Abstract
We have identified and characterized CLARP, a caspase-like apoptosis-regulatory protein. Sequence analysis revealed that human CLARP contains two amino-terminal death effector domains fused to a carboxyl-terminal caspase-like domain. The structure and amino acid sequence of CLARP resemble those of caspase-8, caspase-10, and DCP2, a Drosophila melanogaster protein identified in this study. Unlike caspase-8, caspase-10, and DCP2, however, two important residues predicted to be involved in catalysis were lost in the caspase-like domain of CLARP. Analysis with fluorogenic substrates for caspase activity confirmed that CLARP is catalytically inactive. CLARP was found to interact with caspase-8 but not with FADD/MORT-1, an upstream death effector domain-containing protein of the Fas and tumor necrosis factor receptor 1 signaling pathway. Expression of CLARP induced apoptosis, which was blocked by the viral caspase inhibitor p35, dominant negative mutant caspase-8, and the synthetic caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-(OMe)-fluoromethylketone (zVAD-fmk). Moreover, CLARP augmented the killing ability of caspase-8 and FADD/MORT-1 in mammalian cells. The human clarp gene maps to 2q33. Thus, CLARP represents a regulator of the upstream caspase-8, which may play a role in apoptosis during tissue development and homeostasis.
Apoptosis, a morphologically distinguished form of programmed cell death, is critical not only during development and tissue homeostasis but also in the pathogenesis of a variety of diseases, including cancer, autoimmune disease, viral infection, and neurodegenerative disorders (1–2). Whereas numerous genes that control apoptosis have been identified and partially characterized, the precise mechanisms by which these genes interact to execute the apoptotic program remains poorly understood. In mammals, a family of cysteine proteases (designated caspases) related to the Caenorhabditis elegans gene ced-3 appears to represent the effector arm of the apoptotic program (3–4). The first member of the family identified was interleukin 1β-converting enzyme (5). To date, more than 10 caspases have been identified and partially characterized (3). Each caspase contains characteristic conserved sequences important for proteolytic activity and induction of apoptosis (3–4). Caspase activation is induced by a wide array of death signals and leads to precipitous cleavage of protein substrates and execution of the apoptotic program (1, 2).
Engagement of the surface receptors, Fas/APO-1/CD95 and tumor necrosis factor receptor 1 (TNFR-1), leads to the activation of several caspases and apoptosis (6). Both CD95 and TNFR-1 use the adapter protein FADD/MORT-1 to link cytoplasmic receptor sequences to the upstream FLICE/MACH/Mch5 protease (caspase-8) and Mch4 (caspase-10) (7–14). Both FADD/MORT-1 and caspase-8 interact through conserved death effector domains (DED) located in the N-terminal region of FADD/MORT-1 and pro-domain of caspase-8 (7, 8, 11–13). Upon binding of the corresponding ligands, the CD95 receptor and TNFR-1 recruit caspase-8 to the receptor signaling complex through FADD/MORT-1, leading ultimately to activation of a cascade of caspases and rapid demise of the cell (14). Thus, caspase-8 appears to function as an apical caspase that links upstream death components with downstream caspases (14). The DED domain is not unique to FADD/MORT-1 and caspase-8 in that a group of viruses such as equine herpesvirus type 2 and molluscum contagiosum virus type 1 encode DED-containing viral proteins that function as inhibitors of apoptosis (15–17). Thus, DEDs function as protein-interacting modules that link components and regulatory molecules of death signaling pathways.
Little is known about the regulation of caspases. We reasoned that upstream caspases such as caspase-8 could be targets for the regulation of death signaling pathways. To identify cellular genes that might regulate caspase activity and apoptosis, we searched public databases for expressed sequence tags (ESTs) that encode proteins with homology to caspase-8. We identified a cDNA that encodes a protein designated CLARP (caspase-like apoptosis-regulatory protein) with significant structural and amino acid homology to caspase-8. CLARP interacts with caspase-8 but not with FADD/MORT-1, an adapter molecule of the CD95, TNFR-1, and DR3 death pathways. Unlike caspase-8, CLARP is enzymatically inactive due to the loss of highly conserved residues that are required for proteolytic activity. Remarkably, expression of CLARP induces apoptosis and augments the killing activities of caspase-8 and FADD/MORT-1. These results indicate the CLARP is a regulator of apoptosis that might function in the CD-95, TNFR-1, and DR3 death signaling pathways.
