity were resolved by SDS/PAGE, transferred to nitrocellulose and probed with STAT3 or STAT5 antisera.
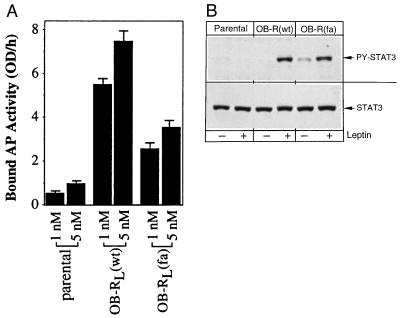
To insure that our observation of ligand-independent signaling by OB-RL(fa) was not due to an artifact arising from transient overexpression of the receptor, and to better approximate receptor expression as would be found in vivo, pooled 293 cells stably expressing OB-RL(wt) or OB-RL(fa) were generated (each 293 cell population represents a pool of >500 G418 resistant colonies). 293 cells were chosen since we have previously found that these cells are capable of supporting a ligand-induced transcription response when transiently transfected with OB-RL (M. Dembski, D.W. White and L.A. Tartaglia, unpublished observations). Cell surface ligand-binding studies demonstrate that the OB-RL(wt) and OB-RL(fa) stable pools exhibit significantly increased ligand binding relative to the parental cell line (Fig. 4A). To determine if the 293 cells stably expressing OB-RL(fa) exhibited constitutive receptor signaling, extracts were prepared and analyzed by immunoblotting using a phosphospecific STAT3 antibody (Fig. 4B). As shown, we find evidence of ligand-independent phosphorylated STAT3 in OB-RL(fa) expressing cells. Thus, under conditions of either transient or stable expression, OB-RL(fa) exhibits constitutive character.
Figure 4.
Endogenous STAT3 is constitutively activated in 293 cells stably expressing OB-RL(fa). Parental or 293 stable pools expressing OB-RL(wt) or OB-RL(fa) were (A) incubated with the listed concentrations of AP-OB fusion protein and bound AP activity was determined (columns represent the average of two binding measurements (and bars reflect differences between the two) or (B) mock stimulated or treated with mouse leptin (100 ng/ml) for 15 min. Extracts were prepared and analyzed by immunoblotting with phospho-specific or control STAT3 antibodies as indicated.
The fatty Mutation Imparts Constitutive Behavior To a Heterologous Intracellular Domain.
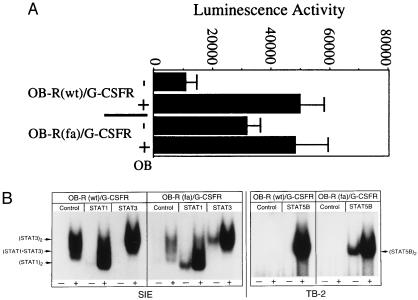
Previously we have described (27) a leptin-responsive chimeric fusion protein containing the extracellular and transmembrane domains of OB-R fused to the intracellular domain of the G-CSF-R (OB-R/G-CSF-R). Therefore, to determine if the fatty mutation could impart constitutive behavior to a heterologous intracellular domain, we generated OB-R(fa)/G-CSF-R and analyzed this molecule for ligand-independent receptor activation. OB-R(fa)/G-CSF-R is constitutively active for transcriptional activation and it’s signaling is enhanced in the presence of ligand (Fig. 5A). In addition, EMSA analysis demonstrated that OB-R(fa)/G-CSF-R constitutively activates STAT proteins (Fig. 5B). Interestingly, we find the mutant receptor chimera constitutively activated STAT1, -3, and -5B. Moreover, the magnitude of ligand-induced STAT activation by this mutant was not appreciably altered from that of the wild-type receptor chimera. Thus, the fatty mutation in the context of the OB-R extracellular domain can impart a constitutive signaling phenotype to the heterologous G-CSF-R intracellular domain, suggesting that receptor activation for each of these molecules may occur by a similar mechanism. However, the different intracellular domains on each of these receptors appears to impart distinct STAT protein activation capabilities (compare Figs. 3A and 5B).
Figure 5.
