Abstract
Activation of the cascade of proteolytic caspases has been identified as the final common pathway of apoptosis in diverse biological systems. We have isolated a gene, termed MRIT, that possesses overall sequence homology to FLICE (MACH), a large prodomain caspase that links the aggregated complex of the death domain receptors of the tumor necrosis factor receptor family to downstream caspases. However, unlike FLICE, the C-terminal domain of MRIT lacks the caspase catalytic consensus sequence QAC(R/Q)G. Nonetheless MRIT activates caspase-dependent death. Using yeast two-hybrid assays, we demonstrate that MRIT associates with caspases possessing large and small prodomains (FLICE, and CPP32/YAMA), as well as with the adaptor molecule FADD. In addition, MRIT simultaneously and independently interacts with BclXL and FLICE in mammalian cells. Thus, MRIT is a mammalian protein that interacts simultaneously with both caspases and a Bcl-2 family member.
Programmed cell death is essential for normal development and maintenance of homeostasis in multicellular organisms. Molecular and genetic characterization of programmed cell death has led to the increasing appreciation of the role of interleukin converting enzyme (ICE) family of death promoting cysteine-proteases (caspases) (1, 2). ICE was first implicated in apoptosis on the basis of its homology to the Caenorhabditis elegans death gene ced-3 (3). Several new members of this family possess a common feature: a C-terminal ICE/CED3 homology domain with a cysteine residue at the active site (1). This active site cysteine is present in a QAC(R/Q)G motif, which is conserved among different members of this family and is involved in substrate binding and catalysis (2). A second common characteristic of this family is their synthesis as an inactive precursor molecule of ≈30–50 kDa, which is subsequently processed to form an active heteromeric enzyme complex with 20 kDa and 10 kDa polypeptides (p20/p10)2 (2, 4). In addition, members of this family possess a small or large N-terminal prodomain. In the case of FLICE (for FADD-like ICE)/MACH (MORT1-associated CED-3 homolog) duplication of a highly conserved motif called death-effector domain (DED) is present (1, 2). FLICE–DEDs interact with a similar domain present at the N terminus of the adaptor molecule FADD (for Fas-associated protein with death domain). FADD also possesses another conserved motif called a death domain (DD) at its C terminus. This domain is responsible for interaction with DD-containing receptors of tumor necrosis factor receptor (TNFR) family either directly or via another DD-containing adaptor molecule TRADD (for TNFR1-associated DD) (10, 18, 19).
In addition to the caspase family, the Bcl-2 family of proteins represents another class of molecules that have been extensively studied for their death-promoting as well as death-inhibiting properties. Bcl-2 is capable of inhibiting apoptotic cell death in diverse biological systems and is related to C. elegans death-antagonist ced-9 (5). Due to its ability to block apoptosis, Bcl-2 has been implicated in the pathogenesis of several malignancies, especially low-grade lymphomas (6).
Although, the caspase and Bcl-2 families of proteins have received much attention, the relationship of these families and mechanisms of interaction at a molecular level was not clear. Recently, C. elegans CED-4 protein was shown to interact with the CED-3 and CED-9 proteins (7–9). In mammalian cells, it was also demonstrated that CED-4 could independently interact with BclXL, an anti-apoptotic member of the Bcl-2 family as well as with ICE and FLICE, two large prodomain caspases (8). Although CED-4 was required as a biochemical link between CED-3 and CED-9, BclXL and FLICE could interact in mammalian cells independent of the presence of CED-4 (8). However, the biochemical linkage between BclXL and FLICE could be attenuated by the presence of a C-terminal deletion mutant of CED-4 that binds CED-3 but not CED-9 (8). This led to the suggestion that interaction between BclXL and FLICE was mediated by an endogenous protein (factor X) and that mutant CED-4 was blocking the recruitment of factor X and thus BclXL to FLICE. The identity of factor X or any other mammalian homolog of CED-4 is not known.
Here, we describe a novel protein, MRIT (MACH-related inducer of toxicity; mrit also means “death” in sanskrit), that simultaneously and independently interacts with BclXL and FLICE. Thus, MRIT may function as a crucial link between cell survival and cell death pathways in mammalian cells.