MATERIALS AND METHODS
Isolation of CLARP and Construction of Expression Plasmids.
The partial nucleotide sequences of cDNAs homologous to human caspase-8 were found in EST and sequence tagged sites (STS) databases of GenBank using the tblastn program. The entire nucleotide sequence of a cDNA containing a 2.0-kb insert corresponding to EST clone 511600 was determined by dideoxy sequencing. The entire ORF of CLARP was tagged at the carboxyl terminus with FLAG sequences and cloned into the expression vector pcDNA3 (Invitrogen) to produce pcDNA3-CLARP-Flag. The human caspase-8 (13) and FADD (EST clone 526925) amino acid sequences each were fused at the carboxyl terminus with AU1 tag sequences and cloned into pcDNA3 to produce pcDNA3-caspase-8-AU1 and pcDNA3-FADD-AU1, respectively. A mutation of caspase-8 (Cys377Ser) was introduced by site-directed PCR methodology (18) and cloned into pcDNA3 to produce caspase-8-mut-AU1. The authenticity of all constructs was confirmed by dideoxy sequencing. DCP2, a Drosophila melanogaster gene homologous to caspase-8, caspase-10, and CLARP was identified by searching the invertebrate GenBank database for caspase-8 homologues using tblastn. The positions of splice junctions in DCP2 gene were calculated according to Brunak et al. (19).
Transfection, Expression, Immunoprecipitation, and Immunodetection of Tagged Proteins.
Human 293T cells (5 × 106) were transfected with expression plasmids by calcium phosphate methods as described previously (20). Briefly, 3 μg of pcDNA3 or pcDNA3-CLARP-Flag was cotransfected with 3 μg of pcDNA3, pcDNA3-caspase-8-AU1, or pcDNA3-FADD-AU1. The total amount of transfected plasmid DNA was always 6 μg. After transfection, 293T cells were harvested at different times and lysed with 0.2% Nonidet P-40 isotonic lysis buffer (21). For immunoprecipitation, 1 mg of soluble protein was incubated with 10 μg/ml of polyclonal anti-Flag, monoclonal anti-AU1, or control antibody overnight at 4°C, and tagged proteins were immunoprecipitated with protein A-Sepharose 4B (Zymed). Immunoprecipitates were subjected to 12% SDS/polyacrylamide electrophoresis and immunoblotted with anti-Flag or anti-AU1 antibodies.
β-Galactosidase Apoptosis Assay and Nuclear Staining of Apoptotic Cells.
293T cells (5 × 105) were cotransfected with 0.3 μg of pcDNA3-β-galactosidase plus 1 μg of each expression plasmid in triplicate by the calcium phosphate method. MCF-7 human breast cancer cells (5 × 105) were transfected with 0.2 μg of pcDNA3-β-galactosidase plus 0 to 1 μg of each expression plasmid with Lipofectamine (GIBCO/BRL). The total amount of transfected plasmid DNA was always 1.7 μg. In some experiments, 20 μM of the caspase inhibitor zVAD-fmk [benzyloxycarbonl-Val-Ala-Asp-(OMe)-fluoromethyl-ketone] (Enzyme Systems Products, Livermore, CA) was added 6 hr after transfection. At 16 and 24 hr after transfection, cells were fixed, stained for β-galactosidase as described (22), and assayed for morphological features of apoptosis. At least 300 blue-staining cells were counted. Staining of nuclei with acridine orange and ethidium bromide was performed as described previously (20). Statistical significance was determined by one-way ANOVA followed by Student-Neuman-Keuls post-hoc comparisons.
Caspase Assay with Fluorogenic Substrates.