OB-R(fa)/G-CSF-R chimera is constitutively active. (A) GT1–7 cells were transfected with the recorder construct pHRRE-SEAP and either OB-R/G-CSF-R (columns 1 and 2) or OB-R(fa)/G-CSF-R (columns 3 and 4). Two days after transfection, cells were mock stimulated or treated with mouse leptin (100 ng/ml) for 24 h. Growth media was harvested and SEAP activity was determined (mean of two separate experiments). (B) COS-1 cells were transfected with expression vectors for OB-R/G-CSF-R or OB-R(fa)/G-CSF-R and the indicated STAT proteins. Controls received empty expression vector. Cells were mock stimulated or treated with mouse leptin (100 ng/ml) for 15 min, extracts were prepared and analyzed by EMSA using the indicated radiolabeled substrate oligonucleotides.
OB-RL(wt) Can Suppress Constitutive Signaling by OB-RL(fa).
Obese fatty rats are homozygous for the fatty allele whereas fatty heterozygotes exhibit minor evidence of body weight dysregulation. Consequently, if constitutive signaling by OB-RL(fa) in fatty/fatty rats was responsible for the development of obesity, the genetics of this animal would predict that OB-RL(wt) activity would be dominant to that of OB-RL(fa). Therefore, experiments were performed to determine whether coexpression of OB-RL(wt) could alter OB-RL(fa) ligand independent activation. In cells cotransfected with OB-RL(fa) and OB-RL(wt), a strong suppression of OB-RL(fa) constitutive reporter gene induction was found (Fig. 6A). However, these cells still have levels of ligand inducible reporter gene activation similar to that of cells expressing only the wild-type receptor.
Figure 6.
OB-RL(wt) can suppress constitutive OB-RL(fa) gene induction. (A) GT1–7 cells were transfected with the recorder construct pHRRE-SEAP and OB-RL(wt) (columns 1–4) or OB-RL(fa) (columns 3–6). Two days after transfection, cells were mock stimulated or treated with mouse leptin (100 ng/ml) for 24 h. Growth media was harvested and SEAP activity was determined (columns represent the average of two binding measurements and the bars reflect differences between the two). (B) GT1–7 cells were transfected with the recorder construct HRRE-SEAP and OB-RL(wt) (columns 5–6), OB-RL(fa) (columns 3–9) or G-CSF-R/OB-R (columns 1–2 and 7–9). Two days after transfection, cells were mock stimulated or treated with either mouse leptin (100 ng/ml) or human G-CSF (100 ng/ml) as indicated for 24 h. Growth media was harvested and SEAP activity was determined (columns represent the average of two measurements and the bars reflect differences between the two).
The results presented above suggest that OB-RL(wt) may complex with OB-RL(fa) and function as a dominant negative to suppress OB-RL(fa) constitutive signaling yet maintain the complex’s ability to respond to ligand induced receptor activation. However, it is also possible that suppression of constitutive OB-RL(fa) signaling could result from the titration by OB-RL(wt) of an essential cytoplasmic cofactor required for constitutive signaling by OB-RL(fa). To test this possibility, we coexpressed OB-RL(fa) and G-CSF-R/OB-R, a previously described (27) receptor chimera containing the extracellular domain of G-CSF-R joined to the transmembrane and intracellular domain of OB-RL, and assayed for effects on OB-RL(fa) constitutive signaling. The G-CSF-R/OB-RL chimera was unable to suppress constitutive signaling by OB-RL(fa) (Fig. 6B). Expression of the chimera was confirmed by G-CSF induced reporter gene activation. Thus, the ability of OB-RL(wt) to repress constitutive signaling by OB-RL(fa) does not appear to be due to cofactor titration by the OB-RL(wt) intracellular domain, and is consistent with a model whereby OB-RL(wt):OB-RL(fa) heterocomplexes are formed that exhibit a wild-type ligand-dependent signaling phenotype.
DISCUSSION
In this report we have characterized signaling activities of murine OB-RL containing a single extracellular domain missense mutation (Gln → Pro), designated OB-RL(fa). Under conditions of stable expression as well as transient transfection, we found that OB-RL(fa) exhibits constitutive signaling as detected by both reporter gene induction and activation of STAT1 and -3. Moreover, we observe that transcriptional induction by this receptor is further enhanced by treatment with ligand (OB). This finding confirms that OB-RL(fa) can still bind its cognate ligand and transduce a signal from the cell surface and is consistent with our ability to measure leptin binding on the surface of OB-RL(fa) transfected cells.
