Crystals of baculovirus-expressed adeno-associated virus serotype 7 capsids have been produced which diffract X-rays to ∼3.0 Å resolution.
Keywords: adeno-associated virus serotype 7
Abstract
Crystals of baculovirus-expressed adeno-associated virus serotype 7 capsids diffract X-rays to ∼3.0 Å resolution. The crystals belong to the rhombohedral space group R3, with unit-cell parameters a = 252.4, c = 591.2 Å in the hexagonal setting. The diffraction data were processed and reduced to an overall completeness of 79.0% and an R merge of 12.0%. There are three viral capsids in the unit cell. The icosahedral threefold axis is coincident with the crystallographic threefold axis, resulting in one third of a capsid (20 monomers) per crystallographic asymmetric unit. The orientation of the viral capsid has been determined by rotation-function searches and is positioned at (0, 0, 0) by packing considerations.
1. Introduction
Adeno-associated viruses (AAVs) are nonpathogenic ssDNA viruses that belong to the Dependovirus genus of the Parvoviridae and are under development for corrective human gene-delivery applications (Muzyczka & Berns, 2001 ▶). AAV7 was the first novel AAV sequence isolated from rhesus monkey heart (Gao et al., 2002 ▶). The AAV serotypes demonstrate differences in cell tropism and transgene expression in specific tissues (Gao et al., 2004 ▶). For example, the efficiency of skeletal muscle transduction with the nonhuman primate AAV7 and AAV8 viruses is significantly greater than that observed with the human AAV2 and AAV5 viruses (Louboutin et al., 2005 ▶). In addition, gene-delivery vectors based on nonhuman primate AAVs such as AAV7 have low reactivity to antibodies directed against human AAVs and are thus considered to be promising vehicles for human gene-therapy applications (Gao et al., 2006 ▶).
AAV capsids are composed of a total of 60 copies of three overlapping capsid viral proteins (VPs), VP1, VP2 and VP3, in a predicted ratio of 1:1:10, arranged with T = 1 icosahedral symmetry. In the crystal structures of AAV2, AAV4 and AAV8 only the common VP3 region of the VPs is observed (Govindasamy et al., 2006 ▶; Nam et al., 2007 ▶; Xie et al., 2002 ▶). A small number of variable amino acids in the overlapping VP3 dictate the recognition of different cell-surface glycans and therefore the differential tropism and transduction properties of the AAVs (Gao et al., 2004 ▶; Kern et al., 2003 ▶; Opie et al., 2003 ▶; Wu et al., 2006 ▶). In addition, comparison of the AAV2, AAV4 and AAV8 structures showed that regions controlling AAV2 receptor recognition, transduction and antigenic reactivity are commonly varied in the other serotypes (Nam et al., 2007 ▶). Thus, understanding the AAV capsid structures and their interactions with cellular factors is a prerequisite for their modification for cell/tissue-specific targeted applications. We report the production, purification, crystallization and preliminary crystallographic analysis of AAV7 capsids as a first step towards structural studies to identify the regions of this virus responsible for its capsid-associated enhanced muscle transduction phenotype.
2. Materials and methods
2.1. Production and purification
A recombinant baculovirus encoding the AAV7 viral capsid open reading frame (ORF) was constructed using the Bac-to-Bac system (Gibco BRL). DH10Bac-competent cells containing the baculovirus genome were transformed with pFastBac transfer plasmids containing the AAV7 component insert. Bacmid DNA purified from recombination-positive white colonies was transfected into Sf9 cells using TransIT Insecta reagent (Mirus). 3 d post-transfection, media containing baculovirus (pooled viral stock) were harvested and a plaque assay was used to prepare independent plaque isolates. Several individual plaques were propagated to passage one (P1) to assay for expression of the AAV7 capsid genes and a selected clone was propagated to P2. A titered P2 stock was used to infect Sf9 insect cells grown in Erlenmeyer flasks at 300 K using Sf-900 II SFM media (Gibco/Invitrogen Corporation). The cells were infected at a multiplicity of infection of 5.0 plaque-forming units per cell for viral capsid production.
