A ternary complex of the proteinase inhibitor (BTCI) with trypsin and chymotrypsin was crystallized and its crystal structure was solved by molecular replacement.
Keywords: proteinase inhibitors, Bowman–Birk inhibitors
Abstract
A ternary complex of the black-eyed pea trypsin and chymotrypsin inhibitor (BTCI) with trypsin and chymotrypsin was crystallized by the sitting-drop vapour-diffusion method with 0.1 M HEPES pH 7.5, 10%(w/v) polyethylene glycol 6000 and 5%(v/v) 2-methyl-2,4-pentanediol as precipitant. BTCI is a small protein with 83 amino-acid residues isolated from Vigna unguiculata seeds and is able to inhibit trypsin and chymotrypsin simultaneously by forming a stable ternary complex. X-ray data were collected from a single crystal of the trypsin–BTCI–chymotrypsin ternary complex to 2.7 Å resolution under cryogenic conditions. The structure of the ternary complex was solved by molecular replacement using the crystal structures of the BTCI–trypsin binary complex (PDB code 2g81) and chymotrypsin (PDB code 4cha) as search models.
1. Introduction
Proteinases are essential in a wide variety of biological processes (Neurath, 1997 ▶), including the digestion of food, the cascade systems of blood clotting and complement, the activation of hormones and the degradation of endogenous proteins. The regulation of proteinase activity is therefore of great importance in vivo for therapeutic intervention (Phillips & Fletterick, 1992 ▶). Proteinase inhibitors are found in microorganisms, plants and animals. They play an important role in the regulation of metabolic pathways, of which proteinases are some of the major components (Joanitti et al., 2006 ▶). Natural inhibitors have been classified into at least 18 different families (Laskowski & Qasim, 2000 ▶), among the best studied of which are the serine proteinase Bowman–Birk inhibitors (BBI) and Kunitz-type inhibitors. BBIs are small proteins with low molecular weight (6–15 kDa) with seven disulfide bonds that are responsible for their thermostability. BBIs have two reactive sites located opposite each other in the structure that enable the simultaneous inhibition of trypsin and chymotrypsin in monocotyledonous plants and elastase in dicotyledonous plants (Mello et al., 2003 ▶). The specificity of inhibition depends primarily on the nature of one unique residue P1 within a loop which interacts with the enzyme, blocking the access of the substrate.
Several three-dimensional structures of BBIs have been reported in the free form (Chen et al., 1992 ▶; Werner & Wemmer, 1992 ▶; Suzuki et al., 1993 ▶; Voss et al., 1996 ▶; Song et al., 1999 ▶; Li de la Sierra et al., 1999 ▶; Catalano et al., 2003 ▶; Debreczeni et al., 2003 ▶) and in complex with trypsin (Tsunogae et al., 1986 ▶; Lin et al., 1993 ▶; Li et al., 1994 ▶; Koepke et al., 2000 ▶; Murthy et al., 2000 ▶; Park et al., 2004 ▶). The crystallization and preliminary crystallographic studies of a crystal of BBI in complex with trypsin and chymotrypsin, without three-dimensional details, has been reported previously (Gaier et al., 1981 ▶). However, no crystal structure of BBI in a ternary complex with trypsin and chymotrypsin has been reported or deposited in the PDB and the only structural studies of this complex on an atomic level were performed using molecular modelling (Freitas et al., 1997 ▶; Li de la Sierra et al., 1999 ▶).
The black-eyed pea trypsin/chymotrypsin inhibitor (BTCI) is a BBI isolated from Vigna unguiculata (cowpea) seeds (Ventura & Xavier Filho, 1966 ▶; Ventura et al., 1971 ▶), a type of bean that represents an important food source for most of the population of the northeast region of Brazil. BTCI is a stable globular protein with 83 amino-acid residues, a molecular weight of 9.1 kDa and seven disulfide bonds. The binding of trypsin and chymotrypsin occurs in two different and independent reactive sites in BTCI (Freitas et al., 1997 ▶; Barbosa et al., 2007 ▶) and was characterized as an endothermic, spontaneous and entropically driven process (Fachetti et al., 1984 ▶; Freitas et al., 1999 ▶). The binary and ternary complexes of BTCI with trypsin and chymotrypsin have been purified and physicochemically characterized on the basis of hydrodynamic methods (Ventura et al., 1975 ▶). Multimeric states of BTCI in monomer–dimer–trimer–hexamer equilibrium with globular ellipsoidal shapes were identified from light-scattering (Ventura et al., 1981 ▶) and atomic force microscopy (AFM) analysis (Silva et al., 2005 ▶). The high disulfide-bond content results in an extremely constrained conformation (Voss et al., 1996 ▶) that may be responsible for the remarkable stability exhibited by this inhibitor (Silva et al., 2001 ▶).