293T cells (5 × 106) were transfected with 3 μg of pcDNA3, pcDNA3-CLARP-Flag, or pcDNA3-Ced-3-Flag (23). At 16 hr after transfection, cells were harvested and lysed with Nonidet P-40 buffer lacking KCl. Flag-tagged proteins were immunoprecipitated with anti-Flag mAb as described, and immune complexes were incubated with the same buffer containing 100 μM of acetyl-Asp-Glu-Val-Asp-AMC (DEVD-AMC) or acetyl-Tyr-Val-Ala-Asp-AMC (Alexis, San Diego, CA) for 1 and 10 hr. Caspase activity was measured with excitation at 360 ± 40 nm and emission at 460 ± 40 nm with a fluorescence spectrophotometer Cytofluor 2300 (Millipore).
Northern Blot Analysis.
A BamHI fragment (probe A, nucleotides 1–870) or BamHI-EcoRI (probe B, nucleotides 870–1,435) of the CLARP cDNA were radiolabeled by random priming using a commercial kit (Boeringer Mannheim) and applied for analysis of human poly(A)+ RNA blots from various lymphoid tissues (CLONTECH) according to the manufacturer’s instructions.
RESULTS
Identification of CLARP, a Protein Homologous to Caspase-8 But Lacking Two Essential Catalytic Residues.
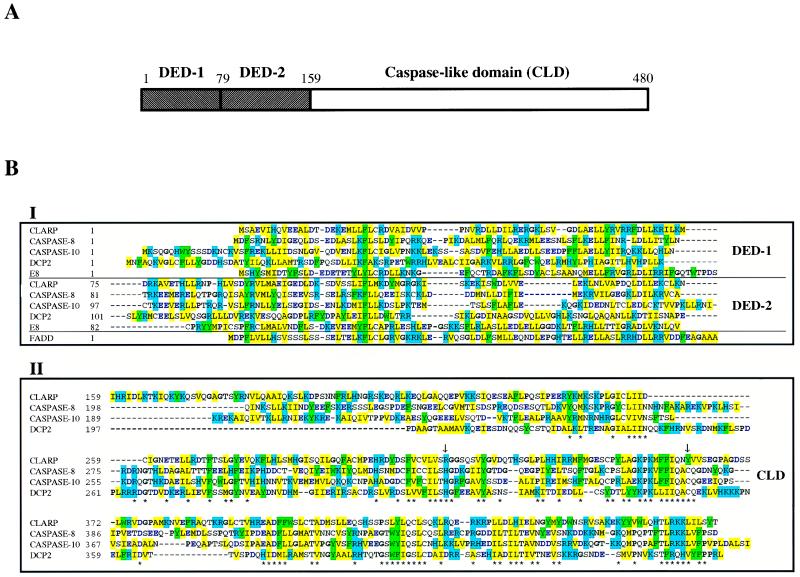
To identify novel apoptosis-regulatory proteins, we screened the GenBank database for cDNAs encoding proteins with homology to caspase-8 sequences by computer homology search. Several ESTs containing overlapping nucleotide sequences with statistically significant amino acid homology to caspase-8 were identified (P = 1.2 × 10−27). The longest cDNA (EST clone 511600) was 2.0 kb, and its nucleotide sequence revealed an ORF that encoded a protein of 480 amino acids with a predicted relative molecular mass of 55,377 (Fig. 1). We designated this protein as CLARP (caspase-like apoptosis-regulatory protein). A blast search revealed that CLARP was a novel molecule with two N-terminal DEDs (residues 1–158) fused to a C-terminal caspase-like domain (residues 159–480). CLARP showed 22% amino acid identity and 51% and 50% similarity with caspase-8 and caspase-10, respectively. In addition, CLARP showed structural and amino acid homology with viral DED-containing apoptosis inhibitory proteins such as E8 (Fig. 1B) and MC159L (15–17). Unlike caspase-8, however, two important amino residues located in the caspase catalytic domain that are predicted to be required for proteolysis were lost in CLARP. To date, all reported caspases have a conserved Cys residue in the catalytic site and a closely positioned His residue that are involved in catalysis (24). Significantly, Arg-315 and Tyr-360 were observed at the corresponding positions of CLARP, respectively (see arrows in Fig. 1B). Replacement of these residues in CLARP was not a cloning artifact or mutation, because three ESTs containing Arg-315 and two containing Tyr-360 were identified in independent cDNA libraries. To elucidate which of these two residues are conserved in other species, we searched the invertebrate GenBank database for sequences homologous to caspase-8. We identified a D. melanogaster gene homologous to caspase-8, designated here as DCP2. Like CLARP, caspase-8, and caspase-10, DCP2 contains two N-terminal DEDs fused to a caspase-like domain (Fig. 1B). Significantly, both Cys and His residues also were conserved in the caspase domain of the Drosophila caspase-8 homologue DCP2 (Fig. 1B). Because both human and D. melanogaster DED-containing caspases contained the critical Cys and His residues in their caspase catalytic domains, it is predicted that human CLARP is catalytically inactive (see below).