The virus capsids were released from the cells by three rapid freeze–thaw cycles in lysis buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100) with the addition of benzonase (Merck KGaA, Germany) in the final cycle. The sample was clarified by two rounds of centrifugation at 10 000 rev min−1 for 15 min at 277 K. The cell lysate was pelleted through a 20%(w/v) sucrose cushion by ultracentrifugation at 45 000 rev min−1 for 3 h at 277 K. The pellet from the cushion was resuspended in lysis buffer by repeated pipetting and left overnight at 277 K for further suspension. The resuspended sample was subjected to a low-speed 2000 rev min−1 spin to remove particulate material and further purified with two rounds of sucrose-step gradients [5–40%(w/v)] by ultracentrifugation at 35 000 rev min−1 for 3 h at 277 K. A visible blue fraction containing empty (no DNA) viral capsids sedimenting at the 20–25% sucrose fractions was extracted after the second gradient and dialyzed against 20 mM Tris–HCl pH 7.5 containing 350 mM NaCl and 2 mM MgCl2 by stirring overnight at 277 K. The approximate concentration of the sample was calculated from optical density measurements assuming an extinction coefficient of 1.7 for calculations in mg ml−1. The concentration was adjusted to ∼10 mg ml−1 using Centricon filters (Amicon Centricons, 100 000 molecular-weight cutoff) at 3000 rev min−1 at 277 K. The purity and integrity of the viral capsids were checked by SDS–PAGE and negative-stain electron microscopy, respectively.
2.2. Electron microscopy
Purified AAV7 viral capsids were viewed using a Joel JEM-100CX II electron microscope (EM). 5 µl purified virus solution at an estimated concentration of 2.0 mg ml−1 was spotted onto a 400 mesh carbon-coated copper grid (Ted Pella, Inc., Redding, California, USA) for 2 min before blotting with filter paper (Whatman No. 5). The sample was then negatively stained with 5 µl 2% uranyl acetate for 17 s, blotted dry and viewed.
2.3. Crystallization
Based on reports for the AAV1 (Miller et al., 2006 ▶), AAV4 (Kaludov et al., 2003 ▶), AAV5 (DiMattia et al., 2005 ▶) and AAV8 (Lane et al., 2005 ▶) viral capsids, AAV7 crystallization conditions were screened against precipitant solutions containing varying polyethylene glycol (PEG) 8000 percentages (4–6%), NaCl (350 mM–1 M) and MgCl2 (10–20 mM) concentrations and pH range (pH 6.0–8.0) at room temperature (RT) and 277 K. A buffer concentration of 20 mM for both bis-Tris (pH 6.0 and 6.5) and Tris–HCl (pH 7.0–8.5) was used for the pH screens. The crystal screens were set up using the hanging-drop vapour-diffusion method (McPherson, 1982 ▶) with 24-well VDX plates and siliconized cover slips (Hampton Research, Laguna Niguel, California, USA). The crystallization drops contained 2 µl sample solution (at ∼10 mg ml−1) and 2 µl precipitant solution and were equilibrated against 1 ml precipitant solution.
2.4. Data collection and reduction
X-ray diffraction data were collected from AAV7 crystals at 100 K at the 22-ID SER-CAT beamline (Advanced Photon Source, Argonne). A crystal-to-detector distance of 400 mm was used with an oscillation angle of 0.3° per image. A total of 271 images were collected from crystals diffracting X-rays to ∼3.0 Å resolution using a wavelength of 0.9724 Å. All the images were indexed, integrated and scaled with the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
2.5. Molecular replacement: particle orientation and position determination
The orientation of the AAV7 viral capsid in the crystal unit cell was calculated using the GLRF self-rotation function program (Tong & Rossmann, 1997 ▶) with observed data between 10.0 and 6.0 Å resolution. The positions of the viral capsid fivefold, threefold and twofold icosahedral symmetry axes were searched with κ = 72, 120 and 180°, respectively. The calculations used ∼10% of the largest amplitudes to represent the second Patterson and a radius of integration of 130 Å. Based on the self-rotation results, a cross-rotation search was conducted with the AMoRe program (Navaza, 1994 ▶) using 20 AAV4 VP subunits (PDB code 2g8g; Govindasamy et al., 2006 ▶) with residues modified to alanines (a polyalanine model). Structure factors were calculated for the AAV4 model using the SFALL subroutine in CCP4 (Collaborative Computational Project, Number 4, 1994 ▶). The AAV4 polyalanine model was oriented into the AAV7 crystal unit cell based on the cross-rotation search results and positioned at (0, 0, 0) based on crystal symmetry constraints to generate initial phases for the observed AAV7 structure factors. Noncrystallographic symmetry (NCS) operators were generated using the 20 AAV4 VP subunit model for further refinement of the AAV7 structure by alternative cycles of phase refinement, density averaging and model building (Brünger et al., 1998 ▶; Emsley & Cowtan, 2004 ▶; Jones et al., 1991 ▶).
