Crotoxin B is a basic phospholipase A2 found in the venom of C. durissus terrificus and is one of the subunits that constitute crotoxin. Here, the crystallization, X-ray diffraction data collection and molecular-replacement solution of a novel tetrameric complex formed by two dimers of crotoxin B isoforms are presented.
Keywords: phospholipase A2, crotoxin B, snake venoms
Abstract
Crotoxin B is a basic phospholipase A2 found in the venom of Crotalus durissus terrificus and is one of the subunits that constitute crotoxin. This heterodimeric toxin, which is the main component of C. d. terrificus venom, is completed by an acidic, nontoxic and non-enzymatic component (crotoxin A) and is involved in important envenomation effects, such as neurological disorders, myotoxicity and renal failure. Although crotoxin was first crystallized in 1938, no crystal structure is currently available for crotoxin, crotoxin A or crotoxin B. In this work, the crystallization, X-ray diffraction data collection to 2.28 Å resolution and molecular-replacement solution of a novel tetrameric complex formed by two dimers of crotoxin B isoforms (CB1 and CB2) is presented.
1. Introduction
Crotoxin, the main protein present in the venom of the South American rattlesnake (Crotalus durissus terrificus), was the first animal toxin to be purified and crystallized (Slotta & Fraenkel-Conrat, 1938 ▶; Faure & Bon, 1988 ▶), but no crystal structure has been made available to date. This toxin exerts its pathophysiological action by blocking neuromuscular transmission (Vital-Brazil & Excell, 1970 ▶), causing modification of neurotransmitter release from nerve termini (Hawgood & Smith, 1977 ▶).
Crotoxin is a heterodimeric complex composed of two subunits: a basic and toxic Asp49-phospholipase A2 (crotoxin B, CB or basic Asp49-PLA2; Hendon & Fraenkel-Conrat, 1971 ▶; Fraenkel-Conrat et al., 1980 ▶; Aird et al., 1986 ▶) and an acidic, nontoxic and non-enzymatic component (crotoxin A, CA or crotapotin; Aird et al., 1985 ▶, 1990 ▶). Several isoforms of both subunits of crotoxin have been isolated from the venom of a single snake (Hendon et al., 1970 ▶; Breithaupt et al., 1974 ▶; Aird & Kaiser, 1985 ▶; Faure & Bon, 1987 ▶, 1988 ▶; Faure et al., 1994 ▶). Several CA and CB isoforms present in a large batch of venom collected from numerous snakes were purified and some were sequenced (Faure & Bon, 1988 ▶; Faure et al., 1994 ▶). The amino-acid composition of the purified basic Asp49-PLA2 isoforms (CB1 and CB2) indicated that they differed at eight amino-acid residues (Faure et al., 1994 ▶). These substitutions resulted in slight modifications in the enzymatic and pharmacological properties of the toxin: one of the crotoxin B isoforms has a low enzymatic activity and a high lethal potency, whereas the other CB isoform was less toxic but more active enzymatically (Faure & Bon, 1988 ▶; Faure et al., 1994 ▶).
The biochemical mechanism of the crotoxin complex constitutes an example of molecular potentiation: the CA subunit hinders the formation of nonspecific interactions between the basic phospholipase A2 and its substrates or other molecules. This complex can be separated only in the presence of 6 M urea or at a pH of below 2 (Faure & Bon, 1988 ▶; Faure et al., 1994 ▶). In isolation, both subunits are reported to be pharmacologically inactive; CA is thought to act as a chaperone protein for CB, increasing the neurotoxic potency of this enzyme (Breithaupt, 1976 ▶; Habermann & Breithaupt, 1978 ▶). When crotoxin interacts with the specific receptors present in the presynaptic nerve endings and possibly in the sarcoplasma and other plasma membranes, the complex dissociates and the CB subunit induces potent neurotoxic, cytotoxic and myotoxic effects (Bon, 1997 ▶).
Despite the harmful effects of this toxin, the crotoxin complex has been successfully tested as an inhibitor of tumour growth and as an efficient analgesic substance (Corin et al., 1993 ▶; Rudd et al., 1994 ▶; Cura et al., 2002 ▶; Zhang et al., 2006 ▶). This fact enhances interest not only in its mechanisms of action but also in the possible practical applications of this toxin as a drug or even as a model for the development of biotechnological products.
