The crystal structure of MnSOD with four dimers per asymmetric unit was refined to an R factor of 21.1% at 1.8 Å resolution. Differences in the monomer fold and dimer interface relative to the human enzyme can be exploited in the design of selective drugs for Bacillus MnSOD.
Keywords: superoxide dismutase
Abstract
The sodA gene of Bacillus subtilis was expressed in Escherichia coli, purified and crystallized. The crystal structure of MnSOD was solved by molecular replacement with four dimers per asymmetric unit and refined to an R factor of 21.1% at 1.8 Å resolution. The dimer structure is very similar to that of the related enzyme from B. anthracis. Larger structural differences were observed with the human MnSOD, which has one less helix in the helical domain and a longer loop between two β-strands and also showed differences in three amino acids at the intersubunit interface in the dimer compared with the two bacterial MnSODs. These structural differences can be exploited in the design of drugs that selectively target the Bacillus enzymes.
1. Introduction
Superoxide dismutases (SODs) are important antioxidant enzymes that are widely distributed in prokaryotic and eukaryotic cells. They catalyze the reduction of the superoxide radical to hydrogen peroxide and dioxygen in a critical reaction that protects aerobic organisms against oxidative damage. SODs have been classified into four families based on their different types of metal centers: copper/zinc, nickel, manganese and iron (Beyer et al., 1991 ▶).
Manganese-containing SODs (MnSODs) are widely present in many bacteria, chloroplasts, mitochondria and the cytosol of eukaryotic cells (Stroupe et al., 2001 ▶). MnSODs in the mitochondria have been suggested to function as tumor suppressors (Hu et al., 2005 ▶) and cancer cells generally have diminished MnSOD activity (Oberley, 2004 ▶).
Several crystal structures of SODs have been determined. The known SOD structures fall into two groups: the Cu/Zn SODs fold into a flattened eight-stranded Greek-key β-barrel, while the MnSODs and FeSODs fold into two-domain structures mainly composed of α-helices (Wuerges et al., 2004 ▶). Crystal structures have been reported of the MnSODs from Homo sapiens (Hsieh et al., 1998 ▶) and many bacteria, including Escherichia coli (Edwards et al., 1998 ▶), Bacillus halodenitrificans (Liao et al., 2002 ▶), B. anthracis (Boucher et al., 2005 ▶), Porphyromonas gingivalis (Sugio et al., 2000 ▶) and Thermus thermophilus (Ludwig et al., 1991 ▶). All MnSOD structures comprise two domains: an α-helical domain and an α/β-domain.
B. anthracis, the causative agent of anthrax, is a Gram-positive rod-shaped spore-forming bacterium. Two genes, BA4499 (sodA1) and BA5696 (sodA2), encode SODs in B. anthracis. B. subtilis is another aerobic and endospore-forming bacterium. The endospore is highly resistant to various stresses, such as heat, oxidants and UV light (Inaoka et al., 1999 ▶). The B. subtilis MnSOD shares high sequence similarity with B. anthracis SodA (77% sequence identity and 86% similarity). The MnSOD encoded by sodA is essential for resistance to oxidative stress in growing and sporulating cells of B. subtilis (Inaoka et al., 1999 ▶). The protein sequences of B. subtilis and human MnSODs share 53% identity and 65% similarity. Any structural differences between human and Bacillus SODs will assist in the development of a potential antibacterial therapy targeting the Bacillus MnSODs. Therefore, we describe the expression in E. coli, purification and determination of the crystal structure of B. subtilis MnSOD and its comparison with the structures of human and B. anthracis MnSODs.
2. Methods
2.1. Expression and purification of B. subtilis 168 MnSOD
In order to study the secretion of MnSOD in B. subtilis, the DNA fragment encoding MnSOD (606 bp) was amplified from total genomic DNA of B. subtilis 168 by PCR using forward primer 5′-GAGGATCTAGAATGGCTTACGAACTTCCAG-3′ and reverse primer 5′-ACCTTGTCGACGCCATTATTTTCGTTCGCT-3′ (Integrated DNA Technologies Inc.). The amplified DNA fragment was cloned into the shuttle vector pDG148 via XbaI and SalI. The positive clone pYHsodA09 was confirmed by restriction maps and sequencing.
