Abstract
The avian erythroblastosis viral oncogene (v-erbB) encodes a receptor tyrosine kinase that possesses sarcomagenic and leukemogenic potential. We have expressed transforming and nontransforming mutants of v-erbB in fibroblasts to detect transformation-associated signal transduction events. Coimmunoprecipitation and affinity chromatography have been used to identify a transformation-associated, tyrosine phosphorylated, multiprotein complex. This complex consists of Src homologous collagen protein (Shc), growth factor receptor binding protein 2 (Grb2), son of sevenless (Sos), and a novel tyrosine phosphorylated form of the cytoskeletal regulatory protein caldesmon. Immunofluorescence localization studies further reveal that, in contrast to the distribution of caldesmon along actin stress fibers in normal fibroblasts, caldesmon colocalizes with Shc in plasma membrane blebs in transformed fibroblasts. This colocalization of caldesmon and Shc correlates with actin stress fiber disassembly and v-erbB-mediated transformation. The tyrosine phosphorylation of caldesmon, and its association with the Shc–Grb2–Sos signaling complex directly links tyrosine kinase oncogenic signaling events with cytoskeletal regulatory processes, and may define one mechanism regulating actin stress fiber disassembly in transformed cells.
In cells undergoing anchorage-dependent growth, the disassembly of actin stress fibers is necessary for progression through the G2/M checkpoint of the cell cycle (1, 2). The cell cycle-dependent reorganization of the actin-based cytoskeleton is controlled by cdc2-mediated serine/threonine phosphorylation of the actin-binding protein caldesmon (3, 4). This regulatory process allows for the orderly disassembly of stress fibers and the rounding up of an anchored cell immediately prior to mitosis. In contrast, fibroblasts transformed by tyrosine kinase oncoproteins undergo anchorage-independent growth. These transformed cells contain reduced numbers of actin stress fibers and focal adhesions, thus displaying altered regulation of cytoskeletal architecture (5–7). There is emerging evidence that such alterations in the organization of the cytoskeleton may be sufficient to establish anchorage-independent growth. Specifically, reduction in expression levels of the actin-binding protein tropomyosin 1 results in actin stress fiber reorganization and anchorage-independent growth in Syrian hamster embryo cells (8). In addition, it has recently been demonstrated that the interaction between the bovine papillomavirus E6 oncoprotein and the focal adhesion protein paxillin is correlated with disruption of the actin-based cytoskeleton and soft agar colony formation in murine C127 cells (9).
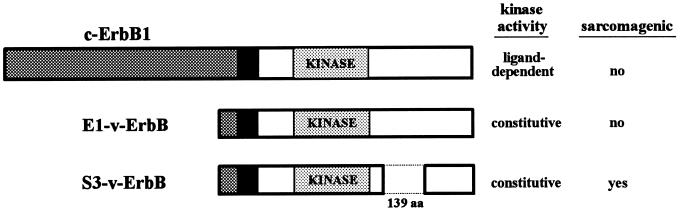
The mechanisms that mediate disruption of the cytoskeleton in cells transformed by tyrosine kinase oncoproteins are unclear, despite the identification of many components of tyrosine kinase signaling pathways. This is due, in part, to the fact that transformation-associated tyrosine phosphorylation events have been difficult to identify in cells transformed by tyrosine kinase oncoproteins. Generally, the tyrosine phosphorylation of specific substrates, such as those identified in v-src- and v-erbB-transformed fibroblasts (10–12), has not been correlated with the transformed phenotype. We recently have shown, however, that transformation-associated tyrosine phosphorylation events can be detected in avian erythroblastosis viral oncogene (v-erbB)-transformed chicken embryo fibroblasts (CEF) (13). Specifically, we have compared the signaling pathways in cells expressing the constitutively active mutants E1-v-ErbB and S3-v-ErbB (Fig. 1). E1-v-ErbB is a transmembrane tyrosine kinase with a truncated extracellular ligand-binding domain that is capable of transforming erythroblasts and producing avian erythroleukemias (14). Similarly, S3-v-ErbB has a truncated ligand-binding domain, but unlike E1, has sustained a 139-aa in-frame deletion within the C terminus that abolishes the protein’s leukemogenic ability while conferring sarcomagenic potential (14). In CEF transformed by S3-v-ErbB, we have identified a multiprotein complex involving the signal adapter proteins Shc (Src homologous collagen protein) and Grb2 (growth factor receptor binding protein 2), and a 75- to 78-kDa phosphoprotein component by coimmunoprecipitation using anti-Shc antibodies (13). This complex does not form in transforming growth factor α-stimulated fibroblasts or in fibroblasts expressing the leukemogenic E1-v-ErbB protein. Furthermore, the 46-kDa isoform of Shc and the 75- to 78-kDa phosphoprotein component of this complex have been shown to be tyrosine phosphorylated in a transformation-associated manner (13). These studies, therefore, suggested that this multiprotein complex plays a vital role in v-ErbB-mediated fibroblast transformation and sarcomagenesis. In this report, we identify a novel tyrosine phosphorylated form of the cytoskeletal regulatory protein caldesmon as the 78-kDa component of this complex. Furthermore, we demonstrate that the formation of this Shc–Grb2–son of sevenless (Sos)–caldesmon complex correlates with v-ErbB transformation and actin stress fiber disassembly.