METHODS
Northern Blot Analysis.
Adult poly(A)+ multiple-tissue Northern blot analysis (CLONTECH) was hybridized with 32P-labeled, full-length MRITα1 probe according to the manufacture’s instruction.
Expression Constructs.
Mammalian expression constructs were made in pCDNA3, pCDNA3-HisA, or B vectors (Invitrogen). AU1–FLICE, AU1–DED–FLICE, and FLAG–BclXL were epitope tagged at the COOH termini. All the MRIT constructs were His-epitope-tagged at the N termini. AU1 and FLAG epitopes were generated by PCR using custom-made primers and confirmed by automated sequencing. Yeast expression plasmids, GAL4 DNA-binding domain vector pAS2, and transactivation vector pGAD 424 were used to in-frame clone MRITα1 and deletion mutants, protease constructs, FADD, BclXL, and BclXS for two-hybrid association studies.
In Vitro Binding Assay.
MRITα1 and -β1 encoding His-tagged proteins were cloned into prokaryotic expression vector pET 28a+ (Novagen). Tagged proteins were expressed, purified, and immobilized to Ni2+ beads according to the manufacturer’s instructions. In vitro translated, 35S-labeled proteins were generated by using TNT-T7 or -T3 reticulocyte lysate systems (Promega). Indicated amounts of labeled proteins (input) were incubated with the beads, washed extensively, and analyzed by SDS/PAGE followed by autoradiography.
Transfection, Coimmunoprecipitation, and Western Blot Analysis.
BHK-CrmA cells were transiently transfected with indicated plasmids using Lipofectamine (GIBCO/BRL), lysed in 1 ml lysis buffer (50 mM Tris, pH 7.6/150 mM NaCl/0.1% Nonidet P40) and incubated with His beads, AU1 beads, or FLAG beads for at least 2 hr. After extensive washing, the beads were eluted in 0.1 M glycine (pH 3.0) for 30 min at room temperature. The eluted samples were then neutralized with 1 M Tris⋅HCl (pH 8.0), boiled in sample buffer, and subsequently analyzed by Western blot analysis.
RESULTS
MRIT Is a Novel DED-Containing Protein.
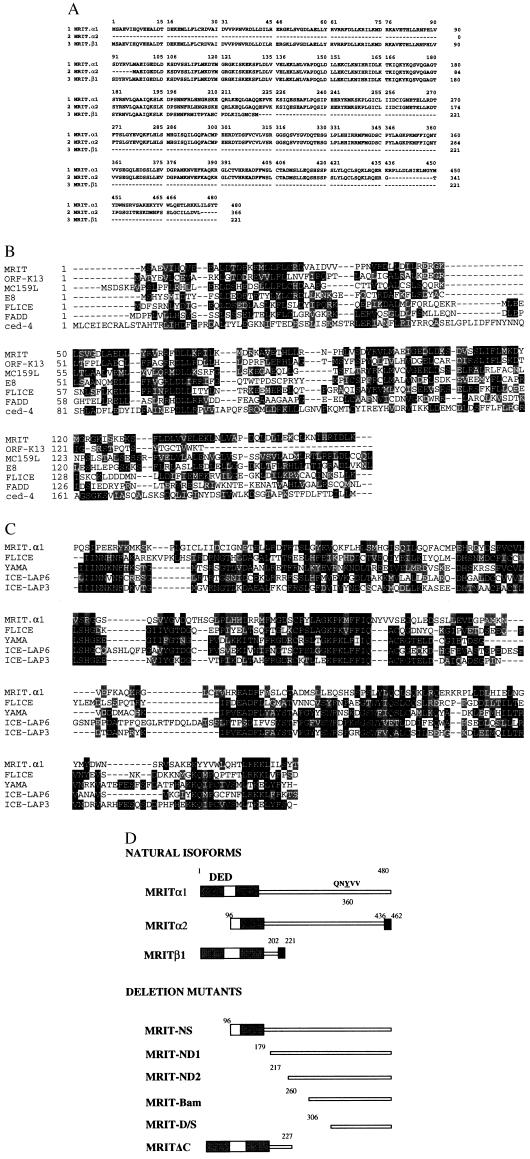
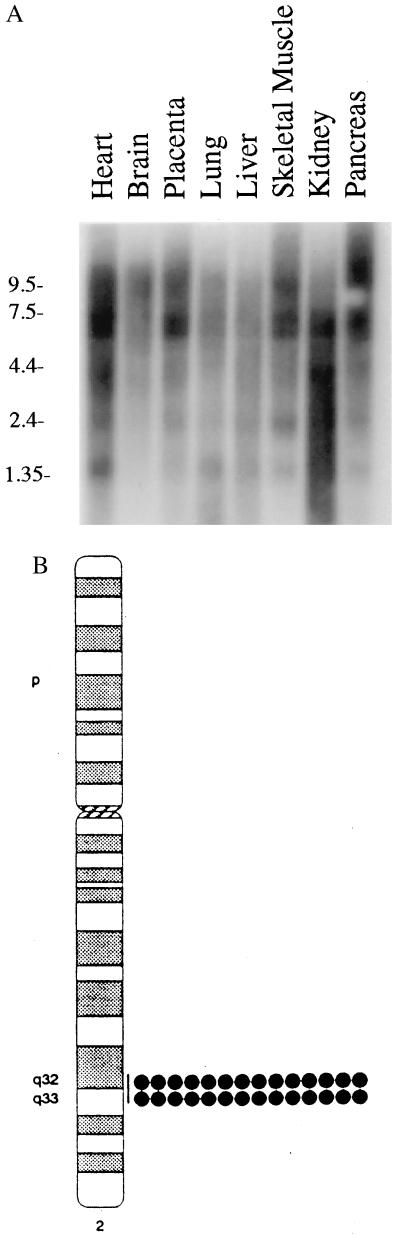