The isolation, biochemical/pharmacological characterization and amino-acid sequence of crotoxin B isoforms (CB1 and CB2) from C. d. terrificus have been reported (Hendon et al., 1970 ▶; Faure & Bon, 1988 ▶; Faure et al., 1994 ▶). Protein sequencing indicated that both CB isoforms are Asp49-PLA2s and consist of 122 amino acids (molecular weights of 14 185 Da for CB1 and 14 245 Da for CB2; UniProtKB/Swiss-Prot data bank codes P62022 and P24027 for CB1 and CB2, respectively; Faure et al., 1994 ▶).
In the present paper, we describe the crystallization, X-ray diffraction data collection and molecular-replacement solution of a tetrameric arrangement of two isoforms of crotoxin B (CB1 and CB2), a novel feature for Asp49-PLA2s. This study may provide insights into the relationship between the myotoxic, cytotoxic and neurotoxic activities and the structural characteristics of this protein. These activities have largely been studied by biochemical techniques; however, there is little structural data available.
2. Materials and methods
2.1. Purification and amino-acid sequences
Crotoxin B (CB) was isolated from C. d. terrificus snake venom by ion-exchange chromatography on a CM-Sepharose column (2 × 20 cm) previously equilibrated with 0.05 M ammonium bicarbonate buffer pH 8.0 (Soares et al., 1998 ▶). The dissociation of crotoxin and the isolation of subunits was performed according to Hendon & Fraenkel-Conrat (1971 ▶). The amino-acid sequences of the two CB isoforms (CB1 and CB2) which formed the asymmetric unit of the crystal used in the X-ray diffraction experiments were the same as those determined by Faure et al. (1994 ▶). The homogeneity of the toxin was analyzed by polyacrylamide gel electrophoresis (PAGE) for basic proteins and SDS–PAGE according to Reisfeld et al. (1962 ▶) and Laemmli (1970 ▶), respectively.
2.2. Crystallization
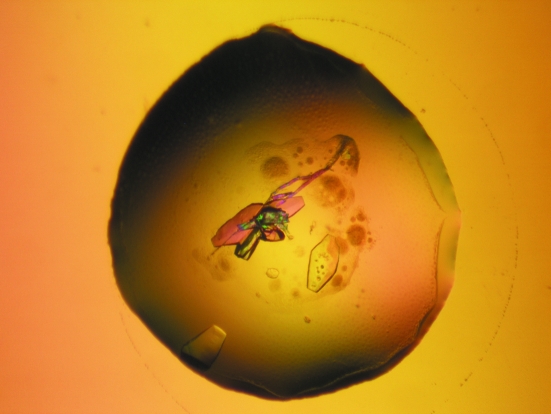
A lyophilized sample of crotoxin B was dissolved in ultrapure water at a concentration of 10 mg ml−1. The sparse-matrix method (Jancarik & Kim, 1991 ▶) was used to perform initial screening of the crystallization conditions. Crystals of CB were obtained by the conventional hanging-drop vapour-diffusion method (McPherson, 1982 ▶) using a 1 µl protein solution:1 µl precipitant ratio; the drop was equilibrated against a reservoir (500 µl) containing 11%(w/v) polyethylene glycol 8000 and 0.1 M Tris–HCl pH 9.0. Crystals measured approximately 0.20 × 0.25 × 0.05 mm after 20 d at 291 K (Fig. 1 ▶).
Figure 1.
Crystal of crotoxin B from C. d. terrificus.
2.3. X-ray data collection and processing
X-ray diffraction data were collected from a single CB crystal at a wavelength of 1.427 Å (at 100 K) using a synchrotron-radiation source (MX1 Station, Laboratório Nacional de Luz Sincrotron, LNLS, Campinas, Brazil) and a MAR CCD imaging-plate detector (MAR Research). The crystal was mounted in a nylon loop and flash-cooled in a stream of nitrogen gas at 100 K using 20%(v/v) glycerol as a cryoprotectant. The crystal-to-detector distance was 85 mm and an oscillation range of 1° was used, resulting in the collection of a total of 103 images. Data processing was carried out using the programs DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶) at 2.28 Å resolution.