Figure 1.
Structure, sequence, and alignment of CLARP with related proteins. (A) Schematic structure of human CLARP. DEDs and caspase-like domain (CLD) are shown as hatched and empty boxes. (B) The amino acid sequence of CLARP is aligned with those of human caspase-8 (accession no. 1401352), human caspase-10 (1498324), DCP-2 (U20542), E8 protein of equine herpesvirus-2 (1360817), and DED of FADD (1082361). DEDs and CLD are shown in box I and box II, respectively. Homologous amino acid residues are highlighted with same colors; black bold for P and G (αβ breaker), blue bold for D and E (acidic residues), blue background for R, K, and H (positive charged residues), green background for W, F, and Y (aromatic residues), and yellow background for I, L, V, A, and M (hydrophobic residues). The two essential catalytic residues are indicated by arrows. Conserved residues among more than three proteins are indicated by stars for box II. The entire nucleotide sequence of CLARP cDNA is available as accession no. AF005774 in GenBank.
CLARP Is Expressed in a Wide Variety of Tissues.
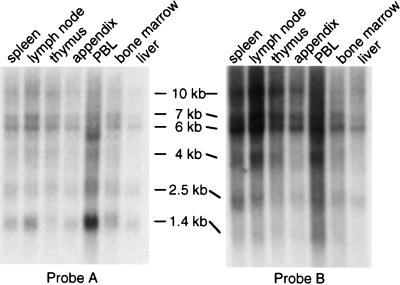
We performed Northern blot analysis to assess the expression of clarp mRNA in various human tissues. Hybridization with two CLARP probes corresponding to the 5′ (probe A) or 3′ (probe B) end of the CLARP cDNA showed several transcripts of 10 kb, 7 kb, 6 kb, 4 kb, 2.5 kb, and 1.4 kb in various human tissues (Fig. 2). The significance and identity of these different mRNA species remains to be determined. They could represent several mRNA forms of CLARP derived by alternative splicing and/or usage of alternative polyadenylation sites. blast search also showed ESTs encoding CLARP from both fetal and adult tissues, including brain, lung, colon, spleen, liver, and pancreas, indicating that CLARP mRNA is expressed in multiple human tissues.
Figure 2.
Expression of CLARP in human tissues by Northern blot analysis. Poly(A)+ RNAs from various immune tissues were hybridized with probe A (nucleotides 1–870) or probe B (nucleotides 870–1,435) of the CLARP cDNA.
CLARP Is Devoid of Intrinsic Caspase Activity When Expressed in Mammalian Cells.
