Abstract
Many syndromes traditionally viewed as individual diseases are heterogeneous in molecular pathogenesis and treatment responsiveness. This often leads to the conduct of large clinical trials to identify small average treatment benefits for heterogeneous groups of patients. Drugs that demonstrate effectiveness in such trials may subsequently be used broadly, resulting in ineffective treatment of many patients. New genomic and proteomic technologies provide powerful tools for the selection of patients likely to benefit from a therapeutic without unacceptable adverse events. In spite of the large literature on developing predictive biomarkers, there is considerable confusion about the development and validation of biomarker based diagnostic classifiers for treatment selection. In this paper we attempt to clarify some of these issues and to provide guidance on the design of clinical trials for evaluating the clinical utility and robustness of pharmacogenomic classifiers.
Keywords: Pharmacogenomics, biomarker, genomics, DNA microarray, clinical trial design, validation
1. Introduction
Physicians need improved tools for selecting treatments for individual patients. For example, many cancer treatments benefit only a minority of the patients to whom they are administered (e.g. Bast and Hortobagyi, 2004; Johnson and Janne, 2005). Being able to predict which patients are most likely to benefit would not only save patients from unnecessary toxicity and inconvenience, but might facilitate their receiving drugs that are more likely to help them. In addition, the current over-treatment of patients results in major expense for individuals and society, an expense which may not be indefinitely sustainable.
Much of the discussion about disease biomarkers is in the context of markers which measure some aspect of disease status, extent, or activity. Such biomarkers are often proposed for use in early detection of disease or as a surrogate endpoint for evaluating prevention or therapeutic interventions. The validation of such biomarkers is difficult for a variety of reasons, but particularly because the molecular pathogenesis of many diseases is incompletely understood and hence it is not possible to establish the biological relevance of a measure of disease status.
A pharmacogenomic biomarker is any measurable quantity that can be used to select treatment; for example, the result of an immunohistochemical assay for a single protein, the abundance of a protein in serum, the abundance of messenger ribonucleic acid (mRNA) transcripts for a gene in a sample of disease tissue or the presence/absence status of a specified germline polymorphism or tumor mutation. A pharmacogenomic classifier is a mathematical function that translates the biomarker values to a set of prognostic categories. These categories generally correspond to levels of predicted clinical outcome. With the advent of gene expression profiling, it is increasingly common to define composite pharmacogenomic classifiers based on the levels of expression of dozens of genes. For a fully specified classifier, however, all of the parameters and cut-points are specified for determining how to weight the different components and how to map the multivariate data into a defined set of categories. A completely defined classifier can be used to select patients and stratify patients for therapy in clinical trials that enable the clinical value of the classifier to be evaluated. Specifying only the genes involved does not enable one to structure prospective clinical validation experiments in which patients are assigned or stratified in prospectively well defined ways.
In this paper we will address some key issues in the development and validation of pharmacogenomic classifiers.
2. Developmental and validation studies
It is important to distinguish the studies which develop pharmacogenomic classifiers from those which evaluate the clinical utility of such classifiers. The vast majority of published prognostic marker studies are developmental and are not adequate for establishing the clinical utility and robustness of a classifier(Simon and Altman, 1994). Developmental studies are often based on a convenience sample of patients for whom tissue is available but who are heterogeneous with regard to treatment and stage. The studies are generally performed in an exploratory manner with no specified eligibility criteria, no primary endpoint or hypotheses and no defined analysis plan. The analysis often includes numerous analyses of different endpoints and patient subsets. Often there are multiple candidate biomarkers to evaluate, multiple ways of measuring and combining the candidate biomarkers. Such an informal approach is appropriate in a developmental study so long as one recognizes that the same study cannot be used to evaluate the clinical value of the resulting biomarkers or classifiers. The developmental study is exploratory and directed to hypothesis formation. The purpose of developmental studies should be to develop completely specified classifiers and completely specified hypotheses that can be tested in subsequent validation studies.
3. Development of Multi-component Classifiers
Four main components to developing a classifier are: (i) Feature selection; (ii) Selecting a prediction model; (iii) Fitting the prediction model to training data; and (iv) Estimating the prediction error that can be expected in future use of the model with independent data.
3.1 Feature selection
Feature selection is often important in developing an accurate classifier. It is well known from the theory of linear regression that including too many “noise variables” in the predictor reduces the accuracy of prediction. A noise variable is a variable that is not related to the thing being predicted. For microarray studies the number of noise variables may be orders of magnitude greater than the number of informative variables.
The most commonly used approach to feature selection is to identify the genes that are differentially expressed among the classes when considered individually. For example, if there are two classes, one can compute a t-test or a Mann-Whitney test for each gene. The log-ratios or log-intensity measurements are generally used as the basis of the statistical significance tests. The genes that are differentially expressed at a specified significance level are selected for inclusion in the class predictor. The stringency of the significance level controls the number of genes that are included in the model. If one wants a class predictor based on a small number of genes, the threshold significance level is made very small. Some statisticians fail to distinguish between “class comparison” problems, where the objective is to identify differentially expressed genes, and “class prediction” problems, where the objective is to do accurate prediction. Class comparison analyses are often appropriate when the objective is understanding biological mechanisms; e.g. what genes get expressed or repressed during wound healing of the kidney. Class prediction analyses are often appropriate for medical problems when the objective is predicting response to a specific treatment. Criteria such as false discovery rate are relevant for class comparison problems because it is useful to know what proportion of the genes reported as differentially expressed among the conditions represent false positives. For class prediction problems, however, the relevant criteria is prediction accuracy. The parameters used for selecting genes to be included in the predictor should merely be viewed as tuning parameters, even if they have the form of nominal significance level thresholds.
A-priori it is not clear what degree of stringency is optimal for feature selection. Dudoit and Fridlyand (Dudoit and Fridlyand, 2003) recommended that the number of genes selected be in the range of 10–100 for most studies. Previous experience with CART classification indicated that being overly stringent and thereby excluding important variables can be more serious than being inadequately stringent and including some noise variables (Breiman, et al., 1984). If there are only a few important variables and they are so differentially expressed that they stand out from among the thousands of noise variables, then high stringency can lead to accurate classification. More often, however, there are a larger number of differentially expressed genes but they do not stand out from among the thousands of noise genes. In this case, a moderate level of stringency may lead to best performance. The parameter controlling stringency of feature selection can be optimized. It is important to recognize, however, that if the selection of the stringency parameter is data dependent, then it must be regarded as part of the model development algorithm for purposes of computing cross-validated estimates of prediction error (see below) (Varma and Simon, 2005).