A sequence (IMAGE Consortium Clone ID 427786) in the National Center for Biotechnology Information GenBank expressed-sequence tag (EST) database was identified as having statistically significant homology to the p20 domain of human ICE-like protease MACH1/FLICE. Several additional EST clones (IMAGE Consortium Clone IDs 338776, 29827, 511600, 119142, 26915, 48465, and GenBank accession no. T30922) were sequenced. Three EST clones (29827, T30922, and 338776) were found to encode full-length ORFs corresponding to three isoforms of a novel gene and were termed MRITα1, -α2, and -β1, respectively (Fig 1A). A blast search revealed that MRIT was a new molecule with two distinct domains: residues 1–165 possessed two DEDs with high homology to known DED-containing proteins including FLICE, FADD, and recently described viral proteins (P < 0.001) (10–14) (Fig. 1B). Weak homology was also detected with the N-terminal region of CED-4, which was recently shown to align with known DED-containing proteins (23). Residues 301–480 also had statistically significant homology with known caspases including FLICE, YAMA/CPP32, ICE–LAP6, and ICE–LAP3 (P < 0.001) (1, 10, 15–17) (Fig. 1C). Although MRITα1 resembled caspases in the COOH-terminal region, it differed from them in one important respect. Instead of having the conserved motif QAC(R/Q)G at the catalytic site, MRIT had the sequence QNYVV. Thus, MRITα1 is not a cysteine protease. Northern blot analysis using the full-length MRITα1 showed multiple transcripts; 6-, 2.5-, and 1.4-kb transcripts were prominent in multiple human poly(A)+ RNA samples (Fig. 2A). Fluorescent in situ hybridization localized MRIT to 2q32–33 region of human metaphase chromosomes (Fig. 2B).
Figure 1.
(On the opposite page.) Sequence characterization of MRIT isoforms. (A) Deduced amino acid sequence of naturally occurring MRIT isoforms. (B) Alignment of MRIT DEDs with known DED-containing proteins. Similar to FLICE, MRIT also has two DEDs (10, 18). Black residues indicate sequence identity and conserved residues are indicated by shaded residues. (C) Alignment of ICE-homology domain of MRIT with FLICE/MACH1 (residues 237–479), YAMA/CPP32 (residues 39–277), ICE-LAP6 (residues 163–416), and ICE-LAP3 (residues 70–303). (D) Diagrammatic representation of MRIT isoforms and deletion mutants. Black boxes in the C termini of MRITα2 and MRITβ1 represent unique 26 and 19 residues, respectively. Gray boxes represent the DED-1 and DED-2.