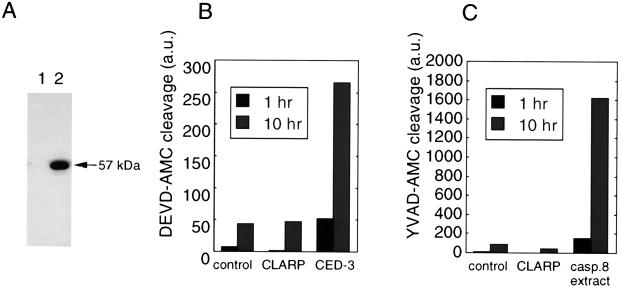
To examine if CLARP has caspase activity, 293T cells were transiently transfected with an expression plasmid producing Flag-tagged CLARP. The tagged CLARP was detected as a 57-kDa protein (Fig. 3A). To determine if CLARP exhibits caspase activity, CLARP was immunoprecipitated with anti-Flag antibody, and the immunoprecipitates were assayed for caspase activity using two fluorogenic peptide substrates. Enzymatic analysis showed that CLARP lacked detectable caspase activity when assayed with two fluorogenic substrates DEVD-AMC and acetyl-Tyr-Val-Ala-Asp-AMC (Fig. 3 B and C). In control experiments, Flag-tagged Ced-3 immunoprecipitates and extracts from cells transfected with a caspase-8 expression plasmid showed significant caspase activities (Fig. 3 B and C). As predicted from the amino acid sequence of CLARP, these results demonstrate that CLARP lacks caspase activity when expressed in mammalian cells.
Figure 3.
CLARP is devoid of significant caspase activity when expressed in cells. (A) Flag-tagged CLARP was detected with anti-Flag antibody in lysates from 293T cells transiently transfected with pcDNA3 (lane 1) and pcDNA3-Flag-CLARP (lane 2). Molecular weight of CLARP was calculated from comparison with protein markers. Protein immunoprecipitates with anti-Flag antibody from cells transfected with pcDNA3-CLARP-Flag were incubated with the fluorogenic DEVD-AMC (B) or acetyl-Tyr-Val-Ala-Asp-AMC (YVAD-AMC) (C). Fluorescence was detected after 1 hr and 10 hr of incubation with CLARP complexes. Protein immunoprecipitates with anti-Flag antibody from cells transfected with pCDNA-CED-3-Flag were used as a positive control for the DEVD-AMC cleavage assay. For the YVAD enzymatic, whole Nonidet P-40 extract from cells transfected with pCDNA-caspase-8-AU1 was used as a positive control.
Overexpression of CLARP Promotes Apoptosis in 293T and MCF7 Cells That is Abrogated by Caspase Inhibitors.
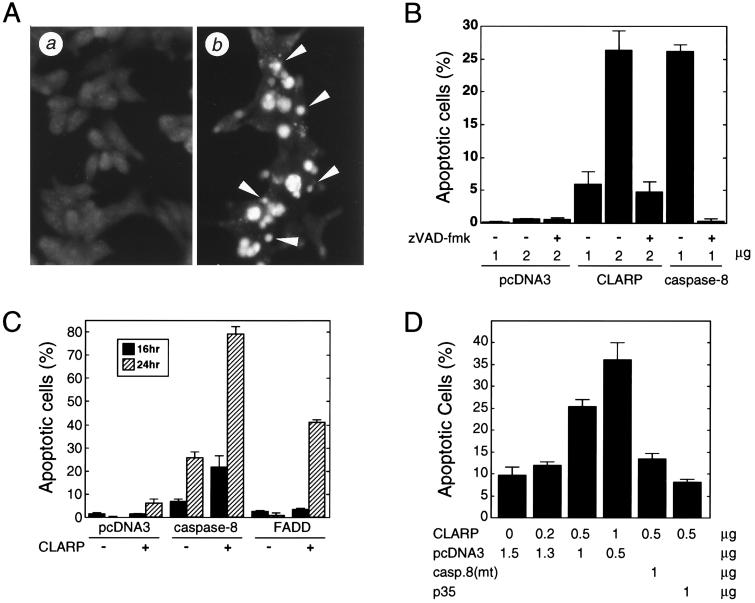
To begin to elucidate the physiological function of CLARP, an expression construct producing CLARP was introduced into 293T and MCF7 breast carcinoma cells and subsequently observed for features of apoptosis. CLARP induced nuclear condensation and fragmentation, a feature characteristic of apoptosis (Fig. 4A). In addition, the CLARP-transfected cells displayed morphological features of adherent cells undergoing apoptosis such as becoming rounded, membrane blebbing, and detachment from the dish (data not shown). Expression of CLARP had a modest, but significant, killing activity when compared with control plasmid. At 24 hr after transfection, about 25% of 293T cells underwent apoptosis compared with less than 3% with empty plasmid (Fig. 4B, P < 0.001). The