Figure 1.
Schematic comparison of the structures and sarcomagenic potential of the c-erbB1 tyrosine kinase growth factor receptor and proteins encoded by the E1- and S3-v-erbB mutants. The intrinsic tyrosine kinase activity and ability to induce fibrosarcomas in vivo is indicated. Deleted amino acids are indicated by dotted lines. ▩, Ligand-binding/extracellular domain; black box, transmembrane domain; KINASE, tyrosine kinase domain.
MATERIALS AND METHODS
Cell Infection, Culture, and Lysis.
Primary cultures of CEF were virally infected with the E1 and S3 mutants of the v-erbB oncogene (15) using a replication-competent avian retroviral vector (16) as described (17). Cells were grown in DMEM (BioWhittaker) supplemented with 10% fetal bovine serum and 1% chicken serum, and screened by anti-ErbB kinase domain immunoblotting as described (13) to verify comparable expression levels of the mutant v-ErbB proteins in fully infected cells. Cells were lysed in a 1% Triton X-100 based lysis buffer containing the phosphatase inhibitors sodium orthovanadate (1 mM) and diisopropyl fluorophosphate (4 mM) as described (13). Protein concentrations were determined using BCA reagents from Pierce.
Protein Purification.
Glutathione S-transferase (GST)–Grb2 in pGEX-3x (Pharmacia) was replicated in Escherichia coli DH5α grown in Luria–Bertani medium supplemented with ampicillin. Induction of the fusion protein in bacteria with isopropyl β-d-thiogalactopyranoside (Sigma) and fusion protein purification on glutathione agarose beads were performed as described (18). Large-scale GST–Grb2 affinity chromatography was performed on 200 mg of whole cell lysate from S3-v-ErbB-transformed fibroblasts. The lysate was incubated with 60 mg GST–Grb2 bound to glutathione beads in precipitation buffer (190 mM NaCl/50 mM Tris/6 mM EDTA/2.5% Triton X-100, pH 7.4] overnight at 4°C. Beads were washed in Tris-buffered saline and eluted with 0.5% SDS at 100°C for 3 min. The eluate was lyophilized, dissolved in standard Laemmli SDS sample buffer plus 200 mM DTT, and heated at 100°C for 3 min; proteins were separated on 10% acrylamide SDS/PAGE.
Micropeptide Sequencing.
The 78- and 75-kDa Coomassie-stained protein bands were excised from the gel and in situ digestion with lysyl endopeptidase (Wako Chemicals, Richmond, VA) was performed using a procedure adapted from Rosenfeld et al. (19) and Fernandez et al. (20). Peptides were extracted and separated on an Applied Biosystems model 130A Separation System (Perkin–Elmer) using a 2.1 × 250 mm column. Fractions corresponding to peptide peaks were adsorbed to polyvinylidene difluoride membranes and sequenced on an Applied Biosystems model 492 Procise Protein Sequencing System. The SWISSPROT database was searched for matching amino acid sequences.
Phosphoamino Acid Analysis.