For the purpose of protein overproduction and crystallography, the DNA fragment encoding MnSOD was amplified by the PCR method using forward primer 5′-CCATGGATGGCTTACGAACTTCCAGAATTACCT-3′, reverse primer 5′-GAATTCTTATTTTCGTTCGCTGTATAGACGAGCCAC-3′ (Integrated DNA Technologies Inc.) and plasmid pYHsodA09 as a template. The product was ligated to the NcoI/EcoRI sites of pBAD-HisA (Invitrogen). The reverse primer contained a stop codon to prevent fusion to the His tag provided by the plasmid. E. coli Top10 harboring pHE25 was cultured in LB medium containing 100 µg ml−1 ampicillin and 1 mM MnCl2 at 310 K. MnSOD expression was induced with 0.2% l-arabinose at an OD600nm of 0.5 and overexpression took place for 4 h. Osmotic shock (Thorstenson et al., 1997 ▶; Ewis & Lu, 2005 ▶) was used to release the overexpressed MnSOD as the first step of purification. A cell pellet (8.5 g wet weight) harvested from 2 l culture was washed twice with 50 ml 50 mM Tris–HCl buffer pH 8.5. The cell pellet was then resuspended in 50 ml 50 mM Tris–HCl pH 8.5 containing 20% sucrose and 2.5 mM EDTA and incubated on ice for 30 min. The cell pellet was collected by centrifugation at 6000g for 15 min, resuspended in 50 ml ice-cold 50 mM Tris–HCl pH 8.5 and incubated overnight with continuous stirring at 277 K. After centrifugation at 6000g for 15 min, the recovered supernatant fraction was applied onto a Mono-Q 26/10 anion-exchange column (Amersham Bioscience Corp.). The protein was eluted with a linear gradient from 0 to 1 M NaCl in 50 mM Tris–HCl pH 8.5 at a flow rate of 1 ml min−1. The protein peak eluted at 0.38 M NaCl was pooled, concentrated by centrifugation in Centricon units (Amicon; 10 kDa cutoff membrane) and loaded onto Superdex G-200 HR (Amersham Bioscience Corp.) in 50 mM Tris–HCl pH 8.5 containing 300 mM NaCl at a flow rate of 0.5 ml min−1. Protein purity was assessed by SDS–PAGE (Fig.1). The purified protein was then concentrated (10 mg ml−1) using Centricon units (Amicon; 10 kDa cutoff membrane) and the elution buffer was replaced by 50 mM Tris–HCl pH 8.5 for crystallization.
2.2. Crystallization and X-ray data collection
MnSOD was crystallized by hanging-drop vapor diffusion at 297 K using 1 µl protein solution and 1 µl mother liquor. Crystallization trials used Hampton Crystal Screen as the initial screen. Crystals grew overnight with 30%(w/v) polyethylene glycol 4000, 0.2 M magnesium chloride hexahydrate and 0.1 M Tris–HCl pH 8.5. Crystals were mounted on a nylon loop and cryoprotected in 25%(v/v) glycerol before flash-cooling in liquid nitrogen. X-ray diffraction data were collected on a MAR CCD 300 detector at the SER-CAT beamline of the Advanced Photon Source (APS) at the Argonne National Laboratory with a wavelength of 1 Å, a crystal-to-detector distance of 220 mm and an oscillation angle of 0.5°. The X-ray data were integrated using HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.3. Structure determination and refinement
The structure of MnSOD was determined by molecular replacement with the CNS package (Brünger et al., 1998 ▶). The structure of the dimer of SodA-1 (PDB code 1xuq) from B. anthracis, which shares 77% sequence identity, was used as a search model (Boucher et al., 2005 ▶). Four dimers per asymmetric unit were predicted, yielding a Matthews coefficient of 2.6 Å3 Da−1 and 52.9% solvent content. Data between 15 and 4 Å were used in the rotation search, resulting in one peak. The rotation solution was then applied in a translation search. After the first translation search, the coordinates of the top translation search were used as the first dimer and merged with the coordinates of the starting model as the second dimer. The location of the first dimer remained fixed, while the location of the second dimer was tested for each rotation peak. The last two dimers were searched for in the same way, yielding a distinct solution with a correction coefficient of 0.19. After rigid-body refinement (CNS) and density modification using the phase probability distribution calculated from the model and the experimental amplitudes and solvent flipping, the figure of merit improved to 0.83. Crystallographic refinement of the MnSOD structure was initially carried out using the CNS package (Brünger et al., 1998 ▶). After iteratively subjecting the model to several rounds of simulated annealing and group and individual B-factor refinement with CNS and manual rebuilding, the R factor and R free dropped to 21.1% and 23.0%, respectively.