pro-apoptotic activity induced by CLARP was inhibited by the broad spectrum caspase inhibitor zVAD-fmk (Fig. 4B). To determine if CLARP could regulate the killing ability of caspase-8 or FADD/MORT-1, these proteins were coexpressed in 293T cells. Expression of CLARP enhanced the killing ability of caspase-8 and FADD/MORT-1 (Fig. 4C, P < 0.001). In addition to 293T cells, expression of CLARP induced apoptotic cell death in MCF-7 cells, which was inhibited by the baculovirus protein p35 (Fig. 4D, P < 0.001). Expression of a dominant negative form of caspase-8 also reduced significantly the killing ability of CLARP (Fig. 4D, P < 0.001), suggesting that the apoptotic activity of CLARP is mediated at least in part by endogenous caspase-8.
Figure 4.
CLARP is a positive regulator of apoptosis. 293T (A–C) and MCF7 (D) cells were transfected with pcDNA3, pcDNA3-CLARP-Flag, pcDNA3-caspase-8-AU1, pcDNA3-FADD-AU1, pcDNA3-caspase-8-mut-AU1, and pcDNA3-p35 as described in Materials and Methods. (B–D) Transfected cells were visualized with β-galactosidase substrate and scored. (A) The nuclei of apoptotic cells were stained with acridine orange and ethidium bromide at 16 hr after transfection with 2 μg of pcDNA3 (a) and pcDNA3-CLARP-Flag (b). Arrows indicate apoptotic cells with condensed and fragmented nuclei. (B) 293T cells transfected with 1–2 μg of each expression plasmid were incubated with 20 μM zVAD-fmk. At 24 hr after transfection, apoptotic cells were counted. (C) 293T cells transfected with 1 μg of each expression plasmid. At 16 and 24 hr after transfection, apoptotic cells were counted. (D) MCF7 cells were transfected with 0.2–1.5 μg of each expression plasmid. At 24 hr after transfection, apoptotic cells were counted.
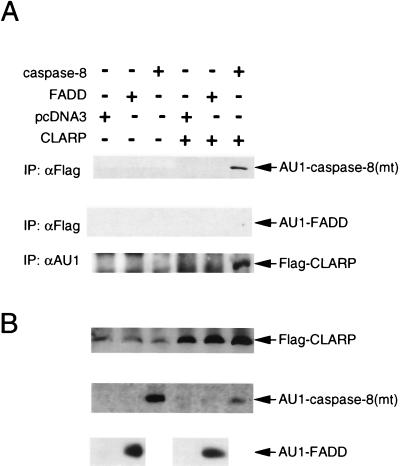
CLARP Interacts with Caspase-8 But Not With FADD/MORT-1.
FADD/MORT-1 interacts with caspase-8 through their corresponding DEDs. The homology between the DEDs of CLARP and caspase-8 and FADD/MORT-1 suggested that CLARP could interact with caspase-8 and/or FADD. Because caspase-8 is a potent killer of mammalian cells, we were unable to achieve high protein expression in cells coexpressing CLARP. Thus, we used an AU1-tagged Ser-377 mutant of caspase-8 (AU1-caspase-8-mt) to study the interaction of CLARP with caspase-8. Immunoblotting with anti-AU1 antibody revealed that AU1-caspase-8-mt coimmunoprecipitated Flag-CLARP (Fig. 5A, Top). To verify the interaction, we performed reciprocal experiments using anti-AU1 antibody to immunoprecipitate AU1-caspase-8-mt with Flag-caspase-8 mt. Consistent with the reciprocal experiment, CLARP was coimmunoprecipitated with AU1-caspase-8-mt (Fig. 5A, Bottom) In contrast, CLARP failed to interact with FADD/MORT-1 (Fig. 5A, Middle).