One 10-cm plate of subconfluent S3-v-ErbB-transformed CEF was washed and grown in DMEM phosphate-free media (Specialty Media, Lavellette, NJ) with 5% fetal bovine serum and incubated with [32P]orthophosphate (Amersham; final concentration 1.7 mCi per ml; 1 Ci = 37 GBq) for 6 h. Lysates were immunoprecipitated with anti-caldesmon antibodies (Sigma, C-0297), and proteins were separated by SDS/PAGE and transferred to Immobilon-P (Millipore). The 78-kDa 32P-labeled caldesmon protein band was acid hydrolyzed in 6 M HCl at 110°C for 60 min, and phosphoamino acid analysis was performed by cellulose thin layer electrophoresis (21).
Immunofluorescence Microscopy.
Cells were grown overnight on glass coverslips in DMEM with 10% fetal bovine serum and 1% chicken serum, fixed in 4% paraformaldehyde, treated with 0.5% Triton X-100, and blocked for 1 h in PBS with 5% goat serum, 0.1% BSA, and 0.1% fish skin gelatin. Cells were incubated at room temperature with primary antibodies [monoclonal α-caldesmon (Sigma C-0297), 1:50 dilution; polyclonal α-Shc (Upstate Biotechnology, Lake Placid, NY), 25 μg/ml] for 4 h, washed in PBS, and then incubated with fluorescein isothiocyanate-conjugated goat α-rabbit or goat α-mouse antibodies for 1 h. F-actin was visualized by incubating cells with rhodamine-phalloidin (1:100 dilution; Molecular Probes) for 2 h at room temperature.
RESULTS
Phosphotyrosyl Protein Complex in v-ErbB-Transformed Cells.
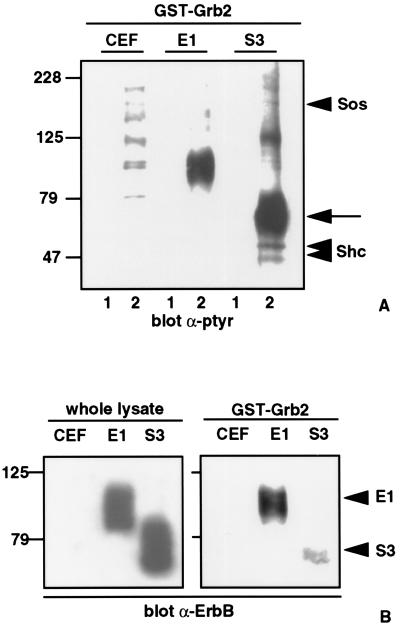
To characterize the transformation-associated Shc-Grb2-pp75-78 multiprotein complex we used recombinant GST–Grb2 fusion proteins to precipitate specific protein components. Tyrosine phosphorylated proteins migrating at 160, 125, 75–78, 52, and 46 kDa are precipitated by GST–Grb2 exclusively in S3-v-ErbB-transformed CEF (Fig. 2A). We previously have shown in S3-v-ErbB-transformed fibroblasts that the 52- and 46-kDa proteins are isoforms of Shc, and that Grb2 associates with these phosphorylated forms of Shc (13). By immunoprecipitating lysates of S3-v-ErbB-transformed CEF with anti-Shc antibodies followed by immunoblotting with anti-Sos antibodies we also have shown that the 160-kDa phosphorylated protein in this complex is Sos (data not shown).
Figure 2.
Transformation-associated, tyrosine phosphorylated, multiprotein complex formation in S3-v-ErbB-transformed fibroblasts. (A) GST–Grb2 binding assay was performed by incubating 500 μg of each cell lysate with either GST bound to glutathione agarose beads (lanes 1) or recombinant GST–Grb2 bound to glutathione beads (lanes 2). Proteins were separated by SDS/PAGE, transferred to nitrocellulose, and immunoblotted with monoclonal antiphosphotyrosine antibodies (α-ptyr). Several major tyrosine phosphorylated proteins precipitate with GST–Grb2 in S3-v-ErbB-transformed fibroblasts as discussed in the text. In contrast, only minor phosphorylated proteins are detected in uninfected CEF, and a single major tyrosine phosphorylated protein migrating at 85–95 kDa is detected in CEF expressing E1-v-ErbB. Lanes: CEF, uninfected chicken embryo fibroblasts; E1, nontransformed CEF expressing the kinase-active E1-v-ErbB; S3, CEF transformed by S3-v-ErbB. Molecular weight standards are indicated in kDa. Arrowheads indicate Shc and Sos, as labeled. The arrow indicates the 75- to 78-kDa phosphotyrosyl protein component. (B) Association of E1- and S3-v-ErbB proteins with Grb2. Infected fibroblasts express equivalent amounts of their respective v-ErbB proteins as demonstrated by immunoblotting 100 μg of whole cell lysates with polyclonal anti-ErbB kinase domain antibodies (α-ErbB) (Left). GST–Grb2 binding assay (Right) was performed as in A, followed by α-ErbB immunoblotting to demonstrate the ability of Grb2 to associate with E1-v-ErbB (E1 arrowhead) or to S3-v-ErbB (S3 arrowhead). Lanes: CEF, E1, and S3 as defined in A.