3. Results and discussion
3.1. Crystallization
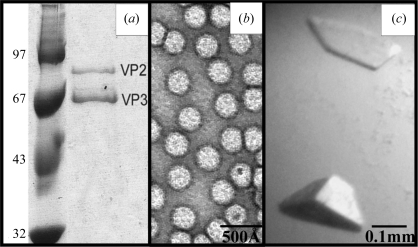
The purity and integrity of the AAV7 capsids were confirmed with SDS–PAGE (Fig. 1 ▶ a) and negative-stain EM (Fig. 1 ▶ b) prior to crystallization. The SDS–PAGE analysis showed that the baculovirus-expressed capsids contained VP2 and VP3 but no detectable levels of the AAV7 VP1, as had been reported for other expressed AAV capsids (Kohlbrenner et al., 2005 ▶). However, this low expression was not detrimental to the assembly of the capsids (Fig. 1 ▶ b), as also previously reported (Kohlbrenner et al., 2005 ▶). Negative-stain EM analysis showed the expressed capsids to be intact. AAV7 crystals grew at RT from conditions consisting of varying NaCl concentrations (350 mM to 1 M NaCl) and 20 mM MgCl2 in 20 mM Tris–HCl pH 7.0 with 5% PEG 8000. The crystals used for X-ray diffraction data collection were selected from 20 mM Tris pH 7.0, 350 mM NaCl, 20 mM MgCl2 and 5% PEG 8000. These crystals were obtained in approximately three weeks and grew to dimensions of ∼0.2 × 0.1 × 0.1 mm (Fig. 1 ▶ c). The crystals were cryoprotected with 30% glycerol in the crystallization solution adjusted to 10% PEG 8000 and flash-cooled prior to data collection.
Figure 1.
Purification and crystallization of AAV7 viral capsids. (a) SDS–PAGE gel showing the AAV7 viral proteins VP2 and VP3 (molecular weights 73 and 62 kDa, respectively) assembled into capsids in the baculovirus expression system in the right lane. VP1 was not present in the capsids. The left lane shows the position of molecular-weight standards (kDa). (b) Transmission electron micrograph of intact AAV7 capsids stained with 2% uranyl acetate. (c) Optical photograph of an AAV7 capsid crystal.
3.2. X-ray data collection and reduction
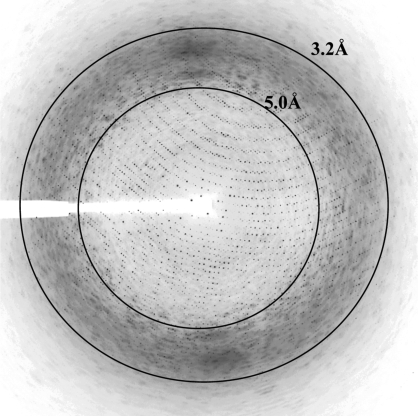
The crystals diffracted to ∼3.0 Å resolution using synchrotron X-rays (Fig. 2 ▶). The crystals belong to the rhombohedral crystal system in space group R3, with unit-cell parameters a = 245.1 Å, α = 62°. The unit cell was converted into the hexagonal setting (a = 252.4, c = 591.2 Å) for further calculations. The data set is 79.0% complete, contains 221 627 unique reflections and scaled with an overall R merge of 12.0%. An average mosaicity of 0.55° was observed during data scaling. The data-processing and scaling statistics are summarized in Table 1 ▶. The Matthews coefficient (V M; Matthews, 1968 ▶) was calculated to be 3.2 Å3 Da−1 assuming a solvent content of ∼60.0% with 20 copies of the AAV7 VP3 monomer present in the crystallographic asymmetric unit as suggested by volume calculations and space-group considerations.
Figure 2.
X-ray diffraction image of an AAV7 crystal: image of a typical 0.3° oscillation photograph. The inner and outer concentric rings indicate the 5.0 and 3.2 Å resolution shells, respectively.
Table 1. Crystal data-collection and processing statistics for AAV7 crystal.