Several authors have developed methods to identify optimal sets of genes which together provide good discrimination of the classes (Bo and Jonassen, 2002, Deutsch, 2003, Kim, et al., 2002, Ooi and Tan, 2003). These algorithms are generally very computationally intensive. Unfortunately, it is not clear whether the increased computational effort of these methods is warranted. In some cases, the claims made do not appear to be based on properly cross-validated calculations; all of the data being used to select the genes and cross-validation used only for fitting the parameters of the model. Thorough studies comparing the performance of such methods to the simpler univariate methods are needed.
Some investigators have used linear combinations of gene expression values as predictors (Khan, et al., 2001, West, et al., 2001). Principal components are the orthogonal linear combinations of the genes showing the greatest variability among the cases. Using principal components as predictive features provides a vast reduction in the dimension of the expression data, but has two serious limitations. One is that the principal components are not necessarily good predictors. The measure of variability used in defining the principal components does not utilize the class membership information; it is total variability. Hence the genes whose expressions have substantial within-class variance but small differences in mean expression among classes may be included in the first few principal components. The second problem is that measuring the principal components requires measuring expression of all the genes. This makes it more difficult to translate the classifier to an alternative assay that does not provide a parallel read-out of all genes as was done in the OncoType Dx classifier (Paik et al. 2004). The method of gene shaving attempts to provide linear combinations with properties similar to the principal components that does not require measuring all of the genes (Hastie, et al., 2000). Partial least squares (Nguyen and Rocke, 2002) attempts to select linear combinations in a manner that utilizes class membership information. This may provide more accurate classifiers but does not reduce the number of genes whose expression needs to be measured. The method of supervised principal components (Bair and Tibshirani, 2004) utilizes principal components of genes selected for their univariate correlations with outcome. This has the advantage of reducing the number of genes whose expressions need to be measured in the future and also tends to avoid over-fitting the training set data.
3.2 Prediction Model
Many algorithms have been used effectively with DNA microarray data for class prediction. Dudoit et al. (Dudoit, et al., 2002, Dudoit and Fridlyand, 2003) compared a wide range of algorithms using publicly available data sets. The algorithms included nearest neighbor classification, linear and quadratic discriminant analysis, diagonal linear and quadratic discriminant analysis, support vector machines, classification trees, and random forest classifiers. Bagging and boosting the classifiers was also evaluated. For two-class problems, a linear discriminant is a function
| (1) |
where xi denotes the log-ratio or log-signal for the i’th gene, wi is the weight given to that gene, and the summation is over the set F of features (genes) selected for inclusion in the class predictor. For a two-class problem, there is a threshold value d, and a sample with expression profile defined by a vector x of values is predicted to be in class 1 or class 2 depending on whether l(x) as computed from equation (1) is less than or greater than d respectively.
Several kinds of class predictors used in the literature have the form shown in (1). They differ with regard to how the weights are determined. The oldest form of linear discriminant is Fisher’s linear discriminant. The weights are selected so that the mean value of l(x) in class 1 is maximally different from the mean value of l(x) in class 2. The squared difference in means divided by the pooled estimate of the within-class variance of l(x) was the specific measure used by Fisher. To compute these weights, one must estimate the correlation between all pairs of genes that were selected in the feature selection step. The study by Dudoit et al. indicated that Fisher linear discriminant analysis did not perform well unless the number of selected genes was small relative to the number of samples; otherwise there are too many correlations to estimate and the method tends to be un-stable and over-fit the data.
Diagonal linear discriminant analysis is a special case of Fisher linear discriminant analysis in which the correlation among genes is ignored. By ignoring such correlations, one avoids having to estimate many parameters, and obtains a method which performs better when the number of samples is small. The weight for the i’th gene is proportional to the difference in sample means for the i’th gene divided by the pooled within-class variance estimate for the i’th gene (Simon, et al., 2003). Golub’s weighted voting method (Golub, et al., 1999) and the compound covariate predictor of Radmacher et al. (Radmacher, et al., 2002) are similar to diagonal linear discriminant analysis and tend to perform very well when the number of samples is small. They compute the weights based on the univariate prediction strength of individual genes and ignore correlations among the genes. For the compound covariate classifier, the weight for the i’th gene is proportional to the pooled variance t statistic for comparing expression of the i’th gene between the two classes (Simon, et al., 2003).
Linear kernel support vector machines use a predictor of the form of equation (1). The weights are determined by maximizing the distance from the closest expression vectors to the hyperplane separating the two classes., instead of a least-squares criterion as in linear discriminant analysis (Ramaswamy, et al., 2001). Linear kernel support vector machines are similar to ridge regression classifiers. Although there are more complex forms of support vector machines, they appear to be inferior to linear kernel SVM’s for class prediction with large numbers of genes (Ben-Dor, et al., 2000). When the number of genes is greater than the number of cases, if the data is not inconsistent, then it is always possible to find a linear function that perfectly separates the classes in the training data. Although this does not directly imply that nonlinear classifiers might not have smaller generalization error, it suggests that most datasets are not large enough to effectively utilize non-linear classifiers with many parameters.
Khan et al. (Khan, et al., 2001) reported accurate class prediction among small, round blue cell tumors of childhood using an artificial neural network. The inputs to the ANN were the first ten principal components of the genes; that is, the l0 orthogonal linear combinations of the genes that accounted for most of the variability in gene expression among samples. Their neural network used a linear transfer function with no hidden layer and hence it was a linear perceptron classifier of the form of equation (1). Most true artificial neural networks have a hidden layer of nodes, use a non-linear transfer function and individual features as inputs. Such a “real” neural network may not perform as well as the principal component perceptron model of Khan et al. because of the number of parameters to be estimated would be too large for the available number of samples.
Nearest neighbor classification is based on a feature set F of genes selected to be informative for discriminating the classes and a distance function d(x, y) which measures the distance between the expression profiles x and y of two samples. The distance function utilizes only the genes in the selected set of features F. Usually, Euclidean distance is the metric used. To classify a sample with expression profile y, compute d(x, y) for each sample x in the training set. The predicted class of y is the class of the sample in the training set which is closest to y with regard to the distance function. A variant of nearest neighbor classification is k-nearest neighbor classification. For example with 3-nearest neighbor classification, you find the three samples in the training set which are closest to the sample y. The class which is most represented among these three samples is the predicted class for y. For microarray data classification, k-nearest neighbor classification is generally used with k equal to one or three because the number of cases is usually limited.
Nearest centroid classification is a variant of nearest neighbor classification. The centroid of the gene expression vectors for cases within a class is the vector containing the mean expression of each component gene for the cases in the class. With nearest centroid classification, a new case is classified into the class whose centroid in the training set it is closest to using the genes selected based on the training data. The method of shrunken centroids (Tibshirani, et al., 2002) is similar to nearest centroid classification but incorporates automatic gene selection by shrinking the class centroids towards the overall mean.