In pursuing the identity of the 75- to 78-kDa phosphotyrosyl protein component in S3-v-ErbB-transformed fibroblasts, the S3-v-ErbB protein itself was a possible candidate because it migrates as a broad tyrosine phosphorylated band at 65–80 kDa and its human homolog, the epidermal growth factor receptor, is known to associate with Grb2 (22). In cells expressing equivalent amounts of their respective v-ErbB proteins (Fig. 2B Left), recombinant Grb2 associates with E1-v-ErbB but binds minimally to S3-v-ErbB (Fig. 2B Right). Therefore, while S3-v-ErbB may be a component of the 75- to 78-kDa phosphoprotein complex, it is not the only protein in this complex.
Transformation-Associated Shc-Grb2-Sos-Caldesmon Complex.
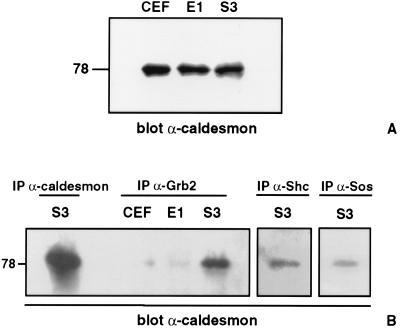
Large-scale GST–Grb2 affinity chromatography and polyacrylamide gel electrophoresis were used to further characterize the additional phosphoproteins in this complex. On Coomassie-stained acrylamide gels, two major protein bands of 75 and 78 kDa, which comigrate with the broad 75- to 78-kDa tyrosine phosphorylated protein band seen in Fig. 2A (S3 lane), were seen consistently. The Coomassie-stained protein migrating at 78 kDa was analyzed by micropeptide amino acid sequencing, resulting in the identification of a 9-aa peptide (KRLQEALER) identical to an internal sequence in the cytoskeletal regulatory protein caldesmon.
Quantitative protein immunoblot analysis of caldesmon in whole cell lysates (Fig. 3A) demonstrates the presence of the 78-kDa nonmuscle isoform of caldesmon (23) in equivalent amounts in uninfected CEF, CEF expressing E1-v-ErbB, and S3-v-ErbB-transformed CEF. These results indicate that caldesmon levels are not altered in S3-v-ErbB-transformed cells, in contrast to previous reports of decreased expression of caldesmon at both the mRNA and protein levels in Src and simian virus 40-transformed cells (23–25). The 78-kDa isoform of caldesmon is known to contain binding domains for myosin, actin, tropomyosin, and calmodulin (26), but previously has not been shown to associate with tyrosine kinase signal adapter proteins. Because our GST–Grb2 affinity chromatography results indicated an association between caldesmon and Grb2, we performed coimmunoprecipitation analyses using antibodies to components of the Shc–Grb2–Sos signal protein complex to corroborate these findings. Fig. 3B demonstrates that caldesmon is present in immunocomplexes formed by Grb2, Shc, and Sos antibodies. A comparison of immunocomplexes from lysates of uninfected CEF, CEF expressing E1-v-ErbB, and S3-v-ErbB-transformed CEF reveals that caldesmon coimmunoprecipitation with antibodies to Grb2 occurs to a markedly enhanced degree in S3-v-ErbB-transformed fibroblasts (Fig. 3B). Thus, the association of the cytoskeletal regulatory protein caldesmon with the Shc–Grb2–Sos signaling complex is a transformation-associated event.
Figure 3.