Figure 2.
(A) Northern blot analysis of poly(A)+ RNA using a full-length cDNA probe reveals that multiple transcripts of MRIT are expressed in human adult tissues. (B) A 1.2-kb EST clone (IMAGE consortium clone 26915) was used to hybridized (at 50 ng/μl) to metaphase cells prepared from phytohemagglutinin-stimulated, BrdU-pulsed, normal male lymphocytes. Fluorescent signals (indicated by dots) were mapped to each of 16 metaphases (signals were seen at 2q32–33 on 28 of 32 homologs). The line indicates the range of locations observed.
MRIT Associates with Large- and Small-Prodomain Caspases and FADD.
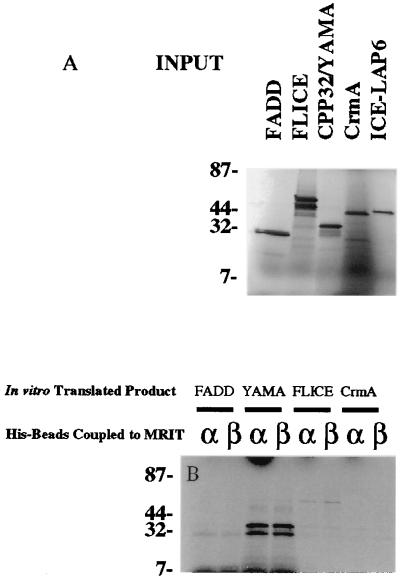
As MRITα1 possessed significant sequence homology to caspases, we tested the possibility that it could interact with them via homophilic interactions. His-tagged recombinant MRITα1 and -β1 associated with in vitro translated FLICE, FADD, and YAMA/CPP32, whereas CrmA did not associate with MRITα1 (Fig. 3).
Figure 3.
MRIT binds to large and small prodomain caspases (FLICE, CPP32/YAMA) in vitro. Recombinant His-tagged MRITα1 or -β1 immobilized on Ni2+ beads were incubated with in vitro translated 35S-labeled FLICE, CPP32/YAMA, or with CrmA. After extensive washing, the bound-labeled proteins were separated by SDS/PAGE and analyzed by autoradiography. The amount of corresponding input radiolabeled protein is shown A. CrmA serves as a negative control for nonspecific binding.
Yeast two-hybrid domain-mapping analysis was used to study the association of MRITα1 and deletion mutants with distinct domains of FLICE. MRITα1 associated with FLICE–DED (prodomain, residues 1–216) and FLICE p20 domain (residues 217–374), but not with FLICE p10 domain (residues 375–479) (Table 1). Although strong interaction of MRIT α1 to FLICE-DED was observed, N-terminal deletion mutants ND1 and ND2 showed weaker interactions, and no interaction was seen with MRIT-D/S (Table 1). Similar results were also found with the full-length FADD construct.
Yeast two-hybrid interaction assay, using FLICE p20 domain with MRITα1 and its deletion mutants, was used to delineate the p20 binding region in MRITα1. FLICE p20 domain interacted with MRITα1, MRIT NS, ND1, and ND2, but not with MRIT D/S (N-terminal 305 residues deleted) or MRITΔC (C-terminal 253 residues deleted) (Table 1). These results suggest that FLICE p20 binding to MRITα1 did not require N-terminal 216 residues. Interestingly, N-terminal deletion constructs MRIT ND1 and ND2 interacted with the FLICE p10 domain, suggesting that the p10 interacting domain was masked by the N-terminal region (Table 1).
Unlike FLICE, YAMA possesses a small prodomain (24 residues). Therefore, two YAMA p20 constructs, one with and one without the prodomain, and a YAMA p10 construct were tested against MRITα1 constructs in the yeast two-hybrid interaction assay. Similar to FLICE p20, YAMA p20 interacted with MRITα1 and MRIT ND1, and ND2, whereas YAMA p10 interacted with MRIT ND1 and ND2 only (Table 1). Thus, MRITα1 possesses multiple interacting domains for FLICE and YAMA (Table 1).