In the studies of Dudoit et al. (Dudoit, et al., 2002, Dudoit and Fridlyand, 2003), the simplest methods, diagonal linear discriminant analysis and nearest neighbor classification, generally performed about as well as more complex methods. Ben-Dor et al. (Ben-Dor, et al., 2000) also compared several methods on several public datasets and found that nearest neighbor classification generally performed as well or better than more complex methods.
3.3 Estimates of predictive accuracy based on developmental studies
Developmental studies are analogous to phase II clinical trials. They should include an indication of whether the genomic classifier is promising and worthy of phase III evaluation. There are special problems in evaluating whether classifiers based on high dimensional genomic or proteomic assays are promising however. The difficulty derives from the fact that the number of candidate features available for use in the classifier is much larger than the number of cases available for analysis. In such situations, it is always possible to find classifiers that accurately classify the data on which they were developed even if there is no relationship between expression of any of the genes and outcome (Radmacher, et al., 2002). Consequently, even in developmental studies, some kind of validation on data not used for developing the model is necessary. This “internal validation” is usually accomplished either by splitting the data into two portions, one used for training the model and the other for testing the model, or some form of cross-validation based on repeated model development and testing on random data partitions. This internal validation should not, however, be confused with the kind of external validation of the classifier utility in a setting simulating broad clinical application.
The most straightforward method of estimating the prediction accuracy is the split-sample method of partitioning the set of samples into a training set and a test set. Rosenwald et al. (Rosenwald, et al., 2002) used this approach successfully in their international study of prognostic prediction for large B cell lymphoma. They used two thirds of their samples as a training set. Multiple kinds of predictors were studied on the training set. When the collaborators of that study agreed on a single fully specified prediction model, they accessed the test set for the first time. On the test set there was no adjustment of the model or fitting of parameters. They merely used the samples in the test set to evaluate the predictions of the model that was completely specified using only the training data. In addition to estimating the overall error rate on the test set, one can also estimate other important operating characteristics of the test such as sensitivity, specificity, positive and negative predictive values.
The split-sample method is often used with so few samples in the test set, however, that the validation is almost meaningless. One can evaluate the adequacy of the size of the test set by computing the statistical significance of the classification error rate on the test set or by computing a confidence interval for the test set error rate. Since the test set is separate from the training set, the number of errors on the test set has a binomial distribution.
Michiels et al. (Michiels, et al., 2005) suggested that multiple training-test partitions be used, rather than just one. The split sample approach is mostly useful, however, when one does not have a well defined algorithm for developing the classifier. When there is a single training set-test set partition, one can perform numerous unplanned analyses on the training set to develop a classifier and then test that classifier on the test set. With multiple training-test partitions however, that type of flexible approach to model development cannot be used. If one has an algorithm for classifier development, it is generally better to use one of the cross-validation or bootstrap resampling approaches to estimating error rate (see below) because the split sample approach does not provide as efficient a use of the available data (Molinaro, et al., 2005).
Cross-validation is an alternative to the split sample method of estimating prediction accuracy (Radmacher, et al., 2002). Molinaro et al. describe and evaluate many variants of cross-validation and bootstrap re-sampling for classification problems where the number of candidate predictors vastly exceeds the number of cases (Molinaro, et al., 2005). Molinaro et al. found that for high dimensional data with small sample sizes, leave-one-out cross-validation and 10-fold cross-validation performed very effectively. Split-sample validation often was quite biased in over-estimating the prediction error. Results for the .632+ bootstrap varied depending on the stability of the classifier and the signal strength of the data. Previous evaluations of resampling methods for estimating prediction error failed to include variable selection and with high dimensional data variable selection influenced performance significantly. The cross-validated prediction error is an estimate of the prediction error associated with application of the algorithm for model building to the entire dataset.
A commonly used invalid estimate is called the re-substitution estimate. You use all the samples to develop a model. Then you predict the class of each sample using that model. The predicted class labels are compared to the true class labels and the errors are totaled. It is well-known that the re-substitution estimate of error is biased for small data sets and the simulation of Simon et al.(Simon, et al., 2003) confirmed that, with an astounding 98.2% of the simulated data sets resulting in zero misclassifications even when no true underlying difference existed between the two groups. Simon et al.(Simon, et al., 2003) also showed that cross-validating the prediction rule after selection of differentially expressed genes from the full data set does little to correct the bias of the re-substitution estimator: 90.2 % of simulated data sets with no true relationship between expression data and class still result in zero misclassifications. When feature selection was also redone in each to cross-validated training set, appropriate estimates of mis-classification error were obtained; the median estimated misclassification rate was approximately 50%.
The simulation results underscore the importance of cross-validating all steps of predictor construction in estimating the error rate. It can also be useful to compute the statistical significance of the cross-validated estimate of classification error. This determines the probability of obtaining a cross-validated classification error as small as actually achieved if there were no relationship between the expression data and class identifiers. A flexible method for computing this statistical significance was described by Radmacher et al. (Radmacher, et al., 2002). It involves randomly permuting the class identifiers among the patients and then re-calculating the cross-validated classification error for the permuted data. This is done a large number of times to generate the null distribution of the cross-validated prediction error. If the value of the cross-validated error obtained for the real data lies far enough in the tail of this null distribution, then the results are statistically significant. This method of computing statistical significance of cross-validated error rate for a wide variety of classifier functions is implemented in the BRB-ArrayTools software (Simon and Lam, 2005). Statistical significance, however, does not imply that the prediction accuracy is sufficient for the test to have clinical utility.
Even if a classifier is developed for a set of patients sufficiently homogeneous and uniformly treated to be therapeutically relevant, it may be important to evaluate whether the classifier predicts more accurately than do standard prognostic factors or adds predictive accuracy to that provided by standard prognostic factors. For example, Rosenwald et al. (Rosenwald, et al., 2002) developed a classifier of outcome for patients with advanced diffuse large B cell lymphoma receiving CHOP chemotherapy. The International Prognostic Index (IPI) is easily measured and prognostically important for such patients, however, and so it was important for Rosenwald et al. to address whether their classifier provided added value. The most effective way of addressing whether a classifier adds predictive accuracy to a standard classification system is to examine outcome for the new system within the levels of the standard system.
3.4 Sample size planning for developmental studies
Sample size planning for development of classifiers when the number of candidate predictors (p) is much larger than the number of cases (n) has not been adequately developed. Most classical methods of sample size planning for developing classifiers require non-singularity of the sample covariance matrix of the covariates and are not applicable to the p>>n setting. Although many classifiers for microarray studies retain fewer than n covariates, proper sample size planning method should account for variability in the selection of covariates, not just for variability in the weights placed on the selected covariates.