Association of caldesmon with Grb2, Shc, and Sos. (A) Equivalent protein expression levels of caldesmon in nontransformed and transformed CEF. A total of 100 μg of each cell lysate was immunoblotted with a monoclonal antibody to caldesmon (Sigma C-0297, 1:1,000 dilution). Lanes: CEF, uninfected chicken embryo fibroblasts; E1, nontransformed CEF expressing the kinase-active E1-v-ErbB; S3, CEF transformed by S3-v-ErbB. (B) Transformation-associated coimmunoprecipitation of caldesmon with Grb2, Shc, and Sos. Equal amounts of each cell lysate were immunoprecipitated with antibodies to caldesmon, Grb2 (monoclonal α-Grb2 antibody, Upstate Biotechnology), Shc (polyclonal α-Shc antibody, Upstate Biotechnology), or Sos (polyclonal α-Sos antibody, Upstate Biotechnology) as indicated, followed by immunoblotting with antibodies to caldesmon. Caldesmon migrates at 78 kDa and coimmunoprecipitates with Grb2, Shc, and Sos in S3-v-ErbB-transformed fibroblasts. Lane designations as in A. The 78-kDa protein that is coimmunoprecipitated by antibodies to Grb2, Shc, and Sos in S3-v-ErbB-transformed fibroblasts comigrates with the 78-kDa isoform of caldesmon immunoprecipitated by anti-caldesmon antibodies and is recognized by three different monoclonal antibodies to caldesmon (Sigma C-0297, C-6542, and C-6292). Caldesmon was not detected in the anti-Grb2 immunoprecipitates of uninfected CEF or CEF expressing E1-v-ErbB. A representative immunoblot of three separate experiments is shown.
Tyrosine Phosphorylation of Caldesmon.
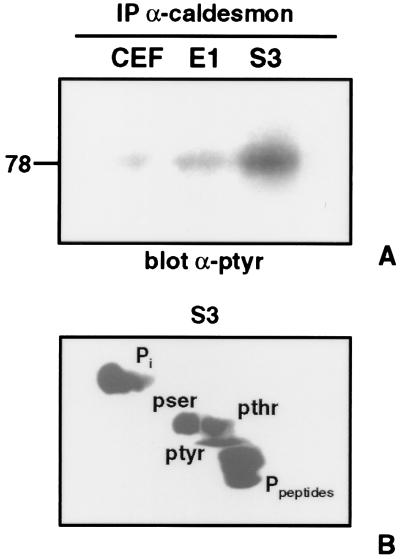
Next we analyzed the phosphorylation status of caldesmon to determine if this protein is one of the tyrosine phosphorylated components of this multiprotein complex. Caldesmon was immunoprecipitated from fibroblasts expressing the sarcomagenic (S3) or leukemogenic (E1) mutants of v-ErbB, followed by antiphosphotyrosine immunoblotting. These studies demonstrate that caldesmon is predominantly tyrosine phosphorylated in S3-v-ErbB-transformed fibroblasts (Fig. 4A). To verify these antiphosphotyrosine immunoblot results caldesmon was immunoprecipitated from [32P]orthophosphate-labeled S3-v-ErbB-transformed fibroblasts, and phosphoamino acid analysis was performed. In S3-v-ErbB-transformed fibroblasts, caldesmon contains phosphotyrosine in addition to phosphoserine and phosphothreonine (Fig. 4B). Caldesmon previously has been reported to be only serine/threonine phosphorylated (3, 4, 27). In this study we report the tyrosine phosphorylation of caldesmon. In this regard, analysis of the avian caldesmon amino acid sequence reveals three potential tyrosine phosphorylation sites: two within the N-terminal myosin-binding domain and one within a C-terminal actin-binding domain (26, 28, 29).
Figure 4.
Tyrosine phosphorylation of caldesmon. (A) Equal amounts of each cell lysate were immunoprecipitated with anti-caldesmon antibodies followed by immunoblotting with antiphosphotyrosine antibodies (α-ptyr). Caldesmon is predominantly tyrosine phosphorylated in S3-v-ErbB-transformed fibroblasts. Lanes: CEF, uninfected chicken embryo fibroblasts; E1, nontransformed CEF expressing the kinase-active E1-v-ErbB; S3, CEF transformed by S3-v-ErbB. (B) Phosphoamino acid analysis of caldesmon in 32P-labeled S3-v-ErbB-transformed CEF by cellulose thin layer electrophoresis. Caldesmon is phosphorylated on tyrosine, serine, and threonine in S3-v-ErbB-transformed fibroblasts. pser, Phosphoserine; pthr, phosphothreonine; ptyr, phosphotyrosine; Pi, inorganic 32P; Ppeptides, phosphopeptides.