2.4. Structural analysis
The structures of MnSOD, B. anthracis SodA-1 (PDB code 1xuq) and human MnSOD (PDB code 2adq) were superimposed on Cα atoms using the program DaliLite (Holm & Park, 2000 ▶). Figures were generated with MolScript (Kraulis, 1991 ▶), BobScript (Esnouf, 1999 ▶), Raster3D (Merritt & Bacon, 1997 ▶) and PyMOL (DeLano, 2002 ▶).
3. Results
3.1. Cloning, expression and purification of MnSOD
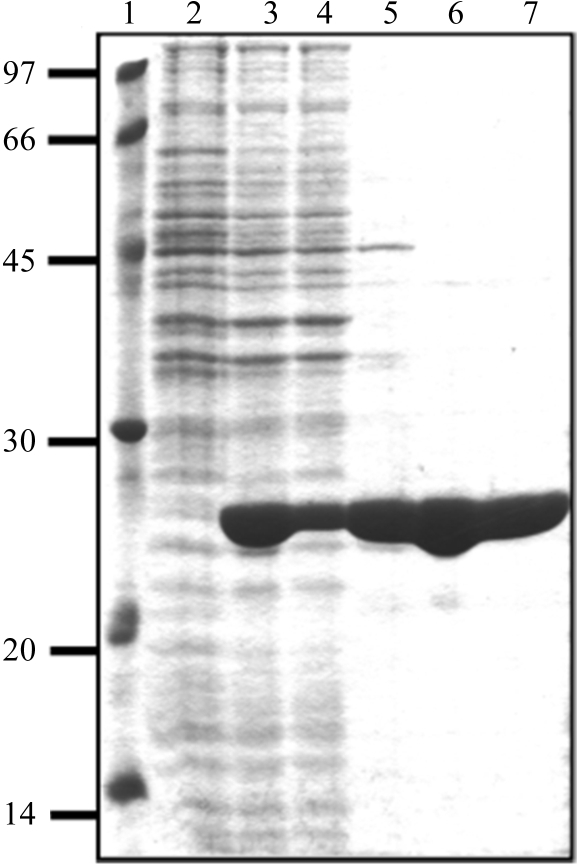
Initial trials to overexpress MnSOD in E. coli DH5α from the shuttle expression clone pHYsodA09 in which the sodA gene was fused to the Pspac promoter were not successful. In order to overexpress MnSOD for crystallographic analysis, the B. subtilis MnSOD protein encoded by the sodA gene was subcloned in pBAD-HisA, expressed in E. coli Top10 and purified by osmotic shock and anion-exchange and gel-filtration chromatography. Inclusion of 1 mM MnCl2 in the culture medium during growth was necessary to maintain enzyme stability and improve overexpression in E. coli Top10. The purified enzyme was the reddish purple color characteristic of MnSODs. An apparent molecular weight of 50 kDa was determined for the purified MnSOD by gel chromatography on a previously calibrated Superdex G-200 HR column. The subunit size was determined to be 25 kDa by SDS–PAGE, indicating that the enzyme consists of two subunits with identical molecular weights (Fig. 1 ▶).