3. Results and discussion
The data-collection statistics are shown in Table 1 ▶. The data set is 90.0% complete at 2.28 Å resolution, with an R merge of 12.7%. The crystals belong to the orthorhombic space group P212121, with unit-cell parameters a = 72.9, b = 81.2, c = 100.0 Å.
Table 1. X-ray diffraction data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Unit-cell parameters (Å) | a = 72.9, b = 81.2, c = 100.0 |
| Space group | P212121 |
| Resolution (Å) | 40–2.28 (2.36–2.28) |
| Unique reflections | 23669 (2423) |
| Completeness (%) | 85.9 (90.0) |
| Rmerge† (%) | 12.7 (54.7) |
| Radiation source | Synchrotron (MX1 Station, LNLS) |
| Data-collection temperature (K) | 100 |
| Average I/σ(I) | 9.57 (2.04) |
| Redundancy | 4.2 (4.2) |
| Matthews coefficient VM (Å3 Da−1) | 2.6 |
| Molecules in the ASU | 4 |
| Solvent content (%) | 52.6 |
R
merge = 
 , where I
i(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and
, where I
i(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and  is the mean intensity of that reflection. Calculated for I > −3σ(I). Data were processed using the HKL suite (Otwinowski & Minor, 1997 ▶).
is the mean intensity of that reflection. Calculated for I > −3σ(I). Data were processed using the HKL suite (Otwinowski & Minor, 1997 ▶).
Packing parameter calculations based on the protein molecular weight indicate the presence of four molecules in the asymmetric unit. This corresponds to a Matthews coefficient V M of 2.60 Å3 Da−1 with a calculated solvent content in the crystals of 52.61%; these values are within the expected range for typical protein crystals (assuming a value of 0.74 cm3 g−1 for the protein partial specific volume; Matthews, 1968 ▶).
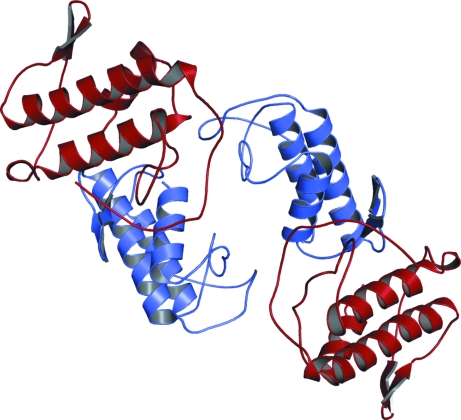
The crystal structure of the basic PLA2 formed by two isoforms of crotoxin B (two molecules of CB1 and two molecules of CB2) was determined using molecular-replacement techniques implemented in the program AMoRe (Navaza, 1994 ▶) using the coordinates of a monomer of native BthTX-I (da Silva-Giotto et al., 1998 ▶), a Lys49-PLA2 from Bothrops jararacussu snake venom (Fig. 2 ▶).
Figure 2.
Tetrameric structure of crotoxin B from C. durissus terrificus shown as a ribbon diagram. The monomers of isoforms CB1 and CB2 are shown in red and blue, respectively. This figure was generated using PyMOL (DeLano, 2002 ▶).
In conclusion, crotoxin B isolated from C. durissus terrificus venom was crystallized and X-ray diffraction data were collected to 2.28 Å. The structure presented a tetrameric arrangement formed by two heterodimers of the CB1 and CB2 isoforms. Initial analysis with PISA (Krissinel & Henrick, 2007 ▶) indicated that the tetrameric structure is stable in solution. Consequently, these data indicate that the tetramer is a feasible biological assembly, supporting the proposal that the interface contacts are not formed as a consequence of crystal packing. Detailed studies of this structure might provide insights into the toxicity mechanisms of crotoxin and other PLA2s.
Acknowledgments
The authors gratefully acknowledge financial support from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação para o Desenvolvimento da UNESP (FUNDUNESP) and Laboratório Nacional de Luz Síncrontron (LNLS, Campinas-SP).