Mukherjee et al. (Mukherjee, et al., 2003) developed a sample size planning method for microarray classifier development based on learning curve estimation methods, but it requires extensive previous data from a study that attempted to distinguish the same classes using the same expression profiles. Fu et al. (Fu, et al., 2005) developed a sequential method for determining when additional samples are no longer based on sequential classifier development after each additional sample is taken.
Most sample size planning methods developed for microarray studies have been for the related objective of identifying genes that are differentially expressed among pre-defined classes. This has been termed class comparison, and it is a component of the class prediction problem considered here. Dobbin and Simon (Dobbin and Simon, 2005) showed that the following approximation can be used for estimating the total number of samples needed for determining whether a given gene is differentially expressed between two equally represented classes:
| (2) |
The quantities tn−2,α/2 and tn−2,β denote percentiles of the t distribution with n−2 degrees of freedom, α is the nominal threshold significance level for declaring a gene differentially expressed based on a gene-by-gene analysis, 1-β is the power for identifying a gene differentially expressed when the class means differ by δ in logarithm of expression and σ is the within class standard deviation of log expression for the gene. Since n occurs on both sides of the equation in (2), the expression must be solved iteratively. For sufficiently large n, the t percentiles can be replaced by normal percentiles.
The probability of incorrect classification based on a single gene whose log expression is normally distributed with common standard deviation σ is Φ(δ/2σ) where δ is the difference in mean log expression between the two classes and Φ denotes the standard normal distribution function. Consequently, the discrimination power of an individual gene is determined by the δ/σ value for that gene. A gene with a δ/σ value of 2 provides about an 84% probability of correct classification when used alone. Genes with smaller δ/σ effect sizes are less valuable for classification of individual patients, even if the research study is large enough to identify them as differentially expressed among the classes.
In the case where all differentially expressed genes are regarded as differentially expressed by the same amount δ, Dobbin and Simon (Dobbin and Simon, 2005) demonstrated the approximate relationship
| (3) |
Where FDR denotes the false discovery rate and π denotes the proportion of the genes that are differentially expressed. The false discovery rate is the proportion of the genes claimed differentially expressed which are false positive findings. Although it is unrealistic to expect that all genes that are differentially expressed have the same mean difference between classes, analysis of (3) is of interest for appreciating the relationships among the parameters. The false discovery rate depends strongly on π/α. If π is large, then a larger value of α will suffice to keep the FDR small. In microarray studies, π is typically in the range of 0.005 to 0.05, with smaller values being more common. With π≥0.005 and β=0.05, a value of α=0.001 is sufficient to limit the FDR to be no greater than 0.17. Using expression (2) with α=0.001, β=0.05, and δ/σ = 2 gives n=29.1 total cases, or 15 cases for each class. The sample size increases substantially if genes with an effect size of less than 2 are of interest, but as noted above, such genes are of less value for classification of individual patients.
Dobbin and Simon (Dobbin and Simon, 2005) also show that for finding genes whose expressions are correlated with a time-to-event variable, an approximation for the number of events required is approximately:
| (4) |
where γ denotes the standard deviation of log expression over the set of samples and h denotes the hazard ratio associated with a one-unit change in log intensity. Using expression profiles to predict risk groups for a time-to-event endpoint may require much larger sample sizes that for class prediction, particularly when the event rate is small.
4. Design of validation studies
Although there is a large literature on prognostic markers, few such factors are used in clinical practice. To a large extent this is due to a lack of adequate validation studies which demonstrate the therapeutic relevance and robustness of pre-specified biomarker classifiers. Prognostic markers are unlikely to be used unless they are therapeutically relevant. Most developmental studies, unless they are based on patients treated in a single clinical trial, are not based on a cohort medically coherent enough to establish therapeutic relevance. Developmental studies also rarely establish the robustness of the classifier and of the underlying assays under conditions that simulate those likely to be found in real world patient management.
The objective of external validation is to determine whether use of a completely specified diagnostic classifier for therapeutic decision making in a defined clinical context results in patient benefit. Patient benefit may represent better efficacy, reduced incidence of adverse events, better convenience or lower costs. The objective is not to repeat the developmental study and see if the same genes are prognostic or if the same classifier is obtained.
Biomarkers are used for very different purposes and validation should relate to fitness for a defined purpose. It is not productive to require validation in some more absolute biological sense for diseases whose molecular pathogenesis is not fully understood. We focus here on the design of validation studies to establish clinical benefit in assisting with treatment selection. For example, the Oncotype-Dx risk score was developed to measure prognosis for node negative, estrogen receptor (ER) positive patients with primary breast cancer receiving Tamoxifen therapy after surgical resection of the primary lesion (Paik, et al., 2004). The validation issue is whether use of this risk score results in clinical benefit. The components of expression signature classifiers need not be valid biomarkers in the sense of the Food and Drug Administration (FDA, 2005). Those criteria require that the role of the biomarker be mechanistically understood and accepted as markers of disease activity. Such criteria are relevant for biomarkers used as surrogate endpoints but not for the components of expression signatures used for tailoring treatments. It is, of course, desirable to understand the mechanistic relationship of the components of an expression signature, but the classifier can be validated without such understanding.
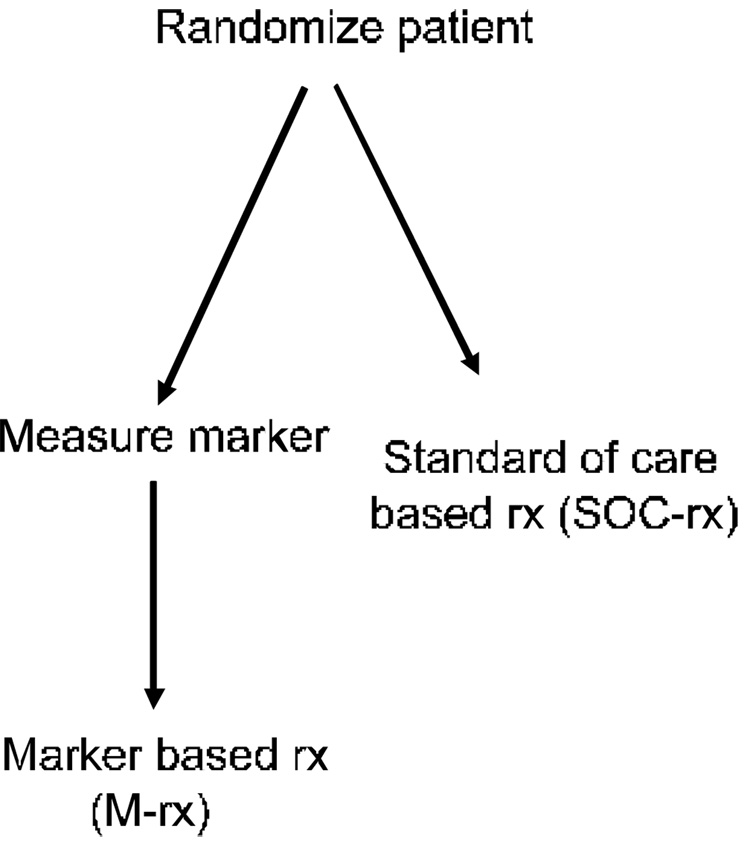
An independent validation study could be a prospective clinical trial in which patients are randomized to treatment assignment without use of the classifier versus treatment assignment with the aid of the classifier. This design requires that the classifier be determined only in half of the patients. Often, however, this design will be inefficient and require a huge sample size because many or most of the patients will receive the same treatment either way they are randomized. For example, consider women with lymph node negative, ER positive breast cancers. About one third of such patients might be expected to be classified as low risk for recurrence based on the Oncotype-DX expression signature based risk score (Paik, et al., 2004). If one wants to test the strategy of withholding cytotoxic chemotherapy (systemic treatment with Tamoxifen alone) from the subset of patients classified as low risk, it would be inefficient to randomize all of the node negative ER positive patients. If one randomizes all the patients and only performs the assay on the half randomized to have classifier based therapy, then the two randomization groups must be compared overall, although two thirds of the patients receive the same treatment in both arms. Designs related to that shown in Figure 1 have been discussed by Sargent et al. (Sargent, et al., 2005).