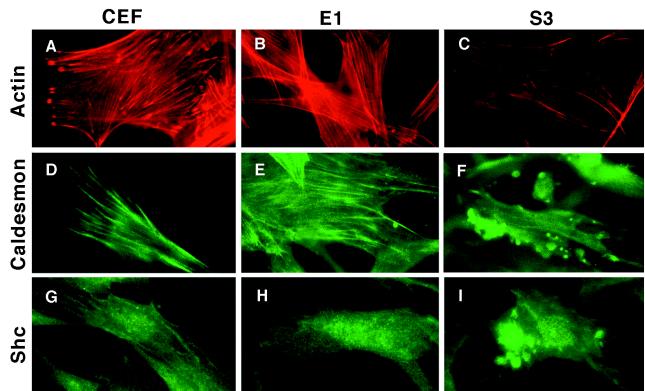
Transformation-Specific Actin Stress Fiber Disassembly and Colocalization of Shc and Caldesmon.
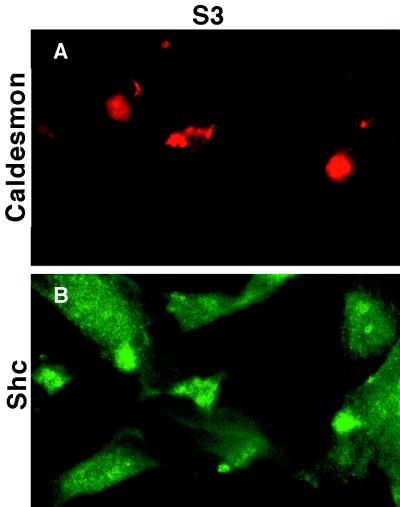
To further evaluate the association of caldesmon with the Shc–Grb2–Sos signaling complex, and to determine the relationship between this complex and actin stress fiber organization, we analyzed the distribution of caldesmon and Shc in v-ErbB-transformed and nontransformed fibroblasts by immunofluorescence microscopy. Actin stress fibers are clearly reduced in S3-v-ErbB-transformed CEF as compared with nontransformed CEF (Fig. 5 A–C). Caldesmon localizes along actin stress fibers in nontransformed CEF (Fig. 5 D and E), whereas in S3-v-ErbB-transformed CEF, caldesmon localizes to the plasma membrane in discrete dorsal cell surface blebs (Fig. 5F). The localization of Shc also differs in transformed versus nontransformed cells. Shc appears in a dispersed, cytoplasmic pattern in nontransformed CEF, but in contrast, localizes to dorsal cell surface blebs in S3-v-ErbB-transformed CEF (Fig. 5 G–I). Double-label immunofluorescent studies with antibodies to both caldesmon and Shc directly demonstrate that these two proteins colocalize to dorsal cell surface blebs in transformed fibroblasts (Fig. 6 A and B). These plasma membrane blebs, which stain intensely with antibodies to both caldesmon and Shc, are distinct from the ventral cell surface structures referred to as “rosette adhesions” or “podosomes” found in Src-transformed fibroblasts (30, 31), based on both their cell surface location and the lack of phalloidin-staining for F-actin in the dorsal cell surface blebs (Fig. 5C). This altered distribution of caldesmon and Shc is correlated with S3-v-ErbB-mediated transformation and actin stress fiber disassembly, and clearly does not occur in CEF expressing the kinase-active, nontransforming E1-v-ErbB. Thus, the in vivo colocalization of caldesmon and Shc corroborates our biochemical data demonstrating the transformation-associated formation of a Shc–Grb2–Sos–caldesmon complex.
Figure 5.
Localization of F-actin, caldesmon, and Shc in S3-v-ErbB-transformed and nontransformed fibroblasts. Immunofluorescence microscopy was used to visualize actin stress fibers, caldesmon, and Shc in uninfected fibroblasts (CEF), nontransformed CEF expressing E1-v-ErbB (E1), and S3-v-ErbB-transformed CEF (S3). (A–C) Marked decrease in actin stress fibers in S3-v-ErbB-transformed fibroblasts. (D–F) Distribution of caldesmon along actin stress fibers in nontransformed fibroblasts, and the localization of caldesmon to dorsal cell surface blebs in S3-v-ErbB-transformed fibroblasts. (G–I) Cytoplasmic distribution of Shc in nontransformed fibroblasts and the localization of Shc to dorsal cell surface blebs in S3-v-ErbB-transformed fibroblasts. Expression of the kinase-active, nonsarcomagenic E1-v-ErbB protein in CEF (E1) does not cause actin stress fiber disassembly or result in the localization of caldesmon and Shc to dorsal cell surface blebs (B, E, and H, respectively).
