Abstract
T-DNA nuclear import is a central event in genetic transformation of plant cells by Agrobacterium. Presumably, the T-DNA transport intermediate is a single-stranded DNA molecule associated with two bacterial proteins, VirD2 and VirE2, which most likely mediate the transport process. While VirE2 cooperatively coats the transported single-stranded DNA, VirD2 is covalently attached to its 5′ end. To better understand the mechanism of VirD2 action, a cellular receptor for VirD2 was identified and its encoding gene cloned from Arabidopsis. The identified protein, designated AtKAPα, specifically bound VirD2 in vivo and in vitro. VirD2–AtKAPα interaction was absolutely dependent on the carboxyl-terminal bipartite nuclear localization signal sequence of VirD2. The deduced amino acid sequence of AtKAPα was homologous to yeast and animal nuclear localization signal-binding proteins belonging to the karyopherin α family. Indeed, AtKAPα efficiently rescued a yeast mutant defective for nuclear import. Furthermore, AtKAPα specifically mediated transport of VirD2 into the nuclei of permeabilized yeast cells.
Nuclear import of nucleic acid molecules occurs in many plant–pathogen interactions. DNA and RNA molecules are transported from the host cell cytoplasm into the nucleus during viral infections. In addition, nuclear import of DNA is a central event in genetic transformation of plant cells by a soil microorganism Agrobacterium tumefaciens. Agrobacterium–plant cell interaction is the only known natural example of DNA transport between kingdoms. In this process, a single-stranded DNA copy of the the portion of the tumor inducing (Ti) plasmid that is transferred to plant cells (T-DNA) is transported from Agrobacterium into the host plant cell nucleus and integrated in the plant genome (1–3). Expression of the wild-type T-DNA elicits the production of growth hormones and, consequently, neoplastic growths (tumors) on the host plant. These tumors then synthesize opines, a major carbon/nitrogen source of Agrobacterium. Because opine import into and catabolism within the bacterial cell require specialized enzymes that are produced by Agrobacterium, practically no other microorganisms can metabolize opines, creating a favorable biological niche for Agrobacterium. (reviewed in refs. 4 and 5).
The T-DNA of Agrobacterium is specified by two 25-bp border repeats. While the wild-type T-DNA carries Ti genes, any DNA placed between the T-DNA borders will be transferred into the host cell and imported into its nucleus. Thus, the T-DNA itself probably does not carry targeting signals. Instead, its nuclear import process is likely mediated by the two Agrobacterium proteins, VirD2 and VirE2, which are thought to directly associate with the transported single-stranded T-DNA molecule (T-strand), forming a transport (T) complex (6–8). In this complex, one molecule of VirD2 is presumed to covalently associate with the 5′ end of the T-strand while VirE2, a single-stranded DNA binding protein, cooperatively coats the rest of the ssDNA molecule (reviewed in ref. 5). Recent studies have demonstrated that both VirE2 and VirD2 contain functional nuclear localization signal (NLS) sequences (9–14). VirE2 has been shown to contain two independently active NLSs located within the central region of the molecule (13) while VirD2 has been demonstrated to carry one functional NLS at its carboxyl terminus (11). VirD2 and VirE2 NLSs belong to the bipartite type sequence characterized by two basic amino acid domains separated by a variable-length linker sequence (15). However, while VirD2 NLS also functioned in animal cells, VirE2 NLSs did not (9). Repositioning of one amino acid residue within the VirE2 NLS enabled it to function in animal cells (9). These observations suggested that VirD2 and VirE2 NLSs are functionally different and that they may be recognized by different receptors within the host plant cell. Identification of such receptors is necessary for understanding the mechanism of T-DNA nuclear uptake. Furthermore, because pathogens often use the existing cellular machinery for their own needs, this information may be relevant to other processes of the nucleic acid nuclear import in plant cells.