Figure 1.
Randomized clinical trial for evaluating whether use of a biomarker based classifier for treatment selection results in improved clinical outcome. All patients with conventional diagnosis are randomized between biomarker based treatment (M-rx) or standard of care based treatment (SOC-rx). This design is often very inefficient.
A more efficient alternative is to perform the assay up front for all patients, and then randomize only those classified as low risk. Those patients would be randomized to either receive Tamoxifen alone or Tamoxifen plus cytotoxic chemotherapy. Randomizing only the patients classified as low risk is much more efficient than randomizing all of the patients.
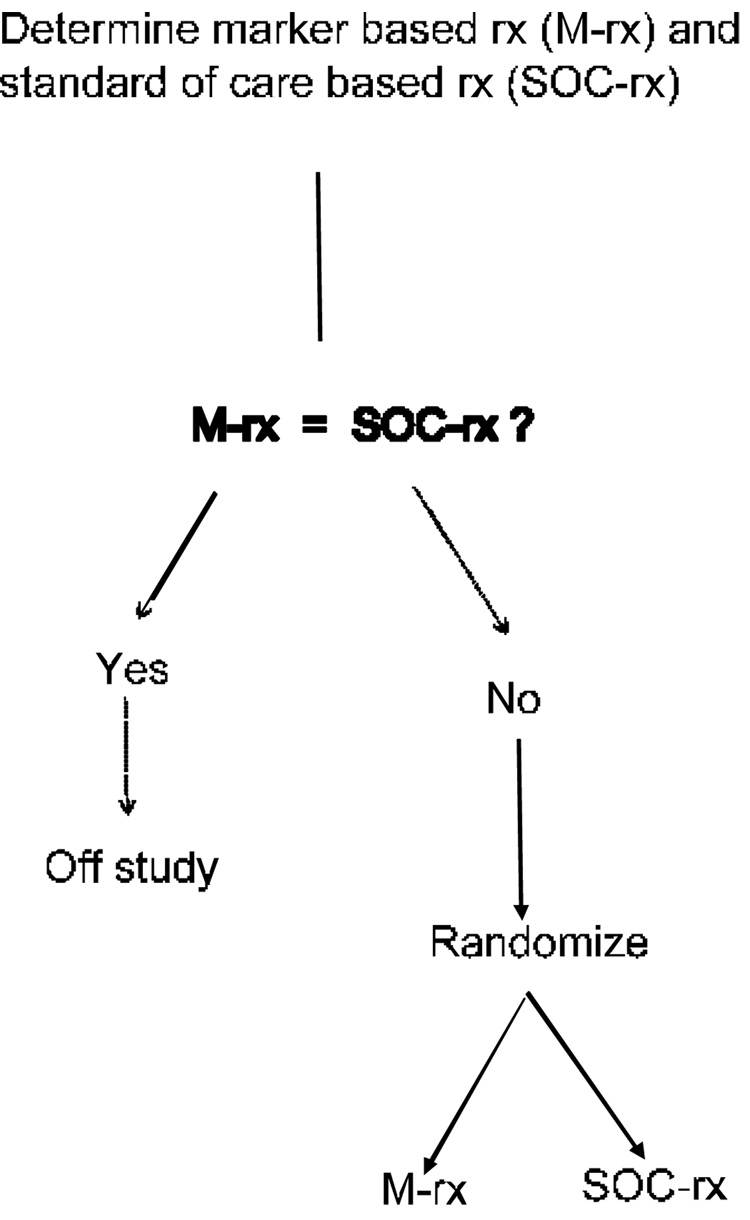
One might argue that treatment determination using a genomic classifier for women with stage I ER positive breast cancer should not be compared to the strategy of giving all such women Tamoxifen plus chemotherapy, because there are practice guidelines available based on tumor size and age that withhold chemotherapy from some patients. Nevertheless, it would still be very inefficient to randomize women to genomic classifier determined therapy or non-genomic practice guidelines determined therapy in which the genomic classifier is measured only on the women randomized to its use. Most of the women will probably receive the same treatment whichever arm they are randomized to. It is much more efficient to perform the assay for measuring the genomic classifier, and then randomize only the women for whom the two treatment strategies differ as indicated in Figure 2. With such a design the magnitude of the difference between randomization groups is not diluted by patients receiving the same treatment in each arm, but it may still require many patients to be screened in order to obtain enough patients to randomize whose treatment strategies differ.
Figure 2.
Improved clinical trial design for evaluating whether use of a biomarker based classifier for treatment selection results in improved clinical outcome. The biomarker classifier based treatment (M-rx) and standard of care based treatment (SOC-rx) are determined before randomization and patients for whom the two treatment strategies agree are not randomized. This design is often much more efficient than that shown in Figure 1.
The null hypothesis for the design of Figure 2 is that the marker based treatment selection strategy is equivalent to the standard care treatment selection strategy. In the breast cancer example described above, the marker based treatment selection strategy called for withholding systemic therapy other than Tamoxifen for patients predicted to be at low risk of recurrence based on the classifier. The standard of care treatment might incorporate decision making based on established predictive markers. For example, the standard of care might be include treatment with Herceptin for patients whose tumors expressed the Her2/neu receptor. Or the standard of care strategy might involve withholding systemic therapy other than Tamoxifen if the tumor size is below a specified threshold. The design of Figure 2 requires that the standard of care treatment and the classifier based treatment for each eligible patient be determined before randomization and only those patients for whom the two treatments differ are randomized.