MRITα1 Independently and Simultaneously Associates with FLICE and BclXL in Mammalian Cells.
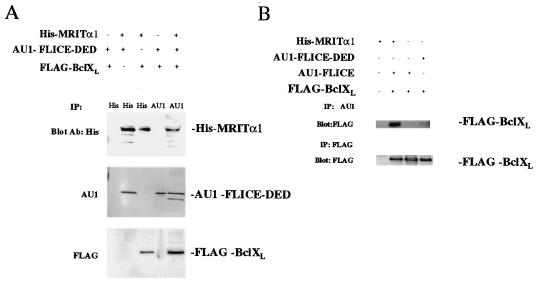
To test the hypothesis that MRIT could biochemically link caspases and Bcl-2 family members in mammalian cells, we cotransfected BHK-CrmA cells with HIS–MRITα1 and FLAG–BclXL or AU1–FLICE–DED. Subsequent precipitation with His beads resulted in coprecipitation of HIS–MRITα1 with FLAG–BclXL or AU1–FLICE–DED (Fig. 4A). Thus, MRITα1 could independently interact with BclXL and FLICE–DED in mammalian cells.
Figure 4.
Direct association of MRIT with FLICE and BclXL in mammalian cells. MRITα1 can directly associates with FLICE–DED–AU1 (A) or FLICE–AU1 (B) and BclXL. BHK-CrmA cells were transiently transfected with indicated expression constructs. Thirty-six hours after transfection, extracts were prepared and immunoprecipitated (IP) with mAb to AU1-l coupled to Sepharose beads (Babco, Richmond, CA), mAb to FLAG coupled to agarose beads (Kodak), or with His beads (Invitrogen). Samples were then blotted with mAb to AU1, FLAG, or 6-His (Babco) and Antiexpress (Invitrogen).
We then examined whether MRITα1 could biochemically link and simultaneously interact with FLAG–BclXL and AU1–FLICE–DED. Unlike the previous report demonstrating strong interaction between FLICE and BclXL in 293T cells (8), nearly undetectable coprecipitation of FLAG–BclXL with AU1–FLICE–DED was seen in BHK cells in the absence of MRITα1 (Fig. 4A). However, triple-transfection with N-terminal epitope-tagged HIS–MRITα1, COOH-terminal epitope-tagged FLAG–BclXL, and AU1–FLICE–DED and subsequent immunoprecipitation with AU1 beads showed coprecipitation of MRITα1 and BclXL with FLICE–DED (Fig. 4A). Thus, MRITα1 could biochemically link and simultaneously interact with both BclXL and FLICE–DED.
The above experiment was repeated using COOH-terminal AU1 epitope-tagged full-length FLICE in place of AU1–FLICE–DED. As before, only a weak interaction between AU1–FLICE and FLAG–BclXL was seen in the absence of MRIT (Fig. 4B). However, triple-transfection with full-length AU1–FLICE, FLAG–BclXL, and HIS-MRITα1, and immunoprecipitation with AU1 beads showed strong coprecipitation of FLAG–BclXL. Thus, MRITα1 could biochemically link and simultaneously interact with full-length FLICE and BclXL.
MRIT DED Isoforms Induce Cell Death in Mammalian Cells.