Values in parentheses are for the highest resolution shell.
| Space group | R3 (hexagonal setting) |
| Unit-cell parameters (Å) | a = 252.4, c = 591.2 |
| VM (Å3 Da−1) | 3.2 |
| Total reflections | 468474 |
| Unique reflections | 221627 (13633) |
| Crystal mosaicity (°) | 0.55 |
| Resolution range (Å) | 50–3.0 (3.11–3.0) |
| Completeness (%) | 79.0 (48.5) |
| Rmerge† (%) | 12.0 (48.7) |
| Redundancy | 2.1 |
| Average I/σ(I) | 8.2 |
R
merge is defined as 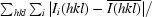
 × 100, where Ii(hkl) is the intensity of an individual reflection and
× 100, where Ii(hkl) is the intensity of an individual reflection and 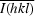 is the average intensity for this reflection; the summation is over all intensities.
is the average intensity for this reflection; the summation is over all intensities.
3.3. Molecular replacement: determination of particle orientation and position
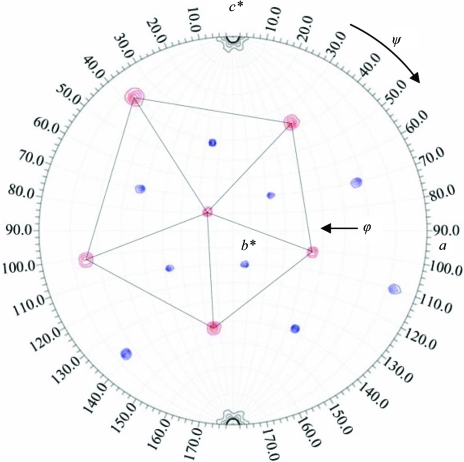
The self-rotation function search identified the orientation of the twofold, threefold and fivefold icosahedral symmetry elements for the AAV7 viral capsid within the R3 unit cell (Fig. 3 ▶). Space-group packing constraints place the AAV7 capsid at the origin (0, 0, 0) of the unit cell with an icosahedral threefold axis along the c axis of the hexagonal setting cell (Fig. 3 ▶). Thus, the crystallographic asymmetric unit contains one third of the 60 monomer subunits (20 VPs) in the T = 1 icosahedral capsid. 20 polyalanine VP subunits of AAV4 were used for the calculation of initial phases for the molecular-replacement structure determination of the AAV7 capsid based on cross-rotation search results in the AMoRe program (Navaza, 1994 ▶) and space-group packing considerations. The R factor and correlation coefficient were 45.4% and 66.5%, respectively, for this initial molecular-replacement procedure.
Figure 3.
AAV7 viral capsid orientation in the H3 hexagonal crystal unit-cell setting. The red and blue contours show the fivefold and threefold icosahedral symmetry elements, respectively, for self-rotation function searches with κ = 72° and 120°. The contours are at 2σ intervals. The peaks representing fivefold positions are delineated by the black pentagon.
This solution was refined with cycles of simulated annealing, individual B-factor refinement and energy-minimization procedures applying NCS operators in the CNS program (Brünger et al., 1998 ▶) interspersed with rounds of manual model building using the program Coot (Emsley & Cowtan, 2004 ▶), but the R work and R free values did not converge to less than 38.6% and 39.3%, respectively. These results therefore called for a re-examination of the X-ray diffraction data with the possibility of partial twinning (Yeates, 1997 ▶). The data were therefore tested for hemihedral twinning using the twinning software in CNS and were shown to have a slight twinning fraction of 0.133 when applying the twinning operator h, −h − k, −l.
The previous structure solution (which had converged to an R work of 38.6% without taking into account that the data were twinned) was then further refined within the CNS software package using the twinning fraction and the twinning operator. The current R factor is 29.4%. Once refined, the AAV7 X-ray structure will be compared with those available for other AAV serotypes, for example, AAV2 (Xie et al., 2002 ▶), AAV4 (Govindasamy et al., 2006 ▶) and AAV8 (Nam et al., 2007 ▶), for further characterization of AAV capsid regions responsible for their differential tissue tropism and transduction efficiencies. Ultimately, structure information on the AAV serotypes will provide a three-dimensional platform for mutagenesis efforts to improve specific cell/tissue-targeting gene-delivery applications for these viral vectors.