Phase III clinical trials generally attempt to utilize an intervention in a manner that it might be used if adopted in broad clinical practice. For evaluating a diagnostic classifier, a multi-center clinical trial provides the challenges of distributed tissue handling and real time assay performance that would be met in general use. The assays might be performed in multiple laboratories and cannot be batched in time with a single set of reagents as might be done in a retrospective study. Consequently, the prospective clinical trial is the gold standard for external validation of a genomic classifier.
Validation based on a new prospective clinical trial will require a long follow-up time for low risk patients. In such circumstances it can be useful to conduct a prospectively planned validation using patients treated in a previously conducted prospective multi-center clinical trial if archived tumor specimens are available for the vast majority of patients. The validation study should be prospectively planned with at least as much detail and rigor as for prospective accrual of new patients. Although assaying procedures probably cannot be distributed over time in the same way as for newly accrued patients, assay reproducibility studies should be conducted to demonstrate that the assay has been standardized and quality controlled sufficiently so that such sources of variation are negligible. A written protocol should be developed to ensure that the study is prospectively planned to evaluate the clinical benefit of a completely specified genomic classifier for a defined therapeutic decision in a defined population in a hypothesis testing manner as it would for a prospective clinical trial.
The study of Paik et al.(Paik, et al., 2004) of the OncoType Dx classifier for women with node negative ER positive breast cancer is an example of careful prospective planning of an independent validation study using archived specimens. Their study was based on the observation that although randomizing only the patients classified as low risk is more efficient than randomizing all of the patients, it still would require many patients. It is a therapeutic equivalence trial in the sense that finding no difference in outcome changes clinical practice; consequently it is important to be able to detect small differences. Since the expected recurrence rate is so low, it would take many patients to detect a difference between the treatment arms. But if the recurrence rate is as low as predicted by the classifier, then the benefit of chemotherapy is necessarily extremely small. Consequently, an alternative design for external validation is a single arm study in which the patients classified as low risk are treated with Tamoxifen alone. If, with long follow-up, these patients have a very low recurrence rate, then the classifier is considered validated for providing clinical benefit because it enabled the identification of patients whose prognosis was so good on Tamoxifen monotherapy that they could be spared the toxicity, inconvenience and expense of chemotherapy. This was the approach used by Paik et al. for validation of the OncoType Dx classifier for patients with node negative, estrogen receptor positive breast cancer (Paik, et al., 2004). The genes that appeared prognostic were initially identified based on published microarray studies. A classifier based on measuring expression of those genes using a different assay that could be performed on the formalin fixed parafin embedded diagnostic biopsy was developed using the archived tissue from studies of the National Surgical Adjuvant Breast and Bowel Project (NSABP) cooperative cancer group. The completely pre-specified classifier was then tested on 668 patients from NSABP B-14 who received tamoxifen alone as systemic therapy. Fifty one percent of the assayed patients fell in the low risk group. They had a distant recurrence rate at 10 years of 6.8 percent (95% confidence interval 4.0 to 9.6). Much higher rates of distant recurrence were seen in the intermediate and high risk groups of the classifier (14.3% and 30.5% respectively).
5. Development of genomic classifiers for experimental drugs
In proposing the introduction of a classifier for improving the utilization of existing therapy, the emphasis should be on validation of the clinical benefit of using the classifier compared to not using it. Classifiers may also be developed, however, to restrict the type of patient/disease in which a new experimental therapy will be evaluated. This is usually only the case for classifiers based on the known molecular target of the drug when the biology is very clear. In this case, the focus would be on evaluating the drug in classifier positive patients and not validating the classifier itself. Simon and Maitournam (Simon and Maitournam, 2004) demonstrated that use of a genomic classifier for focusing a clinical trial in this manner can result in a dramatic reduction in required sample size, depending on the sensitivity and specificity of the classifier for identifying such patients. Not only can such targeting provide a huge improvement in efficiency in phase III development, it also provides an increased therapeutic ratio of benefit to toxicity and results in a greater proportion of treated patients who benefit.
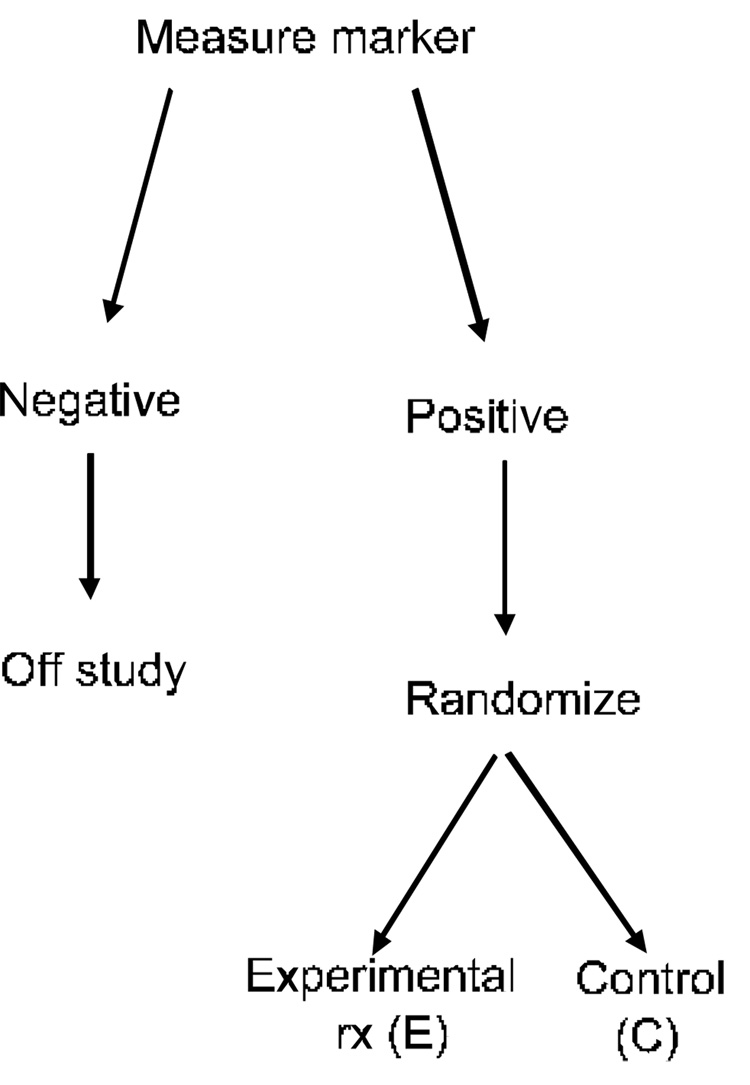
Simon and Maitournam consider use of the Targeted Design shown in Figure 3. During pre-clinical and phase I/II clinical development one identifies a fully specified classifier of which patients have a high probability of responding to the experimental drug. That classifier is then used to select patients for phase III trial. This is a form of enrichment design. Table 1 shows the number of events required for 80% statistical power in the design of Figure 3 for comparing exponential survival times if the treatment results in a halving of the hazard in the patients selected for study using the classifier. The number of events is compared to the number of events required in a standard clinical trial if the classifier is not used to select patients for randomization. The table assumes that the treatment is not effective for the classifier negative patients. More extensive results on relative efficiency of the targeted and untargeted designs are described by Simon and Maitournam (Maitournam and Simon, 2005, Simon and Maitournam, 2004).