The mutation in OB-RL(fa) presumably induces a conformational change that partially mimics the ligand bound state. However, since this complex can further respond to ligand, we believe the conformational change produced by this mutation is not complete. The fatty mutation is in the extracellular region of OB-R, at an amino acid conserved in all known OB-R proteins. Specifically, this glutamine residue is located in the first of two highly conserved C domains of OB-R, a structural motif common to class I cytokine receptors (22). Each of the C domains contain the consensus sequence (Trp-Ser-Xaa-Trp-Ser) postulated to be involved with either ligand binding or protein–protein interaction among receptor chains (11, 36–38). Therefore, we speculate that the fatty mutation may introduce a structural alteration that induces a conformational change to the Trp-Ser-Xaa-Trp-Ser consensus motif, resulting in partial constitutive receptor activation. It will be of great interest to determine if the introduction of other amino acid substitutions in this region of OB-R or to the analogous regions of other cytokine receptor family members can also generate mutants with constitutive character. To our knowledge, the erythropoietin receptor mutant R129C is the only other example of a constitutive homodimerizing cytokine receptor resulting from a single extracellular domain mutation. This mutant contains a cysteine substitution for arginine at codon 129 in the extracellular domain that results in the formation of an interchain disulfide bond between receptor subunits that presumably mimics ligand-induced homodimerization of the erithropoietin receptor (39).
In tissue culture experiments, OB-RL(fa) constitutively activates STAT1 and -3. However, STAT1 appears to be less responsive than STAT3 to both ligand-independent and ligand-induced OB-RL(fa) activation, consistent with a recent report that OB-RL(fa), as compared with OB-RL(wt), exhibits a diminished ability to stimulate enhancer sequences responsive to STAT1 activation (35). Ligand induced activation of STAT1 and -3 by OB-RL involves the box 3 motif (Tyr-Xaa-Xaa-Gln) (amino acids 1141–1144) residing near the C-terminus of the OB-RL intracellular domain (18, 27). Thus, the fatty mutation may trigger a structural change resulting in phosphorylation of the Tyr residue in the box 3 consensus motif, providing receptor recruitment sites for STAT1 and -3. In contrast to its effects on STAT1 and -3, OB-RL(fa) is unable to constitutively activate STAT5B and is highly impaired for ligand-induced STAT5B activation. Thus, OB-RL(fa) appears to adopt a conformation that may prevent efficient interaction and subsequent activation of STAT5B, and is consistent with our previous work demonstrating that distinct OB-RL intracellular domain elements are involved in the activation of STAT5B vs. STAT1 and -3 (18, 27). The OB-R(fa)/G-CSF-R receptor chimera also constitutively activates STAT proteins. However, in contrast to OB-RL(fa), this mutant is capable of ligand-independent activation of STAT1, -3, and -5; moreover, ligand-induced activation of STAT proteins by OB-R(fa)/G-CSF-R chimera is comparable to levels achieved by OB-R/G-CSF-R. Thus, although OB-RL and OB-R/G-CSF-R appear to signal by a similar mechanism, the different intracellular domains of these receptors have distinct capabilities when triggered by the OB-R(fa) extracellular domain.
Accumulating evidence suggests that OB-R, like the receptors for growth hormone (40, 41), erythropoietin (42), and G-CSF (43, 44), signals by a homooligomeric mechanism without participation of an accessory protein (18, 27, 45). The observation that OB-RL(wt) can repress constitutive signaling by OB-RL(fa) suggests that pre-formed OB-RL(fa) and OB-RL(wt) complexes may exist in the absence of ligand. This result implies OB-RL(wt) may also form homodimer or homooligomer complexes, even in the absence of ligand. The role of ligand may therefore be to stabilize the complex or induce a conformational change required for receptor triggering.