Acknowledgments
The authors would like to thank the staff at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. We thank Hyun-Joo Nam for help with X-ray diffraction data collection. Use of APS is supported by the US Department of Energy, Basic Energy Sciences, Office of Science under contract No. W-31-109-ENG-38. This study was funded by NIH project R01 GM082946 to RM, NM, SZ and MA-M.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- DiMattia, M., Govindasamy, L., Levy, H. C., Gurda-Whitaker, B., Kalina, A., Kohlbrenner, E., Chiorini, J. A., McKenna, R., Muzyczka, N., Zolotukhin, S. & Agbandje-McKenna, M. (2005). Acta Cryst. F61, 917–921. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Gao, G. P., Alvira, M. R., Wang, L., Calcedo, R., Johnston, J. & Wilson, J. M. (2002). Proc. Natl Acad. Sci. USA, 99, 11854–11859. [DOI] [PMC free article] [PubMed]
- Gao, G., Lu, Y., Calcedo, R., Grant, R. L., Bell, P., Wang, L., Figueredo, J., Lock, M. & Wilson, J. M. (2006). Mol. Ther.13, 77–87. [DOI] [PubMed]
- Gao, G. P., Vandenberghe, L. H., Alvira, M. R., Lu, Y., Calcedo, R., Zhou, X. & Wilson, J. M. (2004). J. Virol.78, 6381–6388. [DOI] [PMC free article] [PubMed]
- Govindasamy, L., Padron, E., McKenna, R., Muzyczka, N., Kaludov, N., Chiorini, J. A. & Agbandje-McKenna, M. (2006). J. Virol.80, 11556–11570. [DOI] [PMC free article] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kaludov, N., Padron, E., Govindasamy, L., McKenna, R., Chiorini, J. A. & Agbandje-McKenna, M. (2003). Virology, 306, 1–6. [DOI] [PubMed]
- Kern, A., Schmidt, K., Leder, C., Muller, O. J., Wobus, C. E., Bettinger, K., Von der Lieth, C. W., King, J. A. & Kleinschmidt, J. A. (2003). J. Virol.77, 11072–11081. [DOI] [PMC free article] [PubMed]
- Kohlbrenner, E., Aslanidi, G., Nash, K., Shklyaev, S., Campbell-Thompson, M., Byrne, B. J., Snyder, R. O., Muzyczka, N., Warrington, K. & Zolotukhin, S. (2005). Mol. Ther.12, 1217–1225. [DOI] [PMC free article] [PubMed]
- Lane, M. D., Nam, H.-J., Padron, E., Gurda-Whitaker, B., Kohlbrenner, E., Aslanidi, G., Byrne, B., McKenna, R., Muzyczka, N., Zolotukhin, S. & Agbandje-McKenna, M. (2005). Acta Cryst. F61, 558–561. [DOI] [PMC free article] [PubMed]
- Louboutin, J. P., Wang, L. & Wilson, J. M. (2005). J. Gene Med.4, 442–451. [DOI] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals, 1st ed., pp. 96–99. New York: Wiley.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Miller, E. B., Gurda-Whitaker, B., Govindasamy, L., McKenna, R., Zolotukhin, S., Muzyczka, N. & Agbandje-McKenna, M. (2006). Acta Cryst. F62, 1271–1274. [DOI] [PMC free article] [PubMed]
- Muzyczka, N. & Berns, K. I. (2001). Fields Virology, 4th ed., edited by D. M. Knipe & P. M. Howley, pp. 2327–2360. New York: Lippincott, Williams & Wilkins.
- Nam, H.-J., Lane, M. D., Padron, E., Gurda, B., McKenna, R., Kohlbrenner, E., Aslanidi, G., Byrne, B., Muzyczka, N., Zolotukhin, S. & Agbandje-McKenna, M. (2007). J. Virol.81, 12260–12712. [DOI] [PMC free article] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Opie, S. R., Warrington, K. H. Jr, Agbandje-McKenna, M., Zolotukhin, S. & Muzyczka, N. (2003). J. Virol.77, 6995–7006. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed]
- Wu, Z., Asokan, A., Grieger, J. C., Govindasamy, L., Agbandje-McKenna, M. & Samulski, R. J. (2006). J. Virol.80, 11393–11397. [DOI] [PMC free article] [PubMed]
- Xie, Q., Bu, W., Bhatia, S., Hare, J., Somasundaram, T., Azzi, A. & Chapman, M. S. (2002). Proc. Natl Acad. Sci. USA, 99, 10405–10410. [DOI] [PMC free article] [PubMed]
- Yeates, T. O. (1997). Methods Enzymol.276, 345–358.