Figure 3.
Targeted clinical trial design for evaluating a new experimental therapy. A biomarker classifier is developed for identifying those patients most likely to respond to the new treatment (E). Only those patients are randomized to E versus the control treatment. The patients predicted less likely to respond (marker negative) are off study. The targeted design is most useful in cases where the biomarker classifier has a strong biological rationale for identifying responsive patients and where it may not be ethically advisable to expose marker negative patients to the new treatment.
Table 1.
Second column contains the approximate number of events required for 80% power with 5% two-sided log-rank test for comparing arms of targeted design shown in Figure 3. Only marker positive patients are randomized. Treatment hazard ratio for marker positive patients is shown in first column. Time-to-event distributions are exponential and all patients are followed to failure. Last three columns show the approximate number of events required for comparing the arms of a traditional design in which unclassified marker positive and marker negative patients are randomized. Treatment hazard ratio for marker negative patients is assumed to be 1.
| Treatment Hazard Ratio for Marker Positive Patients | Number of Events for Targeted Design | Number of Events for Traditional Design | ||
|---|---|---|---|---|
| Percent of Patients Marker Positive | ||||
| 20% | 33% | 50% | ||
| 0.5 | 74 | 2040 | 720 | 316 |
| 0.67 | 200 | 5200 | 1878 | 820 |
Developing a genomic classifier of which patients are likely to benefit for targeting phase III trials may require larger phase II studies. This depends on the type of drug being developed. For example, if the drug is an inhibitor of a kinase mutated in cancer, then there is a natural diagnostic and no genome-wide screening is needed. For many molecularly targeted drugs, however, the appropriate assay for selecting patients is not known and development of a classifier based on comparing expression profiles for phase II responders versus phase II non-responders may be the best approach. In such instances, one may not have sufficient confidence in the genomic classifier developed in phase II to use it for excluding patients in phase III trials as in Figure 3. It may be better in this case to accept all conventionally eligible patients, and use the classifier in the pre-defined analysis plan.
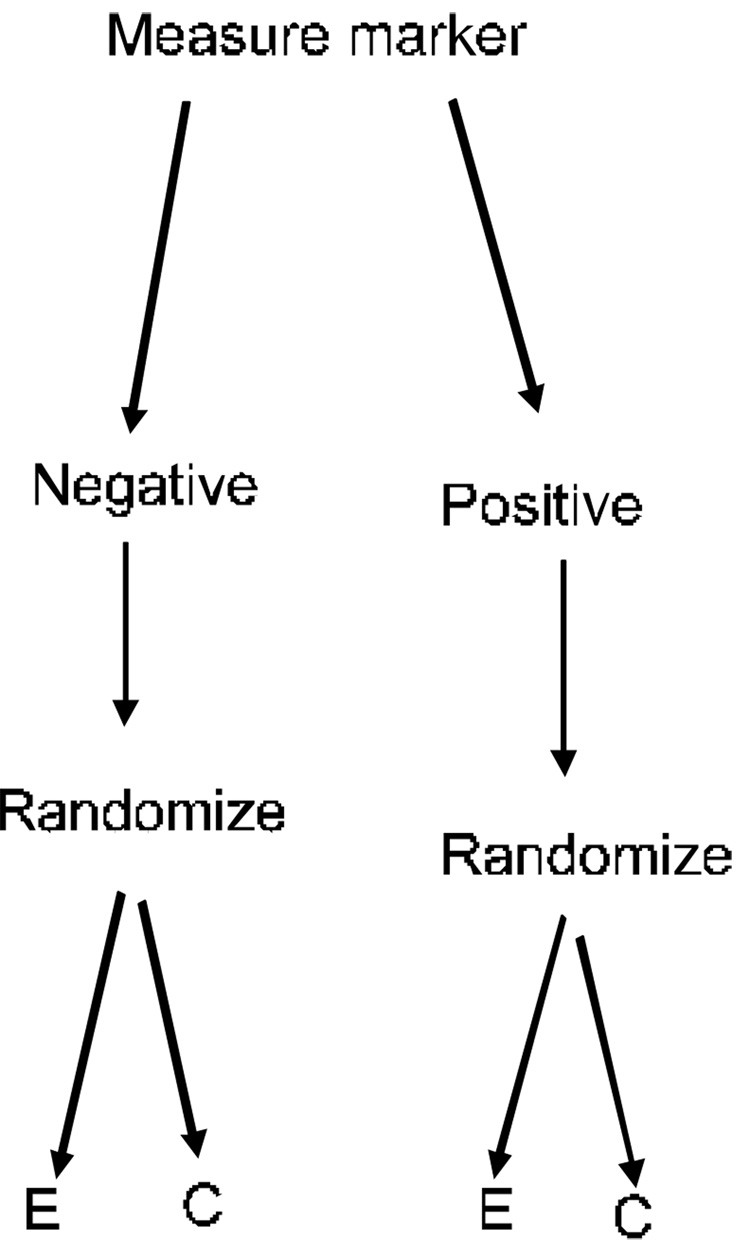
Figure 4 shows the Marker by Treatment Interaction Design discussed by Sargent et al.(Sargent, et al., 2005) and by Pusztai and Hess(Pusztai and Hess, 2004). Both marker positive and marker negative patients are randomized to the experimental treatment or control. The analysis plan either calls for separate evaluation of the treatment difference in the two marker strata or for testing the hypothesis that the treatment effect is the same in both marker strata. When this design is used for development of an experimental drug, an appropriate analysis plan might be to utilize a preliminary test of interaction; if the interaction is not significant at a pre-specified level, then the experimental treatment is compared to the control overall. If the interaction is significant, then the treatment is compared to the control within the two strata determined by the marker. The sample size planning for such a trial and determination of the appropriate significance level for the preliminary interaction test require further study.
Figure 4.
Stratified analysis design for evaluating a new experimental treatment (E) relative to a control (C). The status of a biomarker based classifier of the likelihood of responding to E is utilized in a prospectively specified analysis plan. The biomarker classifier is not just used for stratifying the randomization. Alternative analysis plans are described in the text.