In this article, we present the crystallization, data collection and processing, and molecular-replacement solution of the structure of BTCI in a ternary complex with trypsin and chymotrypsin.
2. Material and methods
2.1. Purification of the BTCI and its ternary complex
Bovine pancreatic trypsin (type I, twice crystallized) and bovine pancreatic α-chymotrypsin (type II, thrice crystallized) were purchased from Sigma (St Louis, Missouri, USA). BTCI was purified from V. unguiculata seeds as reported previously (Ventura & Xavier Filho, 1966 ▶; Ventura et al., 1971 ▶). Briefly, the crude extract from triturated seeds was obtained by two steps of precipitation with 2.5%(v/v) trichloroacetic acid (TCA) and 70%(w/v) ammonium sulfate, respectively. The lyophilized crude extract was dissolved in 50 mM Tris–HCl pH 7.3 and applied onto an anion-exchange DEAE-cellulose column equilibrated with the same buffer. BTCI was eluted with a linear salt gradient from 0 to 1.0 M NaCl in the same buffer. Fractions were dialyzed against water, lyophilized and stored at 253 K.
The ternary complex (chymotrypsin–BTCI–trypsin) was obtained in two steps. Firstly, BTCI and trypsin were dissolved in a 1:1 ratio in 50 mM Tris–HCl, 0.2 M KCl pH 7.5 for 30 min at room temperature. The α-chymotrypsin was then added to this solution, which was incubated for an additional 30 min. All three proteins were directly dissolved in the buffer to give a 1:1:1 ratio at a concentration of 500 µM. The resulting ternary complex was purified from the mixture by size-exclusion chromatography using a Sephadex G-75 with a flow rate of 25 ml h−1 previously equilibrated with the same buffer as indicated above (Ventura et al., 1975 ▶). The purified complex was dialyzed against water and lyophilized for storage at 253 K and crystallization assays.
2.2. Crystallization and data collection and processing
Crystallization was achieved by screening with commercially available crystallization kits from Hampton Research, Molecular Dimensions, Jena Bioscience and Emerald BioStructures using the sitting-drop vapour-diffusion method (McPherson, 1990 ▶) at 294 K and Cryschem HR3160 plates from Hampton Research. The reservoir volume was 300 µl and the drops were prepared by adding 1 µl reservoir solution to 1 µl protein solution (lyophilized protein dissolved in Milli-Q water) at a concentration of 8 mg ml−1. Crystals of the chymotrypsin–BTCI–trypsin ternary complex grew under several conditions and the crystal used for data collection was grown in 0.1 M HEPES pH 7.5, 10%(w/v) polyethylene glycol (PEG) 6000, 5%(v/v) 2-methyl-2,4-pentanediol (MPD), corresponding to condition No. 12 of Structure Screen 2 from Molecular Dimensions.