Figure 6.
Colocalization of caldesmon and Shc in S3-v-ErbB-transformed fibroblasts. Double-label immunofluorescence, using monoclonal antibodies to caldesmon and polyclonal antibodies to Shc, was performed on S3-v-ErbB-transformed CEF (S3). Primary antibodies to caldesmon and Shc are the same as used in Fig. 5. The secondary antibodies are rhodamine-conjugated goat anti-mouse and fluorescein isothiocyanate-conjugated goat α-rabbit. A and B are representative images of the same cells showing colocalization of caldesmon (A) and Shc (B) to dorsal cell surface blebs.
DISCUSSION
In conclusion, we have demonstrated the transformation-associated coupling of a tyrosine kinase downstream signaling complex with the cytoskeletal regulatory protein caldesmon. Our coimmunoprecipitation studies reveal the enhanced association of caldesmon with the Shc–Grb2–Sos signaling complex in v-ErbB-transformed fibroblasts. Likewise, the immunofluorescent localization studies demonstrate the transformation-specific disassembly of actin stress fibers, dissociation of caldesmon from actin microfilaments, and in vivo colocalization of caldesmon and Shc to plasma membrane blebs. The exact structure and function of this Shc–Grb2–Sos–caldesmon complex in vivo is unknown. Although the Shc–Grb2–Sos complex, and its activation of Ras, have been extensively studied (32, 33), the nature of the association between these signaling molecules and caldesmon is unclear. In fact, caldesmon may associate with multiple protein–protein interaction domains within this complex with varying degrees of stability. Specifically, in its tyrosine phosphorylated state, caldesmon may interact with the phosphotyrosine binding (PTB) or SH2 domains of Shc, or caldesmon may interact with the SH3 domains of Grb2 via a putative PxxP (P, proline; x, any amino acid) SH3-binding motif in its proline-rich C terminus.
We speculate that the tyrosine phosphorylation of caldesmon and its association with the Shc–Grb2–Sos complex mimics the cdc2 kinase-mediated regulation of caldesmon. Specifically, tyrosine phosphorylated caldesmon may mediate the disassembly of actin stress fibers during interphase in transformed cells, just as serine/threonine phosphorylated caldesmon directs this process during prophase in normal cells. Other potential mechanisms of regulating caldesmon in a transformation-specific manner previously have been reported. These include the reduction of caldesmon expression in Src and simian virus 40 transformed cells (23–25), and the disruption of genes encoding caldesmon-like molecules by chromosomal translocation in certain human tumors (34). Therefore, caldesmon may be a common target of oncogenic processes, resulting in the transformation-specific reorganization of the actin-based cytoskeleton and establishment of anchorage-independent cell growth. Further investigation of the Shc–Grb2–Sos–caldesmon complex, including identification of the tyrosine kinase responsible for phosphorylating caldesmon, will allow us to more precisely define the signaling network that governs anchorage-independent growth in cells transformed by tyrosine kinases.
Acknowledgments
We thank Benjamin Madden, Greg Poynter, and Lynn Cordes for their technical contributions; Daniel McCormick and David Toft for helpful discussions; and Paul Findell (Syntex Research, Palo Alto, CA) for providing the GST-Grb2 pGEX plasmid. We thank Diane Jelinek and Michael Getz for critical review of the manuscript. This work was supported by the Mayo Foundation and National Institutes of Health grants CA01627 and CA51197.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: v-erbB, avian erythroblastosis viral oncogene; Shc, Src homologous collagen protein; Grb2, growth factor receptor binding protein 2; Sos, son-of-sevenless protein; GST, glutathione S-transferase; CEF, chicken embryo fibroblasts.