Freidlin and Simon (Freidlin and Simon, 2005) proposed an alternative analysis plan for the design of Figure 4. They suggested that the overall null hypothesis for all randomized patients is tested at the 0.04 significance level. A portion, e.g. 0.01, of the usual 5 percent false positive rate is reserved for testing the new treatment in the subset predicted by the classifier to be responsive. The analysis starts with a test of the overall null hypothesis, without a preliminary test of interaction. If the overall null hypothesis is rejected, then one concludes that the treatment is effective for the randomized population as a whole and that the classifier is not needed. If the overall null hypothesis is not rejected at the 0.04 level, then a single subset analysis is conducted; comparing the experimental treatment to the control in the subset of patients predicted by the classifier as being most likely to be responsive to the new treatment. If the null hypothesis is rejected, then the treatment is considered effective for the classifier determined subset. This analysis strategy provides sponsors an incentive for developing genomic classifiers for targeting therapy in a manner that does not unduly deprive them of the possibility of broad labeling indications when justified by the data. Although this analysis strategy does not ensure that the statistical significance of an overall treatment effect is not driven by treatment benefit for the classifier positive subset, the design provides data for evaluating treatment effect in classifier negative patients.
6. Conclusions
Physicians need improved tools for selecting treatments for individual patients. The genomic technologies available today are sufficient to develop such tools. There is not broad understanding of the steps needed to translate research findings of correlations between gene expression and prognosis into robust diagnostics validated to be of clinical utility. This paper has attempted to identify some of the major steps needed for such translation.
Acknowledgements
Thanks to Dr. Wenu Jiang for the computing of Table 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biology. 2004;2:511–522. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC, Hortobagyi GN. Individualized care of patients with cancer: A work in progress. New England Journal of Medicine. 2004;351:2865–2867. doi: 10.1056/NEJMe048300. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A, Bruhn L, Friedman N, et al. Tissue classification with gene expression profiles. Journal of Computational Biology. 2000;7:536–540. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- Bo TH, Jonassen I. New feature subset selection procedures for classification of expression profiles. Genome Biology. 2002;3:0017.1–0017.11. doi: 10.1186/gb-2002-3-4-research0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Belmont, CA: Wadsworth International Group; 1984. [Google Scholar]
- Deutsch JM. Evolutionary algorithms for finding optimal gene sets in microarray prediction. Bioinformatics. 2003;19:45–54. doi: 10.1093/bioinformatics/19.1.45. [DOI] [PubMed] [Google Scholar]
- Dobbin K, Simon R. Sample size determination in microarray experiments for class comparison and prognostic classification. Biostatistics. 2005;6:27–38. doi: 10.1093/biostatistics/kxh015. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for classification of tumors using gene expression data. Journal of American Statistical Association. 2002;97:77–87. [Google Scholar]
- Dudoit S, Fridlyand J. In: Statistical analysis of gene expression microarray data. Speed T, editor. New York: Chapman & Hall/CRC; 2003. pp. 93–158. [Google Scholar]
- FDA. Food and Drug Administration. Rockville MD: U.S. Department of Health and Human Services; 2005. Guidance for Industry: Pharmacogenomic Data Submissions. [Google Scholar]
- Freidlin B, Simon R. Adaptive signature design: An adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clinical Cancer Research. 2005;11:7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- Fu WJ, Dougherty ER, Mallick B, Carroll RJ. How many samples are needed to build a classifier: a general sequential approach. Bioinformatics. 2005;21:63–70. doi: 10.1093/bioinformatics/bth461. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Dowing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression modeling. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Hastie R, Tibshirani R, Eisen M, et al. Gene shaving as a method for identifying distinct sets of genes with similar expression patterns. Genome Biology. 2000;1:1–0003.21. doi: 10.1186/gb-2000-1-2-research0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Janne PA. Selecting patients for epidermal growth factor inhibitor treatment: A FISH story or a tale of mutations? Journal of Clinical Oncology. 2005;23:6813–6816. doi: 10.1200/JCO.2005.97.008. [DOI] [PubMed] [Google Scholar]
- Khan J, Wei JS, ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nature Medicine. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Dougherty ER, Barrera J, Chen Y, Bittner ML, Trent JM. Strong feature sets from small samples. Journal of Computational Biology. 2002;9:127–146. doi: 10.1089/10665270252833226. [DOI] [PubMed] [Google Scholar]
- Maitournam A, Simon R. On the efficiency of targeted clinical trials. Statistics in Medicine. 2005;24:329–339. doi: 10.1002/sim.1975. [DOI] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. The Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: A comparison of resampling methods. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Tamayo P, Rogers S, Rifkin R, Engle A, Campbell C, Golub TR, Mesirov JP. Estimating dataset size requirements for classifying DNA microarray data. Journal of Comutational Biology. 2003;10:119–142. doi: 10.1089/106652703321825928. [DOI] [PubMed] [Google Scholar]
- Nguyen DV, Rocke DM. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics. 2002;18:39–50. doi: 10.1093/bioinformatics/18.1.39. [DOI] [PubMed] [Google Scholar]
- Ooi CH, Tan P. Genetic algorithms applied to multi-class prediction for the analysis of gene expression data. Bioinformatics. 2003;19:37–44. doi: 10.1093/bioinformatics/19.1.37. [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner fL, Walker MG, Watson D, Park T, Hiller W, fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New England Journal of Medicine. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Pusztai L, Hess KR. Clinical trial design for microarray predictive marker discovery and assessment. Annals of Oncology. 2004;15:1731–1737. doi: 10.1093/annonc/mdh466. [DOI] [PubMed] [Google Scholar]
- Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. Journal of Computational Biology. 2002;9:505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Load M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proceedings of the National Academy of Science U.S.A. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. New England Journal of Medicine. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Conley BA, Allegra C, Collette L. Clinical trial designs for predictive marker validation in cancer treatment trials. Journal of Clinical Oncology. 2005;23:2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- Simon R, Altman DG. Statistical aspects of prognostic factor studies in oncology. British Journal of Cancer. 1994;69:979–985. doi: 10.1038/bjc.1994.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the analysis of DNA microarray data: Class prediction methods. Journal of the National Cancer Institute. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clinical Cancer Research. 2004;10:6759–6763. doi: 10.1158/1078-0432.CCR-04-0496. [DOI] [PubMed] [Google Scholar]
- Simon R, Lam AP. Bethesda (MD): National Cancer Institute; Biometric Research Branch Technical Report 28. 2005 http://linus.nci.nih.gov/brb.
- Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and analysis of DNA microarray investigations. New York: Springer; 2003. [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proceedings of the National Academy of Science U.S.A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2005 doi: 10.1186/1471-2105-7-91. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H. Predicting the clinical status of human breast cancer by using gene expression profiles. Proceedings of the National Academy of Science. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]