The X-ray diffraction data sets were collected at the MX1-D03B beamline of the Brazilian Synchrotron Light Laboratory (LNLS) with a wavelength of 1.427 Å and an oscillation of 1.0° on a MAR CCD detector with a circular X-ray-sensitive surface of 165 mm in diameter combined with a MAR DTB goniostat. Multiple crystals were attached to each other and were separated in order to provide a diffraction pattern from a monocrystal. The largest piece, corresponding to a monocrystal measuring about 225 × 100 × 15 µm, was soaked in mother solution containing 25%(w/v) PEG 6000 as a cryoprotectant for less than 30 s and mounted in a loop for data collection at 100 K. Crystals were cryocooled directly in a stream of nitrogen vapour at 100 K in order to minimize radiation damage. The diffraction data set was integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.3. Molecular replacement
A molecular-replacement solution was found in two steps: we first used the crystallographic structure of the BTCI–trypsin complex (Barbosa et al., 2007 ▶; PDB code 2g81) as a search model; then, with the BTCI–trypsin model fixed, the chymotrypsin (Tsukada & Blow, 1985 ▶; PDB code 4cha) structure was used as a search model. This procedure was performed with the MOLREP program (Vagin & Teplyakov, 1997 ▶) within the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
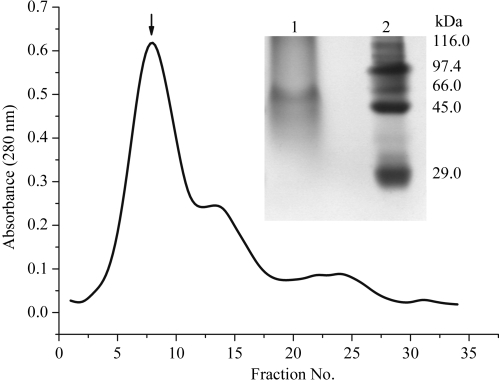
The chymotrypsin–BTCI–trypsin ternary complex was purified by size-exclusion chromatography (Fig. 1 ▶) as described by Ventura et al. (1975 ▶) and corresponds to a band of approximately 55 kDa on native polyacrylamide gel electrophoresis (inset in Fig. 1 ▶). This ternary complex is formed with one molecule of both trypsin and chymotrypsin, as previously reported (Ventura & Xavier Filho, 1966 ▶; Ventura et al., 1975 ▶). By determining the inhibition of one enzyme in the presence of high concentrations of the other, using specific synthetic substrates, these authors showed that the trypsin-reactive site was distinct from the chymotrypsin-reactive site in BTCI. The binding of trypsin and chymotrypsin occurs in these two different and independent reactive sites in BTCI (Freitas et al., 1997 ▶; Barbosa et al., 2007 ▶) and was characterized as an endothermic, spontaneous and entropically driven process (Fachetti et al., 1984 ▶; Freitas et al., 1999 ▶).
Figure 1.
Size-exclusion chromatography of the chymotrypsin–BTCI–trypsin ternary complex, corresponding to the results of Ventura et al. (1975 ▶). The arrow indicates the eluted ternary complex. Inset, Coomassie-stained (12%) polyacrylamide gel of the chymotrypsin–BTCI–trypsin ternary complex electrophoresed under nondenaturing conditions. Lane 1, chymotrypsin–BTCI–trypsin ternary complex (∼55 kDa); lane 2, high-molecular-weight markers from Sigma–Aldrich: E. coli β-galactosidase (116 kDa), rabbit muscle phosphorylase b (97.4 kDa), bovine albumin (66 kDa), egg albumin (45 kDa) and bovine erythrocyte carbonic anhydrase (29 kDa).
Several structures of BBIs in free and complexed forms (see §1) have been solved by crystallographic and nuclear magnetic resonance techniques. Nevertheless, no structures of BBIs in ternary complexes with both chymotrypsin and trypsin have been deposited in the PDB to date. We thus present the chymotrypsin–BTCI–trypsin ternary complex crystallized in space group P21, with unit-cell parameters a = 49.56, b = 93.26, c = 70.81 Å, β = 95.91° (Fig. 2 ▶). The crystal diffracted to a resolution of 2.7 Å and data-collection and processing statistics are presented in Table 1 ▶. Considering the molecular weight of 55 kDa (Ventura et al., 1975 ▶) for the ternary complex and the unit-cell parameters obtained, the Matthews coefficient (Matthews, 1968 ▶) for one complex per asymmetric unit is 2.9 Å3 Da−1, with a solvent content of 57.9%.
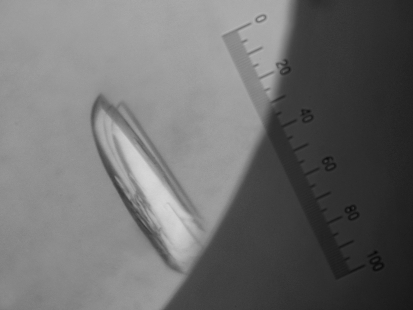
Figure 2.