As detailed above, we have characterized multiple biochemical defects of OB-R containing the fatty mutation. These include reduced cell surface expression of both short and long receptor isoforms, constitutive activation of STAT1 and -3, and greatly impaired ligand-induced activation of STAT5B. However, which of these defects is responsible for the loss of body weight homeostasis in these animals remains to be conclusively resolved. Obese fatty rats have been reported to have a 10–50-fold increase in circulating leptin levels relative to lean controls (46, 47). We have previously suggested that reduced cell surface expression of short OB-R isoform with the fatty mutation in the rat choroid plexus may result in aberrant transport of circulating leptin into the central nervous system (18). However, more recent data indicate these animals have concentrations of cerebral spinal fluid leptin equal to nonobese controls (47), suggesting that the fatty rat is obese due to diminished leptin responsiveness in hypothalamic neurons. Consistent with this possibility, fatty rats respond to intracerebroventricular injections of leptin only at greatly increased doses relative to lean controls (48). In this report we have measured only a 2–3-fold decrease in cell surface expression of OB-RL(fa) relative to OB-RL(wt). Although we cannot exclude the importance of this small difference in expression to the development of obesity in these animals, we speculate fatty rats may be obese primarily as a consequence of leptin resistance generated by altered OB-RL(fa) signaling.
The obesity of fatty rats may arise due to the inability of OB-RL(fa) to properly induce STAT5. However, Vaisse et. al. (19) have reported STAT3 activation in the hypothalamus of leptin treated wild-type but not db/db mice, suggesting an important role for this factor in body weight regulation. We have observed and ligand-independent activation of STAT3 by OB-RL(fa). A possible reconciliation of these observations is to postulate that in fatty rats constitutive activation of STAT3 by OB-RL(fa) may result in desensitization of the leptin signaling pathway. Alternatively, leptin’s effects may require cycling between low level basal and high level ligand-induced OB-RL signaling in the hypothalamus. A high baseline degree of activation due to constitutive signaling by OB-RL(fa) may result in this differential only being achieved at extremely high doses of leptin, consistent with reports that high dose intracerebroventricular leptin is required to induce a leptin response in the fatty rat (48). Moreover, if fatty rats are obese as a consequence of constitutive OB-RL(fa) signaling, this is not inconsistent with the recessive nature of the fatty mutation since we have found that coexpression with OB-RL(wt) results in suppression of constitutive OB-RL(fa) signaling.
In conclusion, we have described multiple biochemical defects of OB-RL(fa). Constitutive activity introduced by the Gln → Pro extracellular domain mutation will provide important insights into the requirements of OB-R triggering. Moreover, evidence of both constitutive and defective signaling by OB-RL(fa) generates important questions regarding mechanisms that can generate leptin resistance and loss of mammalian body weight homeostasis.
Acknowledgments
We wish to thank Drs. J. Rippenger and G.H. Fey (Friedrich Alexander University, Erlangen, Germany) for STAT1, -3, and -5B cDNA, Karen K. Kuropatwinski, Jean Schoening and Nanhua Deng for technical assistance and Dr. Ruth Gimeno for critical reading of the manuscript. This work was financed by Hoffmann–LaRoche (Nutley, NJ), National Cancer Institute Grant CA26122 (to H.B.) and National Institutes of Health Grants DK52431 and DK47473 (to R.L. and S.C.).
ABBREVIATIONS
- AP
alkaline phosphatase
- EMSA
electrophoretic mobility-shift assay
- G-CSF
granulocyte colony-stimulating factor
- G-CSF-R
G-CSF receptor