Figure 5.
CLARP interacts with caspase-8 but not with FADD. (A) Lysates were immunoprecipitated with anti-Flag antibody (Top and Middle) or anti-AU1 antibody (Bottom). Immunoprecipitates were immunoblotted with anti-AU1 antibody (Top and Middle) or anti-Flag antibody (Bottom). (B) One hundred micrograms of total lysate were immunoblotted with anti-Flag (Top) and anti-AU1 (Middle and Bottom) antibodies. A weak background band comigrating with CLARP was detected with polyclonal anti-Flag antibody.
The Human clarp Gene Is Located in Chromosome 2 at 2q33.
We searched the human genomic database for the chromosomal localization of the clarp gene. A STS WI-12098 tag (EST clone 47260, GenBank accession no. H11002) was found to contain overlapping nucleotide sequences with the coding region of CLARP. The genomic locus of the STS WI-12098 has been mapped to human chromosome 2 between the markers D2S116 and D2S307. This places the human clarp gene in chromosome 2 at 2q33 (200–206 cM).
DISCUSSION
The structure of CLARP is intriguing in that it resembles that of caspase-8 and caspase-10, but unlike these upstream caspases, CLARP is catalytically inactive. The lack of caspase activity exhibited by CLARP appears to be due to the loss of two critical residues Cys and His that are highly conserved in the caspase domain of all reported active caspases. Based on the x-ray crystal structure of caspase-1 (24), the corresponding amino acid residues His-237 and Cys-285 of caspase-1 are involved in catalysis, and mutations of these residues abrogate the enzymatic activity of caspase-1 and other caspases (24). The critical role of these residues is further supported by the presence of corresponding His and Cys amino acids in the caspase domain of DCP2, a Drosophila caspase-8 homologue identified in this study. A role for DCP2 in programmed cell death of Drosophila remains to be established. However, the conservation of a caspase-8 homologue in flies suggests an important role of DED-containing caspases in both vertebrate and invertebrate apoptosis. We would suggest that like its structurally related mammalian counterparts, caspase-8 and caspase-10, DCP2 is likely to link death signaling pathways with downstream effector caspases such as DCP1 in flies (25).
Caspase-8 and FADD/MORT-1 are two DED-containing molecules that are part of death signaling pathways that mediate Fas/Apo-1/CD95 and TNFR-1-induced apoptosis (7–10). Expression of CLARP promoted apoptosis and augmented the killing activity of caspase-8 and FADD/MORT-1. While this manuscript was in the reviewing process, three groups reported independently, identical proteins to CLARP, Casper (26), c-FLIPL (27), and I-FLICE (28). Although the precise mechanism(s) by which CLARP promotes apoptosis remains to be further investigated, at least two nonexcluding possibilities could explain these observations. First, CLARP could activate caspase-8 and/or other DED-containing caspases via homophilic interactions through the corresponding DED of CLARP. This mechanism would be similar to that previously proposed for the FADD/MORT-1-caspase-8 interaction (7–10). An alternative, nonexcluding possibility is that CLARP could promote apoptosis by interacting with an inhibitor of caspase-8 and/or FADD/MORT-1. Several viral proteins such as E8 and MC159L have been shown to inhibit Fas/Apo-1/CD95-induced apoptosis by interaction with caspase-8 or FADD/MORT-1 (15–17). Thus, CLARP could bind and inactivate cellular inhibitory proteins of caspase-8 or FADD/MORT-1 via DED interactions. Because CLARP-induced apoptosis was inhibited by the caspase inhibitors p35 and zVAD-fmk, these results suggest that the death pathway induced by CLARP ultimately leads to the activation of caspases and is regulated by a caspase-sensitive checkpoint. The lack of caspase activity exhibited by CLARP would have predicted that this protein also could function as an inhibitor of apoptosis. However, we failed to generate any evidence that supports a role for CLARP as a negative regulator of apoptosis under several cell culture conditions. However, we cannot formally rule out that CLARP could function as an inhibitor of apoptosis in other culture systems or during physiological cell death in vivo.