Figure 1.
Purification steps of MnSOD. Lane 1, low-molecular-weight markers (kDa). Lane 2, crude extract of uninduced E. coli Top10 cells harboring pHE25 plasmid. Lane 3, crude extract of E. coli Top10 harboring pHE25 plasmid induced with 0.2% arabinose for 4 h in LB medium containing 1 mM MnCl2. Lane 4, cytoplasmic extract from cells after osmotic shock. Lane 5, periplasmic protein extract resulting from osmotic shock, showing that approximately 90% of MnSOD was recovered during this step. Lane 6, fraction eluted from Q-Sepharose containing partially purified MnSOD. Lane 7, purified MnSOD from the final purification step (Superdex G-200 HR column).
3.2. Overall structure of MnSOD
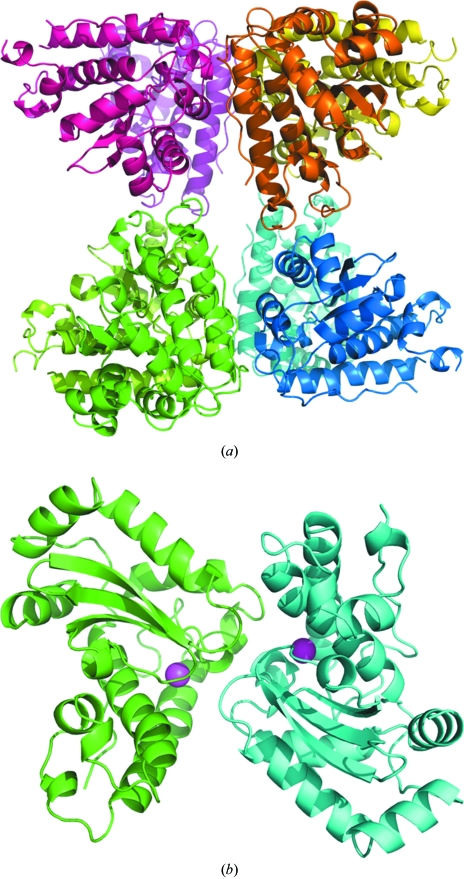
The crystal structure of MnSOD was solved at 1.8 Å resolution by molecular replacement and refined to an R factor of 21.1%. The crystallographic data and refinement statistics are given in Table 1 ▶. The MnSOD structure was determined in space group P1, with four homodimers in the asymmetric unit (Fig. 2 ▶ a). There are no significant structural differences between the main-chain atoms of the four dimers. Superimposing each of the two chains in the four dimers resulted in r.m.s.d. values of 0.2–0.3 Å for all Cα atoms.
Table 1. Crystallographic data-collection and refinement statistics.
Values in parentheses are for the last shell.
| Data collection | |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 68.4, b = 84.0, c = 91.9, α = 99.1, β = 105.9, γ = 105.6 |
| Unique reflections | 243111 |
| Rmerge | 13.3 (30.5) |
| Redundancy | 2.2 |
| Completeness (%) | 90.0 (51.3) |
| 〈I/σ(I)〉 | 17.6 |
| Refinement | |
| Resolution (Å) | 50–1.6† |
| R factor/Rfree (%) | 21.1/23.0 |
| No. of residues | 1591 |
| No. of water molecules | 910 |
| No. of Mn atoms | 8 |
| R.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.1 |
| Ramachandran plot, residues in | |
| Most favored regions (%) | 91.8 |
| Additional allowed regions (%) | 7.0 |
| Averaged B values (Å2) | |
| Main-chain atoms | 17.8 |
| Side-chain atoms | 20.5 |
| Solvent atoms | 25.4 |
The effective resolution is 1.8 Å for 95.4% completeness in the highest resolution shell.
Figure 2.