- OB
leptin
- OB-R
OB receptor
- OB-RL
long OB-R isoform
- OB-R(wt)
wild-type OB-R
- OB-R(fa)
OB-R containing the fatty mutation
- SEAP
secreted alkaline phosphatase
- STAT
signal transducer and activator of transcription
- HRRE
hematopoietin receptor response element
References
- 1.Zhang Y, Pronca R, Maffei M, Barone M, Leopold L, Friedman J M. Nature (London) 1994;372:425–431. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 2.Campfield L A, Smith F J, Guisez Y, Devos R, Burn P. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- 3.Halaas J L, Gajiwala K S, Mafei M, Cohen S L, Chait B T, Rabinowitz D, Lallone R L, Burley S K, Friedman J M. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 4.Pellymounter M A, Cullen M J, Baker M B, Hecht R, Winters D, Boone T, Collins F. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 5.Stephens T W, Bashinski M, Bristow P K, Bue-Vallesky J M, Burgett S G, Craft L, Hale J, Hoffmann J, Hsiung H M, Kriauciunas A, Mackellar W, Rosteck P R, Schoner B, Smith D, Tinsley F C, Zhang X Y, Helman M. Nature (London) 1995;377:530–534. doi: 10.1038/377530a0. [DOI] [PubMed] [Google Scholar]
- 6.Campfield L A, Smith F J, Burn P. Horm Metab Res. 1996;28:619–632. doi: 10.1055/s-2007-979867. [DOI] [PubMed] [Google Scholar]
- 7.Spiegelman B M, Flier J S. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 8.White D W, Tartaglia L A. Cytokine Growth Factor Rev. 1996;7:303–309. doi: 10.1016/s1359-6101(96)00040-8. [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia L A, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J, Muir C, Sanker S, Moriarty A, Moore K J, Smutko J S, Mays G G, Woolf E A, Monroe C A, Tepper R I. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 10.Heldin C-H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 11.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 12.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Thierfelder W E, Kreider B, Silvennoinen O. Trends Biol Sci. 1994;19:222–227. doi: 10.1016/0968-0004(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Charlot O, Tartaglia L A, Woolf E A, Weng X, Ellis S J, Lakey N D, Culpepper J, Moore K J, Breitbart R E, Duyk G M, Tepper R I, Morgenstern J P. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 14.Lee G H, Proenca R, Montez J M, Carroll K M, Darvishzadeh J G, Lee J I, Friedman J M. Nature (London) 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 15.Devos R, Richards J G, Campfield L A, Tartaglia L A, Guisez Y, Van der Heyden J, Travernier J, Plaetinck G, Burn P. Proc Natl Acad Sci USA. 1996;93:5668–5673. doi: 10.1073/pnas.93.11.5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer J G, Hoggard N, Williams N, Lawrence C B, Hannah L T, Trayhurn P. FEBS Lett. 1996;387:113–116. doi: 10.1016/0014-5793(96)00473-5. [DOI] [PubMed] [Google Scholar]
- 17.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim M H, Skoda R C. Proc Natl Acad Sci USA. 1996;93:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumann H, Morella K K, White D W, Dembski M, Bailon P S, Kim H, Lai C-F, Tartaglia L A. Proc Natl Acad Sci USA. 1996;93:8374–8378. doi: 10.1073/pnas.93.16.8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaisee C, Halaas J L, Horvath C M, Darnell J E, Jr, Stoffel M, Friedman J M. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 20.Zucker L M, Zucker T F. J Hered. 1961;52:275–278. [Google Scholar]
- 21.Chua S C, Jr, Chung W K, Wu-Peng X S, Zhang Y, Liu S-M, Tartaglia L A, Leibel R L. Science. 1996;271:994–996. doi: 10.1126/science.271.5251.994. [DOI] [PubMed] [Google Scholar]
- 22.Phillips M S, Liu Q, Hammond H A, Dugan V, Hey P J, Caskey C T, Hess J F. Nat Genet. 1996;13:18–19. doi: 10.1038/ng0596-18. [DOI] [PubMed] [Google Scholar]
- 23.Iida M, Murakami T, Ishida K, Mizuno A, Kuwajima M, Shima K. Biochem Biophys Res Commun. 1996;222:19–26. doi: 10.1006/bbrc.1996.0691. [DOI] [PubMed] [Google Scholar]