Crystal of chymotrypsin–BTCI–trypsin complex obtained by sitting-drop vapour diffusion at 294 K. One division of the ruler corresponds to 5 µm.
Table 1. Chymotrypsin–BTCI–trypsin complex data-collection and processing statistics.
Values in parentheses are for the last resolution shell.
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 49.56, b = 93.26, c = 70.81, β = 95.91 |
| Mosaicity (°) | 2.5 |
| Temperature (K) | 100 |
| Wavelength (Å) | 1.427 |
| Frame oscillation (°) | 1 |
| Crystal-to-detector distance (mm) | 120.0 |
| No. of frames | 146 |
| Resolution limits (Å) | 30.00–2.70 (2.80–2.70) |
| 〈I/σ(I)〉 before merging | 6.1 (1.1) |
| 〈I/σ(I)〉 after merging | 8.1 (1.3) |
| Completeness (%) | 97.4 (90.3) |
| Multiplicity | 2.7 (2.2) |
| Rmerge† | 0.145 (0.436) |
| No. of reflections | 45399 |
| No. of unique reflections | 16907 (1558) |
R
merge = 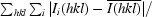
 , where Ii(hkl) is the intensity of reflection hkl,
, where Ii(hkl) is the intensity of reflection hkl, 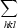 is the sum over all reflections and
is the sum over all reflections and 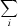 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
A molecular-replacement solution was found using two search models independently; the first was the structure of the trypsin–BTCI complex (PDB code 2g81; Barbosa et al., 2007 ▶) and the second was that of chymotrypsin (PDB code 4cha; Tsukada & Blow, 1985 ▶). The correct rotation-function and translation-function peaks were clearly marked with a correlation coefficient of 0.65 and produced a solution with one full ternary complex in the asymmetric unit, consisting of one BTCI molecule, one trypsin molecule and one chymotrypsin molecule. The resulting orientation matrix was applied to the search models, which were then submitted to rigid-body refinement, yielding an R cryst of 0.423. The possibility that the inhibitor might be bound to two molecules of the same enzyme was verified in two ways. Firstly, the initial electron-density map at regions of insertions/deletions in the alignment of the two enzymes was inspected. Secondly, different molecular replacements searching for two molecules of trypsin or two of chymotrypsin with the BTCI molecule fixed were performed. The map clearly shows the identity of the each enzyme and the parameters of the molecular-replacement trials using two molecules of the same enzyme are poorer than that using one molecule of each enzyme.
At present, the model is being refined and new crystallization trials are being carried out to improve crystal quality, particularly mosaicity and resolution. This improvement will be important in order to clearly define the waters involved in the complex interfaces and also to more precisely define the interactions between the enzymes and the inhibitor. Of special interest is the comparison with other known structures of BBI inhibitors and inhibitor–enzyme complexes in order to address the possible conformational changes that take place in the enzymes’ active sites or inhibitor-reactive sites upon complexation.
Acknowledgments
This work was supported in part by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Associação Brasileira de Tecnologia de Luz Síncrotron (ABTLuS).
References
- Barbosa, J. A., Silva, L. P., Teles, R. C., Esteves, G. F., Azevedo, R. B., Ventura, M. M. & Freitas, S. M. (2007). Biophys. J.92, 1638–1650. [DOI] [PMC free article] [PubMed]
- Catalano, M., Ragona, L., Molinari, H., Tava, A. & Zetta, L. (2003). Biochemistry, 42, 2836–2846. [DOI] [PubMed]
- Chen, P., Rose, J., Love, R. C., Wei, H. & Wang, B.-C. (1992). J. Biol. Chem.267, 1990–1994. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Debreczeni, J. É., Bunkóczi, G., Girmann, B. & Sheldrick, G. M. (2003). Acta Cryst. D59, 393–395. [DOI] [PubMed]
- Fachetti, H. C. S., Mizuta, K. & Ventura, M. M. (1984). An. Acad. Bras. Cienc.56, 311–317.