Overall structure of B. subtilis MnSOD. (a) The asymmetric unit of the MnSOD structure is composed of four dimers, which are colored dark and light magenta, gold, blue and green. (b) The MnSOD dimer backbone colored by subunit with Mn atoms shown as magenta spheres.
The monomer has the typical two-domain fold of MnSODs comprising the helical domain and the α/β domain, as shown in Fig. 2 ▶(b). One manganese ion is present in each subunit. The helical domain is composed of three α-helices, α1 (Lys21–Val43), α2 (Val54–Ser63) and α3 (Glu66–Ser87), with several connecting loops. The loop between residues 43 and 50 is partially disordered in three of the subunits. The α/β-domain starts with two α-helices, α4 (Gly99–Phe109) and α5 (Phe112–Gly125), followed by three β-strands, β1 (Gly130–Asn139), β2 (Lys140–Pro147) and β3 (Thr158–Asp164), which form a β-sheet, and three α-helices, α6 (Glu167–Gln175), α7 (Arg178–Phe185) and α8 (Trp191–Glu200).
The dimer interface has extended symmetric interactions that mainly involve residues from the loop containing Phe127, Gly128 and Ser129 and the α6 helix (Trp166, Glu167, His168, Tyr171 and Leu172) of each subunit. The side chains of Phe127, Glu167 and Tyr171 protrude into the other subunit and form major intersubunit interactions. Intersubunit hydrogen bonds are formed between the side chains of Glu167 and His168, and between Tyr171 and His31. Phe127 makes hydrophobic interactions with Trp166, Tyr35, Asn74 and Asn148 from the other subunit. Tyr171 also makes hydrophobic contacts with Ile26 and Tyr171, while Leu172 contacts Leu172 from the other subunit.
The four dimers are organized in the crystallographic asymmetric unit as illustrated in Fig. 2 ▶(a). The interface and interactions between pairs of dimers are less extensive than those between the two subunits in each dimer. The AD (red/green) and BC (blue/gold) dimer pairs have an interface that includes a hydrogen-bond interaction between the carbonyl O atom of Glu44 and the side chain of Lys144, as well as hydrophobic contacts between Pro65 in one subunit and Ser107 and Val108 in the other subunit. The interactions between the C and D dimers (red/gold and blue/green) involve hydrogen bonds between the side chains of Lys30 and Ser183 and between the side chain of Lys21 and the carbonyl O atom of Glu16 and hydrophobic contacts of Pro17 with Lys21 and Thr25, and between the guanidinium groups of Arg177 from both dimers. The relative paucity of interactions at the dimer–dimer interfaces in the four dimers of the asymmetric unit implies that the arrangement of the four dimers is primarily a consequence of packing in the crystal lattice with little physiological relevance.
3.3. Structural comparison with B. anthracis SodA-1 and human MnSOD
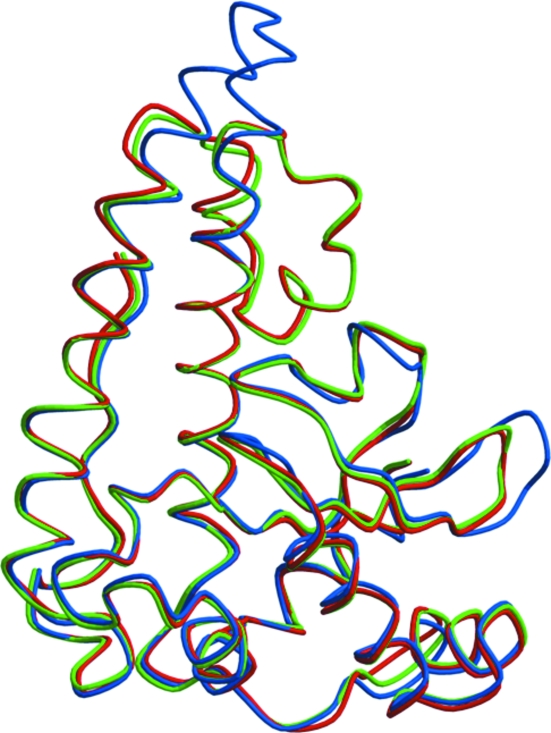
B. subtilis MnSOD and B. anthracis SodA-1 share 77% sequence identity and 86% similarity. Both proteins are dimers with high similarity in their overall structures. Superimposing the Cα atoms of the B. subtilis MnSOD dimer structure with the B. anthracis SodA-1 structure (PDB code 1xuq) gives an r.m.s.d. of 0.5 Å, while the monomers also can be superimposed with an r.m.s.d. of 0.5 Å (Fig. 3 ▶). The largest differences between the two structures are in the loop regions between residues 44–51 and residues 95–101, although the difference between the positions of equivalent Cα atoms is no more than 1.5 Å (Fig. 3 ▶).