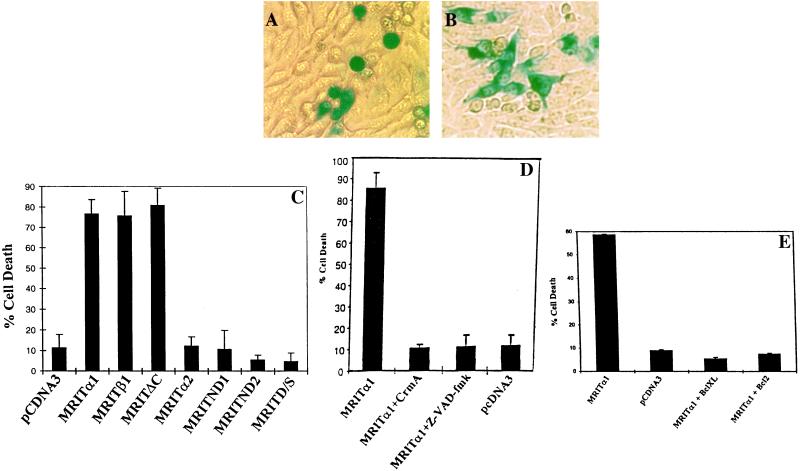
To test whether MRIT isoforms can induce apoptosis, we cotransfected BHK or HeLa cells with various MRIT expression constructs along with LacZ expression plasmid. Cells were subsequently fixed and stained with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal). Overexpression of MRITα1, -β1, and -ΔC isoforms in BHK and HeLa cells induced rapid apoptosis as measured by cellular and nuclear morphology (Fig. 5 A, C, and D). Induction of apoptosis was confirmed by nuclear staining with Hoechst 33342 (D.K.M.H., unpublished data), and by inhibition with z-VAD-fmk and CrmA, two inhibitors of caspases (Fig. 5D). However, MRITα2 or N-terminal DED deletion mutants, ND1, ND2, and D/S failed to induce cell death in these cells (Fig. 5 C and D) (D.K.M.H., unpublished data). Thus, MRIT mediates apoptosis via activation of ICE-like cysteine proteases. Next, we tested the ability of various members of Bcl-2 family to block MRIT induced cell death in MCF7 cell line. As shown in Fig. 5E, Bcl-2 and BclXL could effectively block MRIT induced apoptosis.
Figure 5.
MRIT induces cell death in BHK cells. (A) Cell death mediation by MRITα1 and protection by cotransfection with CrmA (B). (C) Quantitation of cell death induced by MRIT isoforms. BHK cells were cotransfected with indicated MRIT isoforms (1 μg) along with a β-galactosidase expression construct (0.3 μg) using Superfect (Qiagen, Chatsworth, CA). Positive cells showing β-galactosidase activity and apoptotic (round and condensed) morphology were scored after 36–48 hr. Representative results from at least five independent experiment is shown. (D) Overexpression of MRIT (0.5 μg) in BHK cells induces apoptosis that is blocked by CrmA (0.5 μg) and z-VAD-fmk (10 μM). (E) Cell death mediated by overexpression of MRITα1 (0.1 μg) in MCF7 cells is also blocked by Bcl-2 (0.5 μg) and BclXL (0.5 μg).
DISCUSSION
MRIT Resembles FLICE in DEDs.
MRIT is a new member of the caspase family with several unique properties. The molecule was identified based on its sequence homology to FLICE, a caspase believed to be a proximal protease that triggers activation of downstream death proteases. FLICE is recruited to the DD-containing receptors such as Fas, TNFR1 and DR3 via cytosolic adaptors such as FADD and TRADD (for TNFR1-associated DD). This recruitment is responsible for activation of FLICE and subsequent amplification of proteolytic cascade. MRIT possesses several structural and functional similarities with FLICE. Both these proteins have long prodomains consisting of two DEDs. Also, naturally occurring isoforms consisting only of such prodomains (β forms) have been identified for both proteins (18). The β1 isoform of FLICE (MACHβ1) was found to be significantly cytotoxic when expressed alone in HeLa cells (18) and we have seen similar results in BHK cells (P.M.C., unpublished observation). Like the β1 form of FLICE, the MRITβ1 isoform showed significant cytotoxicity when overexpressed in BHK cells. Both MRITβ1 and MACHβ1 interacted with FADD, an adaptor molecule with both a DED and a DD, which links the DD receptors TNFR1, Fas, and DR3 to caspases (10, 18, 19). It is likely that the MRITβ1 isoforms mediate its toxicity by first binding to FADD and subsequently binding to and activating endogenously expressed MRITα isoform, as has been proposed for the mechanism of cytotoxicity of the β1 isoform of FLICE (MACHβ1) (18). Alternatively, MRITβ1 may mediate its toxicity by directly binding and activating endogenous FLICE and/or other caspases.
MRIT Resembles FLICE in the Protease Domain.