- Freitas, S. M., Ikemoto, H. & Ventura, M. M. (1999). J. Protein Chem.18, 307–313. [DOI] [PubMed]
- Freitas, S. M., Mello, L. V., Silva, M. C., Vriend, G., Neshich, G. & Ventura, M. M. (1997). FEBS Lett.409, 121–127. [DOI] [PubMed]
- Gaier, J. R., Tulinsky, A. & Liener, I. E. (1981). J. Biol. Chem.256, 11417–11419. [PubMed]
- Joanitti, G. A., Freitas, S. M. & Silva, L. P. (2006). Curr. Enzyme Inhib.2, 199–217.
- Koepke, J., Ermler, U., Warkentin, E., Wenzl, G. & Flecker, P. (2000). J. Mol. Biol.298, 477–491. [DOI] [PubMed]
- Laskowski, M. & Qasim, M. A. (2000). Biochim. Biophys. Acta, 1477, 324–337. [DOI] [PubMed]
- Li, Y., Huang, Q., Zhang, S., Liu, S., Chi, C. & Tang, Y. (1994). J. Biochem. (Tokyo), 116, 18–25. [DOI] [PubMed]
- Li de la Sierra, I., Quillien, L., Flecker, P., Gueguen, J. & Brunie, S. (1999). J. Mol. Biol.285, 1195–1207. [DOI] [PubMed]
- Lin, G., Bode, W., Huber, R., Chu, C. W. & Engh, R. A. (1993). Eur. J. Biochem.212, 549–555. [DOI] [PubMed]
- McPherson, A. (1990). Eur. J. Biochem.189, 1–23. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mello, M. O., Tanaka, A. S. & Silva-Filho, M. C. (2003). Mol. Phylogenet. Evol.27, 103–112. [DOI] [PubMed]
- Murthy, H. M., Judge, K., DeLucas, L. & Padmanabhan, R. (2000). J. Mol. Biol.301, 759–767. [DOI] [PubMed]
- Neurath, H. (1997). Trends Biochem. Sci.14, 268–271. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Park, E. Y., Kim, J. A., Kim, H. W., Kim, Y. S. & Song, H. K. (2004). J. Mol. Biol.343, 173–186. [DOI] [PubMed]
- Phillips, M. A. & Fletterick, R. J. (1992). Curr. Opin. Struct. Biol.2, 713–720.
- Silva, L. P., Azevedo, R. B., Morais, P. C., Ventura, M. M. & Freitas, S. M. (2005). Proteins, 61, 642–648. [DOI] [PubMed]
- Silva, L. P., Leite, J. R. S. A., Bloch, C. Jr & Freitas, S. M. (2001). Protein Pept. Lett.8, 33–38.
- Song, H. K., Kim, Y. S., Yang, J. K., Moon, J., Lee, J. Y. & Suh, S. W. (1999). J. Mol. Biol.293, 1133–1144. [DOI] [PubMed]
- Suzuki, A., Yamane, T., Ashida, T., Norioka, S., Hara, S. & Ikenazka, T. (1993). J. Mol. Biol.234, 722–734. [DOI] [PubMed]
- Tsukada, H. & Blow, D. M. (1985). J. Mol. Biol.184, 703–711. [DOI] [PubMed]
- Tsunogae, Y., Tanaka, I., Yamane, T., Kikkawa, J., Ashida, T., Ishikawa, C., Watanabe, K., Nakamura, S. & Takahashi, K. (1986). J. Biochem. (Tokyo), 100, 1637–1646. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Ventura, M. M., Martin, C. O. & Morhy, L. (1975). An. Acad. Bras. Cienc.47, 335–346.
- Ventura, M. M., Mizuta, K. & Ikemoto, H. (1981). An. Acad. Bras. Cienc.53, 195–201.
- Ventura, M. M. & Xavier Filho, J. (1966). An. Acad. Bras. Cienc.38, 553–566.
- Ventura, M. M., Xavier Filho, J., Moreira, R. A., Aquino, A. M. & Pinheiro, P. A. (1971). An. Acad. Bras. Cienc.43, 233–242. [PubMed]
- Werner, M. H. & Wemmer, D. E. (1992). Biochemistry, 31, 999–1010. [DOI] [PubMed]
- Voss, R. H., Ermler, U., Essen, L. O., Wenzl, G., Kim, Y. M. & Flecker, P. (1996). Eur. J. Biochem.242, 122–131. [DOI] [PubMed]