Figure 3.
Superposition of MnSOD subunits from B. subtilis (green), B. anthracis (red) and H. sapiens (blue).
Human MnSOD has a different oligomeric structure: it forms a homotetramer of 22 kDa subunits. At the protein-sequence level, B. subtilis MnSOD shares 53% identity and 65% similarity with human MnSOD (PDB code 2adq). The tertiary structures are similar; superimposing the two monomer structures gives an r.m.s.d. of 1.3 Å for equivalent Cα atoms. The major difference lies in the helical domain (Fig. 3 ▶ b). In contrast to human MnSOD, which only contains two long α-helices, the two Bacillus MnSODs have a third short helix inserted between the two conserved α-helices. The inserted helix is bent at an angle of 120° away from the other two helices, forming a more compact domain than that present in the human enzyme (Fig. 3 ▶). There is also a two-residue insertion in the human enzyme between residues Asn138 and Gly139 that form the turn between two antiparallel β-strands in the B. subtilis MnSOD.
3.4. Comparison of manganese-binding sites and dimer interfaces
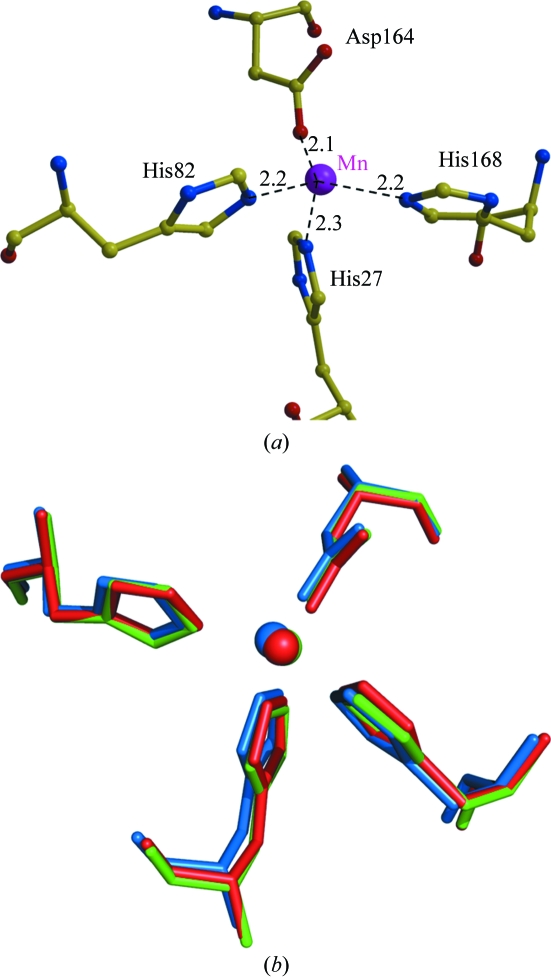
The manganese ion in each monomer of MnSOD is stabilized by close coordination with three histidines (His27, His82 and His168), one aspartic acid (Asp164) and one water molecule (Fig. 4 ▶ a). The residues in the Mn-binding site of B. subtilis MnSOD are in almost identical positions in the other two enzymes (Fig. 4 ▶ b) and are conserved in the structures of other SODs (Boucher et al., 2005 ▶; Quint et al., 2006 ▶). An outer layer around the Mn-binding site is formed by the aromatic side chains of Trp131, Trp166, Tyr35, Phe85, Trp86 and His31, which are 5–6 Å distant from the Mn. The Mn-binding site is close to the intersubunit interface in the dimer. The coordinating His168 forms a hydrogen bond with Glu167 from the other subunit in the dimer. The coordinating Asp164 and His168 are at either end of the loop containing Trp166 and Glu167, which contributes intersubunit interactions. These residues at the subunit interface in the dimer are conserved in the sequences of both B. anthracis and human MnSODs. The conserved network of interactions between the Mn-binding site and the subunit interface suggests its importance for the stability and activity of the enzyme. Only three amino acids at the subunit interface of the dimer differ between the Bacillus and human enzymes: Phe127 is substituted by Gln in the human MnSOD, Asn74 becomes Phe and Ile26 undergoes a conservative change to Leu. Ile26 is adjacent to the coordinating His27 and makes hydrophobic intersubunit contacts; however, little change is expected for the interactions of Leu at the equivalent position in the human enzyme. The substitution of the polar Gln for the hydrophobic Phe127 and the hydrophobic Phe for Asn74 in the human enzyme provide larger changes that might be exploited in the design of antibacterial agents. However, the two side chains (Phe127/Gln and Asn74/Phe) retain similar hydrophobic intersubunit contacts with each other in both the bacterial and human enzymes.