Because CLARP can interact with and enhance caspase-8-mediated apoptosis, it is likely that this protein can function in caspase-8-mediated signaling pathways. These will include those pathways induced by engagement of the CD95 and TNFR-1 surface receptors. Caspase-8 and FADD/MORT-1 are also components of the cell death pathway mediated by the DR3 receptor (29), suggesting that CLARP also might modulate this apoptotic signaling pathway. Furthermore, CLARP could function in the regulation of additional cell death pathways via interaction with molecules containing DED modules. Future studies need to address a role for CLARP in the regulation of physiological cell death during tissue development and homeostasis.
Acknowledgments
We thank M. Benedict, D. Wu, H. Wallen, M. González-García, and L. del Peso for critical review of the manuscript, E. Alnemri for caspase-8 plasmid, and V. Dixit for MCF-7 cells and pcDNA3-p35. This work was supported by Grant R01 CA64556-01 from the National Institutes of Health. T.K. was supported by a fellowship from the Japan Science and Technology Corporation. Y. H. was supported by Postdoctoral Immunopathology Training Grant T32A107413-03 from the National Institutes of Health. G. N. was supported by Research Career Development Award K04 CA64421-01 from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CLARP, caspase-like apoptosis-regulatory protein; AMC, amino-4-methylcoumarin; DED, death effector domain; TNFR-1, tumor necrosis factor receptor 1; EST, expressed sequence tag; AU1-caspase-8-mt, AU1-tagged Ser-377 mutant of caspase-8; zVAD-fmk, benzyloxycarbonyl-Val-Ala-Asp-(OMe)-fluoromethylketone; DEVD-AMC, acetyl-Asp-Glu-Val-Asp-AMC.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF005774).
References
- 1.Vaux D L, Strasser A. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S. Trends Biochem Sci. 1995;20:198–202. doi: 10.1016/s0968-0004(00)89007-6. [DOI] [PubMed] [Google Scholar]
- 5.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 6.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 8.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 9.Kischkel F C, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnaiyan A M, Tepper C G, Seldin M F, O’Rourke K, Kischkel F C, Hellbardt S, Krammer P H, Peter M E, Dixit V M. J Biol Chem. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- 11.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 12.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Nature (London) 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 17.Hu S, Vincenz C, Buller M, Dixit V M. J Biol Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 18.Simonian P L, Grillot D A M, Merino R, Núñez G. J Biol Chem. 1996;271:22764–22772. doi: 10.1074/jbc.271.37.22764. [DOI] [PubMed] [Google Scholar]
- 19.Brunak S, Engelbrecht J, Knudsen S. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 20.Inohara N, Ding L, Chen S, Núñez G. EMBO J. 1997;16:1686–1694. doi: 10.1093/emboj/16.7.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 22.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 23.Wu D, Wallen H D, Núñez G. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 24.Wilson K P, Black J A, Thomson J A, Kim E E, Griffith J P, Navia M A, Murcko M A, Chambers S P, Aldape R A, Raybuck S A, Livingston D J. Nature (London) 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 25.Song Z, McCall K, Steller H. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 26.Shu H B, Halpin D R, Goeddel D V. Immunity. 1997;6:751–763. doi: 10.1016/s1074-7613(00)80450-1. [DOI] [PubMed] [Google Scholar]
- 27.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J L, Schroter M, Burns K, Mattmann C, Rimoldi D, French L E, Tschopp J. Nature (London) 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 28.Hu S, Vincenz C, Ni J, Gentz R, Dixit V M. J Biol Chem. 1997;272:17255–17257. doi: 10.1074/jbc.272.28.17255. [DOI] [PubMed] [Google Scholar]
- 29.Chinnaiyan A M, O’Rourke K, Yu G L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]