In addition to its homology to FLICE in the prodomain, MRIT possesses significant sequence homology to FLICE (as well as other caspases) in the C-terminal protease domain. However, MRIT does not possess the consensus caspase catalytic motif QAC(R/Q)G and therefore is unlikely to act as a cysteine protease. Based on the ability of MRITα1 to interact with small and large prodomain caspases, it is likely that MRITα1 mediates its cytotoxicity by activating these caspases. Finally, the C-terminal isoforms of both MRIT and FLICE, lacking the complete prodomains (i.e., MRITα2 and MACHα3), are inactive as mediators of apoptosis (Fig. 5C) (18).
MRIT Has Unique Properties.
Despite its overall structural and functional homology to FLICE, MRIT has several distinct features which are analogous to C. elegans CED-4 protein. First, both MRIT and CED-4 possess weak sequence homology in the DEDs. Although MRIT can induce apoptosis when overexpressed in mammalian cells, whether CED-4 can do the same is a hot topic of debate in the field. There has been one published report of CED-4-mediated apoptosis in mammalian cells (8), and we have seen weak apoptosis induced by CED-4 overexpression in MCF7 cells. Second, both MRIT and CED-4 have the ability to bind to caspases through their prodomains and the protease domain (8). Although MRIT can bind to both short- and long-prodomain caspases, CED-4 can bind only to long-prodomain caspases (8). However, this difference is not surprising, as there are no known naturally occurring short-prodomain caspases in C. elegans. Also, caspase inhibitors z-VAD-fmk and CrmA (8) could inhibit apoptosis mediated by both of these proteins. Therefore, both these proteins are likely to mediate apoptosis indirectly by activating caspases. Third, both proteins have the ability to bind to members of Bcl-2 family (7–9). In addition, both proteins can simultaneously bind caspases and members of the Bcl-2 family (8). Such interactions have been posited based on genetic evidence in C. elegans that CED-4 can couple the caspase dependent death pathway, represented by CED-3, and the anti- or pro-death functions of the C. elegans homologs of Bcl-2, CED-9 (20, 21). Recently, studies in mammalian cells showed that CED-4 can link CED-3 and CED-9 when they are overexpressed and suggested the presence of a mammalian protein, factor X, which is able to perform a similar function for the mammalian homologs FLICE and BclXL (8). In the present study we have demonstrated that MRIT can also interact simultaneously with BclXL and FLICE. Finally, anti-apoptotic members of Bcl-2 family can block apoptosis mediated by both MRIT and CED-4 (8).
In summary, although the in vivo function of MRIT is not known, MRIT can simultaneously interact with a member of both the caspase family of death effectors and the Bcl-2 family of death-regulating molecules. Thus, MRIT is a mammalian protein that may play an important role in regulating cell death.
Table 1.
Interaction of MRIT isoforms and deletion mutants with FLICE, CPP32/YAMA, and FADD in yeast two-hybrid assay
| FLICE prodomain (1–216) | FLICE p20 (217–374) | FLICE p10 (375–479) | YAMA pro+p20 (1–175) | YAMA p20 (25–175) | YAMA p10 (176–277) | FADD (1–208) | |
|---|---|---|---|---|---|---|---|
| MRITα1 (1–480) | +++ | + | − | + | ++ | − | +++ |
| MRIT–NS (96–480) | ND | ++ | − | − | ++ | − | + |
| MRITND 1 (179–480) | + | ++ | ++ | ++ | ++ | + | + |
| MRITND 2 (217–480) | + | ++ | + | + | + | +++ | ND |
| MRIT D/S (306–480) | − | − | − | − | − | − | − |
| MRITΔC (1–227) | ND | − | − | − | − | + | +++ |
MRIT isoforms and deletion mutants cloned in-frame in the GAL4 DNA binding domain construct (pAS2) were cotransfected with plasmids encoding GAL4 transactivating domain (pGAD424) containing FLICE prodomain (residues 1–216), FLICE p20 (residues 217–374), FLICE p10 (residues 375–479), CPP32/YAMA prodomain and p20 (residues 1–175), YAMA p20 (residues 25–175), YAMA p10 (residues 176–277), and full-length FADD (residues 1–208). The results were assessed by two-hybrid, β-galactosidase filter assay (22). Strong association (+++) was defined by the appearance of blue color within 2 hr; ++, 12 hr; +, 24 hr; +/−, 36 hr; and −, no detectable association after 48 hr. Controls included empty vectors only, single plasmid controls for self-transactivation, negative controls of self association of MRIT isoforms, positive control for strong association (Fas cytosolic domain with Fas cytosolic domain), negative control (Fas cytosolic domain with death domain deleted Fas cytosolic domain).