Figure 4.
Active site of MnSOD. (a) Coordination of the Mn atom of B. subtilis MnSOD. Interatomic interactions are shown as broken lines with distances in angstroms. (b) Superposition of the active site of MnSODs from B. subtilis (green), B. anthracis (red) and H. sapiens (blue).
4. Discussion
MnSOD (encoded by sodA) is essential for resistance to oxidative stress in growing and sporulating cells of B. subtilis (Inaoka et al., 1999 ▶). sodA mutants also exhibit decreased transformation efficiency as a result of superoxide inhibition of the ComQXP quorum-sensing circuitry (Ohsawa et al., 2006 ▶). Although MnSOD is classified as an intracellular protein, it is also consistently found in extracellular protein samples of many bacilli (Gohar et al., 2005 ▶). As a secreted protein, MnSOD was identified as a spore-coat protein and was proposed to play a role in spore-coat assembly (Henriques et al., 1998 ▶). It is not clear how MnSOD is secreted in B. subtilis as it lacks a classical signal peptide at the N-terminus or other known secretion-signature sequences. Elucidation of the unknown mechanism for nonconventional protein secretion will benefit from this new crystal structure of MnSOD. MnSOD was purified from E. coli using an osmotic shock technique, which can facilitate the purification of some recombinant proteins expressed in bacteria, as described by Ewis & Lu (2005 ▶). The new crystal structure of the MnSOD from B. subtilis is very similar to the structure of the related enzyme from B. anthracis. However, the two bacterial MnSODs show significant structural differences from the human MnSOD. The human MnSOD has one less helix in the helical domain, a larger turn between antiparallel β-strands and differences in three residues at the intersubunit interface. These structural and sequence differences can be exploited in the design of potential antibacterial agents that selectively target the Bacillus enzymes. However, inhibitors that bind in the highly conserved enzyme active sites are unlikely to be effective drugs. Potential drugs that bind to distinct sites on the subunit interface and disrupt the quaternary structure and enzyme activity are more likely to be selective for Bacillus enzymes.
Supplementary Material
PDB reference: MnSOD, 2rcv, r2rcvsf
Acknowledgments
This research was supported in part by the Georgia State University Molecular Basis of Disease Program, the Georgia Research Alliance, the Georgia Cancer Coalition and National Institutes of Health award GM034776. We thank the staff at the SER-CAT beamline at the Advanced Photon Source, Argonne National Laboratory for assistance during X-ray data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357.