References
- 1.Sanger J M, Mittal B, Dome J S, Sanger J W. Cell Motil Cytoskeleton. 1989;14:201–219. doi: 10.1002/cm.970140207. [DOI] [PubMed] [Google Scholar]
- 2.Nurse P. Nature (London) 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 3.Yamashiro S, Yamakita Y, Hosoya H, Matsumura F. Nature (London) 1991;349:169–172. doi: 10.1038/349169a0. [DOI] [PubMed] [Google Scholar]
- 4.Yamakita Y, Yamashiro S, Matsumura F. J Biol Chem. 1992;267:12022–12029. [PubMed] [Google Scholar]
- 5.Edelman G M, Yahara I. Proc Natl Acad Sci USA. 1976;73:2047–2051. doi: 10.1073/pnas.73.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boschek C B, Jockusch B M, Friis R R, Back R, Grundmann E, Bauer H. Cell. 1981;24:175–184. doi: 10.1016/0092-8674(81)90513-4. [DOI] [PubMed] [Google Scholar]
- 7.Button E, Shapland C, Lawson D. Cell Motil Cytoskeleton. 1995;30:247–251. doi: 10.1002/cm.970300402. [DOI] [PubMed] [Google Scholar]
- 8.Boyd J, Risinger J I, Wiseman R W, Merrick B A, Selkirk J K, Barrett J C. Proc Natl Acad Sci USA. 1995;92:11534–11538. doi: 10.1073/pnas.92.25.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong X, Howley P M. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds A B, Roesel D J, Kanner S B, Parsons J T. Mol Cell Biol. 1989;9:629–638. doi: 10.1128/mcb.9.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer S, LaBudda K, McGlade J, Hayman M J. Mol Cell Biol. 1994;14:3253–3262. doi: 10.1128/mcb.14.5.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu H G, Chang C M, Ravi L, Ling L, Castellano C M, Walter E, Pelley R J, Kung H-J. Mol Cell Biol. 1994;14:6868–6878. doi: 10.1128/mcb.14.10.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McManus M J, Connolly D C, Maihle N J. J Virol. 1995;69:3631–3638. doi: 10.1128/jvi.69.6.3631-3638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maihle N J, Kung H-J. Biochim Biophys Acta. 1988;948:287–304. doi: 10.1016/0304-419x(89)90003-6. [DOI] [PubMed] [Google Scholar]
- 15.Raines M A, Maihle N J, Moscovici C, Moscovici M G, Kung H-J. J Virol. 1988;62:2444–2452. doi: 10.1128/jvi.62.7.2444-2452.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly D C, Toutenhoofd S L, Maihle N J. J Virol. 1994;68:6804–6810. doi: 10.1128/jvi.68.10.6804-6810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez J, DeMott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 21.Kamps M P, Sefton B M. Anal Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 22.Buday L, Downward J. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 23.Owada M K, Hakura A, Iida K, Yahara I, Sobue K, Kakiuchi S. Proc Natl Acad Sci USA. 1984;81:3133–3137. doi: 10.1073/pnas.81.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novy R E, Lin J L-C, Lin J J-C. J Biol Chem. 1991;266:16917–16924. [PubMed] [Google Scholar]
- 25.Yamashiro S, Yoshida K, Yamakita Y, Matsumura F. Adv Exp Med Biol. 1994;358:113–122. doi: 10.1007/978-1-4615-2578-3_11. [DOI] [PubMed] [Google Scholar]
- 26.Matsumura F, Yamashiro S. Curr Opin Cell Biol. 1993;5:70–76. doi: 10.1016/s0955-0674(05)80010-9. [DOI] [PubMed] [Google Scholar]
- 27.Litchfield D W, Ball E H. J Biol Chem. 1987;262:8056–8060. [PubMed] [Google Scholar]
- 28.Bryan J, Imai M, Lee R, Moore P, Cook R G, Lin W G. J Biol Chem. 1989;264:13873–13879. [PubMed] [Google Scholar]
- 29.Wang C L A, Wang L C, Xu S, Lu R C, Saavedra-Alanis V, Bryan J. J Biol Chem. 1991;266:9166–9172. [PubMed] [Google Scholar]
- 30.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Reynolds A P, Kanner S B, Vines R R, Parsons J T. Mol Cell Biol. 1991;11:5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egan S E, Giddings B W, Brooks M W, Buday L, Sizeland A M, Weinberg R A. Nature (London) 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 33.Pawson T, Schlessinger J. Curr Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 34.Zani V J, Asou N, Jadayel D, Heward J M, Shipley J, Nacheva E, Takasuki K, Catovsky D, Dyer M J. Blood. 1996;87:3124–3134. [PubMed] [Google Scholar]