- 24.Chua S C, Jr, White D W, Wu-Peng X S, Liu S-M, Okada N, Kershaw E E, Chung W K, Power-Kehoe L, Chua M, Tartaglia L A, Leibel R L. Diabetes. 1996;45:1141–1143. doi: 10.2337/diab.45.8.1141. [DOI] [PubMed] [Google Scholar]
- 25.Baumann H, Prowse K R, Marinkovic S, Won K-A, Jahreis G P. Ann NY Acad Sci. 1989;557:280–297. doi: 10.1111/j.1749-6632.1989.tb24021.x. [DOI] [PubMed] [Google Scholar]
- 26.Mellon P L, Windle J J, Goldsmith P C, Padula C A, Roberts J L, Weiner R I. Neuron. 1990;5:1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- 27.White D W, Kuropatwinski K K, Devos R, Baumann H, Tartaglia L A. J Biol Chem. 1996;272:4065–4072. doi: 10.1074/jbc.272.7.4065. [DOI] [PubMed] [Google Scholar]
- 28.Lai C-F, Ripperger J, Morella K K, Wang Y, Gearing D P, Horseman N D, Campos S P, Fey G H, Baumann H. J Biol Chem. 1995;270:23254–23257. doi: 10.1074/jbc.270.40.23254. [DOI] [PubMed] [Google Scholar]
- 29.Ripperger J A, Fritz S, Richter K, Hocke G M, Lottspeich T, Frey G H. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- 30.Higuchi R, Krummel B, Saiti R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morella K K, Lai C-F, Kumaki S, Kumaki N, Wang Y, Bluman E M, Witthuhn B A, Ihle J N, Giri J, Gearing D P, Cosman D, Zeigler S F, Tweardy D J, Campos S P, Baumann H. J Biol Chem. 1995;270:8298–8310. doi: 10.1074/jbc.270.14.8298. [DOI] [PubMed] [Google Scholar]
- 32.Lopata M A, Cleveland D W, Sollner-Webb H. Nucleic Acids Res. 1984;12:5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H J, Flanagan J G. Cell. 1994;79:157–168. doi: 10.1016/0092-8674(94)90408-1. [DOI] [PubMed] [Google Scholar]
- 35.Rosenblum C I, Tota M, Cully D, Smith T, Collom R, Qureshi S, Hess J-F, Phillips M S, Hey P J, Vongs A, Fong T M, Xu L, Chen H Y, Smith R G, Schindler C, Van der Ploeg L H T. Endocrinology. 1996;137:5178–5181. doi: 10.1210/endo.137.11.8895396. [DOI] [PubMed] [Google Scholar]
- 36.D’Andrea A D, Fasman G D, Lodish H F. Cell. 1989;58:1023–1024. doi: 10.1016/0092-8674(89)90499-6. [DOI] [PubMed] [Google Scholar]
- 37.Bazan J F. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazaki T, Marayama M, Yamada G, Hatakeyama M, Taniguchi T. EMBO J. 1991;10:3191–3197. doi: 10.1002/j.1460-2075.1991.tb04881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimura A, Longmore G, Lodish H F. Nature (London) 1990;348:647–649. doi: 10.1038/348647a0. [DOI] [PubMed] [Google Scholar]
- 40.Cunningham B C, Ultsh M, de Vos A M, Mulkerrin M G, Clausner K R, Wells J A. Science. 1991;254:821–825. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 41.de Vos A M, Ulsch M, Kossiakoff A A. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- 42.Watowich S S, Yoshimura A, Longmore G D, Hilton D J, Yoshimura Y, Lodish H F. Proc Natl Acad Sci USA. 1992;89:2140–2144. doi: 10.1073/pnas.89.6.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukunaga R, Ishizaka-Ikeda E, Pan C-X, Seto Y, Nagata S. EMBO J. 1991;10:2855–2865. doi: 10.1002/j.1460-2075.1991.tb07835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishizaka-Ikeda E, Fukuinaga R, Wood W I, Goedell D V, Nagata S. Proc Natl Acad Sci USA. 1993;90:123–127. doi: 10.1073/pnas.90.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett B D, Solar G P, Yuan J Q, Mathias J, Thomas G R, Mathews W. Curr Biol. 1996;6:1170–1180. doi: 10.1016/s0960-9822(02)70684-2. [DOI] [PubMed] [Google Scholar]
- 46.Maffei M, Halaas J, Ravussin E, Pratley R E, Lee G H, Zhang Y, Fei H, Kim S, Lallone R, Ranganathan S, Kern P A, Friedman J M. Nat Med. 1995;1:1155–1161. doi: 10.1038/nm1195-1155. [DOI] [PubMed] [Google Scholar]
- 47.Wu-Peng X S, Chua S C, Jr, Okada N, Liu S-M, Nicolson M, Leibel R L. Diabetes. 1996;46:513–518. doi: 10.2337/diab.46.3.513. [DOI] [PubMed] [Google Scholar]
- 48.Cusin I, Rohner-Jeanrenaud F, Stricken-Krongrad A, Jeanrenaud B. Diabetes. 1996;45:1146–1450. doi: 10.2337/diab.45.10.1446. [DOI] [PubMed] [Google Scholar]