References
- Beyer, W., Imlay, J. & Fridovich, I. (1991). Prog. Nucleic Acid Res. Mol. Biol.40, 221–253. [DOI] [PubMed]
- Boucher, I. W., Kalliomaa, A. K., Levdikov, V. M., Blagova, E. V., Fogg, M. J., Brannigan, J. A., Wilson, K. S. & Wilkinson, A. J. (2005). Acta Cryst. F61, 621–624. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Edwards, R. A., Baker, H. M., Whittaker, M. M., Whittaker, J. W., Jameson, G. B. & Baker, E. N. (1998). J. Biol. Inorg. Chem.3, 161–171.
- Esnouf, R. M. (1999). Acta Cryst. D55, 938–940. [DOI] [PubMed]
- Ewis, H. E. & Lu, C.-D. (2005). FEMS Microbiol. Lett.253, 295–301. [DOI] [PubMed]
- Gohar, M., Gilois, N., Graveline, R., Garreau, C., Sanchis, V. & Lereclus, D. (2005). Proteomics, 5, 3696–3711. [DOI] [PubMed]
- Henriques, A. O., Melsen, L. R. & Moran, C. P. J. (1998). J. Bacteriol.180, 2285–2291. [DOI] [PMC free article] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Hsieh, Y., Guan, Y., Tu, C., Bratt, P. J., Angerhofer, A., Lepock, J. R., Hickey, M. J., Tainer, J. A., Nick, H. S. & Silverman, D. N. (1998). Biochemistry, 37, 4731–4739. [DOI] [PubMed]
- Hu, Y., Rosen, D. G., Zhou, Y., Feng, L., Yang, G., Liu, J. & Huang, P. (2005). J. Biol. Chem.280, 39485–39492. [DOI] [PubMed]
- Inaoka, T., Matsumura, Y. & Tsuchido, T. (1999). J. Bacteriol.181, 1939–1943. [DOI] [PMC free article] [PubMed]
- Kraulis, P. J. (1991). J. Appl. Cryst.24, 946–950.
- Liao, J., Li, M., Liu, M. Y., Chang, T., Le Gall, J., Gui, L. L., Zhang, J. P., Liang, D. C. & Chang, W. R. (2002). Biochem. Biophys. Res. Commun.294, 60–62. [DOI] [PubMed]
- Ludwig, M. L., Metzger, A. L., Pattridge, K. A. & Stallings, W. C. (1991). J. Mol. Biol.219, 335–358. [DOI] [PubMed]
- Merritt, E. A. & Bacon, D. J. (1997). Methods Enzymol.277, 505–524. [DOI] [PubMed]
- Oberley, T. D. (2004). Antioxid. Redox Signal.6, 483–487. [DOI] [PubMed]
- Ohsawa, T., Tsukahara, K., Sato, T. & Ogura, M. (2006). J. Biochem. (Tokyo), 139, 203–211. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Quint, P., Reutzel, R., Mikulski, R., McKenna, R. & Silverman, D. N. (2006). Free Radic. Biol. Med.40, 453–458. [DOI] [PubMed]
- Stroupe, M. E., DiDonato, M. & Tainer, J. A. (2001). In Handbook of Metalloproteins, edited by A. Messerschmidt, R. Huber, K. Weighardt & T. Poulos. Chichester: Wiley.
- Sugio, S., Hiraoka, B. Y. & Yamakura, F. (2000). Eur. J. Biochem.267, 3487–3495. [DOI] [PubMed]
- Thorstenson, Y. R., Zhang, Y., Olson, P. S. & Mascarenhas, D. (1997). J. Bacteriol.179, 5333–5339. [DOI] [PMC free article] [PubMed]
- Wuerges, J., Lee, J. W., Yim, Y. I., Yim, H. S., Kang, S. O. & Carugo, K. D. (2004). Proc. Natl Acad. Sci. USA, 101, 8569–8574. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: MnSOD, 2rcv, r2rcvsf