Acknowledgments
We thank Vishva Dixit for his generous gift of CPP32/YAMA, ICE-LAP3, ICE-LAP6, and FLICE, and CrmA cDNAs in pCDNA3. We also thank Craig Thompson for BclXL and BclXS in pBluescript, Stan Korsemeyer for pSFFV–Bcl-2 and Bax, David Hockenbery and Jason O’Neil for purified His-tagged Bcl-2. This research was supported by the Stowers Institute for Medical Research, Cardiovascular Training Grant (D.K.M.H.), Damon Runyon–Walter Winchell Foundation Postdoctoral Fellowship (P.M.C), Howard Hughes Medical Institute predoctoral fellowship (M.E.W.), National Institutes of Health Grants GM37905 and RR00166 (L.E.H), Department of Energy Grant DE-FG06-93ER62173 (L.E.H), and National Institutes of Health Grant HL03174 (S.M.S).
ABBREVIATIONS
- ICE
interleukin converting enzyme
- FADD
Fas-associated protein with death domain
- FLICE
FADD-like ICE
- MACH
MORT1-associated CED-3 homolog
- DED
death-effector domain
- DD
death domain
- TNFR
tumor necrosis factor receptor
- MRIT
MACH-related inducer of toxicity
- EST
expressed-sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U85059).
References
- 1.Alnemri E S, Livingston D J, Nichloson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1997;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 2.Miller D K. Semin Immunol. 1997;9:35–49. doi: 10.1006/smim.1996.0058. [DOI] [PubMed] [Google Scholar]
- 3.Yuan J, Shaham S, Ledoux S, Ellis H, M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 4.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 5.Hengartner M O, Horvitz H R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 6.Vaux D, L, Strasser A. Proc Natl Acad Sci USA. 1996;93:2239–2243. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spector M A, Desnoyer S, Hoeppner D J, Hengartner M O. Nature (London) 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 8.Chinnaiyan A M, O’Rourke K, Lane B, Dixit V M. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 9.Wu D, Wallen H D, Nunez G. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 10.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 11.Chinnaiyan A, M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 12.Hu S, Vincenz C, Buller M, Dixit V M. J Biol Chem. 1997;272:9621–9628. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 13.Bertub J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thome M, Schneider P, Hofman K, Fickenscher H, Meinl E, Neipel F, Mattman C, Burns K, Bodmer J, Schroeter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Nature (London) 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 15.Tewari M, Quan L T, O’Rourke K, Desnoyer S, Zeng Z, Beidler D, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–808. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 16.Duan H, Hudson P L, Wing J P, He W W, Dixit V M. J Biol Chem. 1996;271:1621–1625. doi: 10.1074/jbc.271.3.1621. [DOI] [PubMed] [Google Scholar]
- 17.Duan H, Orth K, Chinnaiyan A M, Poirier G G, Froelich C J, He W W, Dixit V M. J Biol Chem. 1996;271:16720–16724. doi: 10.1074/jbc.271.28.16720. [DOI] [PubMed] [Google Scholar]
- 18.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 19.Chinnaiyan A M, O’Rourke K, Yu G, Lyons R H, Garg M, Duan H, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 20.Ellis H M, Horvitz H R. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 21.Shaham S, Horvitz H R. Cell. 1997;86:201–208. doi: 10.1016/s0092-8674(00)80092-6. [DOI] [PubMed] [Google Scholar]
- 22.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 23.Bauer M K A, Wesselborg S, Schulze-Osthoff K. FEBS Lett. 1997;402:256–258. doi: 10.1016/s0014-5793(96)01497-4. [DOI] [PubMed] [Google Scholar]