References
- Aird, S. D. & Kaiser, I. I. (1985). Toxicon, 23, 361–374. [DOI] [PubMed]
- Aird, S. D., Kaiser, I. I., Lewis, R. V. & Kruggel, W. G. (1985). Biochemistry, 24, 7054–7058. [DOI] [PubMed]
- Aird, S. D., Kaiser, I. I., Lewis, R. V. & Kruggel, W. G. (1986). Arch. Biochem. Biophys.249, 296–300. [DOI] [PubMed]
- Aird, S. D., Yates, J. R., Martino, P. A., Shabanowitz, J., Hunt, D. F. & Kaiser, I. I. (1990). Biochim. Biophys. Acta, 1040, 217–224. [DOI] [PubMed]
- Bon, C. (1997). Venom Phospholipase A2 Enzymes: Structure, Function and Mechanism, edited by R. M. Kini, pp. 269–285. Chichester: Wiley & Sons.
- Breithaupt, H. (1976). Toxicon, 14, 221–233. [DOI] [PubMed]
- Breithaupt, H., Rübsamen, K. & Habermann, E. (1974). Eur. J. Biochem.49, 333–345. [DOI] [PubMed]
- Corin, R. E., Viskatis, L. J., Vidal, J. C. & Etcheverry, M. A. (1993). Invest. New Drugs, 11, 11–15. [DOI] [PubMed]
- Cura, J. E., Blanzaco, D. P., Brisson, C., Cura, M. A., Cabrol, R., Larrateguy, L., Mendez, C., Sechi, J. C., Silveira, J. S., Theiller, E., de Roodt, A. R. & Vidal, J. C. (2002). Clin. Cancer Res.8, 1033–1041. [PubMed]
- da Silva Giotto, M. T., Garratt, R. C., Oliva, G., Mascarenhas, Y. P., Giglio, J. R., Cintra, A. C. O., de Azevedo, W. F. Jr, Arni, R. K. & Ward, R. J. (1998). Proteins, 30, 442–454. [DOI] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. San Carlos: DeLano Scientific.
- Faure, G. & Bon, C. (1987). Toxicon, 25, 229–234. [DOI] [PubMed]
- Faure, G. & Bon, C. (1988). Biochemistry, 27, 730–738. [DOI] [PubMed]
- Faure, G., Choumet, V., Bouchier, C., Camoin, L., Guillaume, J.-L., Monegier, B., Vuilhorgne, M. & Bon, C. (1994). Eur. J. Biochem.223, 161–164. [DOI] [PubMed]
- Fraenkel-Conrat, H., Jeng, T. W. & Hsiang, M. (1980). Natural Toxins, edited by D. Eaker & T. Wadström, pp. 561–567. Oxford: Pergamon.
- Habermann, E. & Breithaupt, H. (1978). Toxicon, 16, 19–30. [DOI] [PubMed]
- Hawgood, B. J. & Smith, J. W. (1977). Br. J. Pharmacol.61, 597–606. [DOI] [PMC free article] [PubMed]
- Hendon, R. A. & Fraenkel-Conrat, H. (1971). Proc. Natl Acad. Sci. USA, 68, 1560–1563. [DOI] [PMC free article] [PubMed]
- Hendon, R. A., Roy, D. & Fraenkel-Conrat, H. (1970). Toxicon, 8, 135.
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol.372, 774–797. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: Wiley.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Reisfeld, R. A., Lewis, J. & Williams, D. A. (1962). Nature (London), 195, 281–283. [DOI] [PubMed]
- Rudd, C. L., Viskatis, L. J., Vidal, J. C. & Etcheverry, M. A. (1994). Invest. New Drugs, 12, 183–184. [DOI] [PubMed]
- Slotta, K. H. & Fraenkel-Conrat, H. (1938). Ber. Dtsch. Chem. Ges.71, 1076–1081.
- Soares, A. M., Rodrigues, V. M., Homsi-Brandeburgo, M. I., Toyama, M. H., Lombardi, F. R., Arni, R. K. & Giglio, J. R. (1998). Toxicon, 36, 503–514. [DOI] [PubMed]
- Vital-Brazil, O. & Excell, B. J. (1970). J. Physiol. (Lond.), 212, 34–35.
- Zhang, H. L., Han, R., Chen, Z. X., Chen, B. W., Gu, Z. L., Reid, P. F., Raymond, L. N. & Oin, Z. H. (2006). Toxicon, 48, 175–182. [DOI] [PubMed]