Presently, most of our knowledge about the process of Agrobacterium–plant cell interaction derives from studies of its bacterial components (5). No plant cell factors involved in such central steps of Agrobacterium infection as T-DNA nuclear transport and integration have been identified. Here, we used the yeast two-hybrid system to isolate the plant cell receptor for VirD2. Amino acid sequence analysis indicated that the VirD2 receptor is homologous to animal karyopherin α and yeast SRP1 proteins. Furthermore, the VirD2 receptor complemented a yeast mutant defective in nuclear import and transported the fluorescently labeled VirD2 protein into the nuclei of permeabilized yeast cells.
MATERIALS AND METHODS
Plasmid Construction.
The pGS377 plasmid containing the ORF of VirD2 under the T7 polymerase promoter (16) was used for overexpression of VirD2 in Escherichia coli. The pGS377ΔNLS plasmid was constructed by exchanging the EcoRI–BamHI fragment of the wild-type VirD2 sequence in pGS377 with the EcoRI–BamHI fragment of pGD-A119 containing a VirD2 mutant with deleted carboxyl-terminal NLS (del1, 2 construct) (11).
The pGBTD2, pGBTD2ΔNLS, and pGADAtKAPα (Arabidopsis thaliana karyopherin α) plasmids containing the GAL4 DNA binding domain fused in-frame to the full-length VirD2 protein of Agrobacterium, VirD2 with deleted carboxyl-terminal NLS (11), or AtKAPα, respectively, were constructed as follows. The protein ORFs were amplified by PCR, using a high-fidelity VentR DNA polymerase (New England Biolabs) with an error rate 5- to 15-fold lower than that of Taq DNA polymerase. BamHI restriction sites were introduced at the 5′ and 3′ ends of each ORF to maintain the reading frame of the GAL4 DNA binding domain. The BamHI fragments were then cloned into the corresponding site of pGBT9 (CLONTECH). To verify that PCR amplification introduced no unintended mutations into the VirD2 NLS domain, the DNA sequence of the corresponding regions of pGBTD2 and pGBTD2ΔNLS constructs was determined using the dideoxynucleotide sequencing protocol (17).
The pLAM5 plasmid containing amino acids 66–320 of the human lamin C protein fused to the GAL4 activation domain (18) was kindly provided by Rolf Sternglanz (State University of New York, Stony Brook).
For construction of pGEMAtKAPα and pGEMD2, the PCR-amplified BamHI fragments of the AtKAPα and VirD2 ORFs (see above) were subcloned into the BamHI site of pGEM7Zf+ (Promega).
For yeast expression of the unfused AtKAPα, the GAL4 activation domain was removed from pGADAtKAPα by oligonucleotide-directed mutagenesis (19), producing the pAtKAPα construct. For expression of the SRP1 from Saccharomyces cerevisiae, PCR was used to introduce BamHI restriction sites at both 5′ and 3′ ends of the SRP1 ORF (20). This fragment was then subcloned into the BamHI-digested pAtKAPα plasmid in place of the AtKAPα gene, producing pSRP1. For negative control, the vector was self-ligated, producing the pGADΔG plasmid.
Construction of Arabidopsis cDNA Library in pGAD424.
Poly(A)+ mRNA was prepared using the Stratagene messenger RNA isolation kit (Stratagene) from the above-ground parts of mature flowering Arabidopsis thaliana (ecotype Columbia) plants. The cDNA was made using the Stratagene ZAP-cDNA synthesis kit in which the first strand synthesis was primed with the poly(dT)–XhoI linker-primer. After the second strand synthesis, EcoRI adapters were ligated to the blunt ends of the cDNA followed by digestion with XhoI and EcoRI restriction endonucleases and size fractionation. The cDNA was then subcloned unidirectionally as EcoRI and XhoI fragments into EcoRI and SalI sites of the pGAD424 plasmid containing the yeast GAL4 activation domain (CLONTECH). The resulting unamplified library contained 5 × 106 independent clones with an insertion rate of 95% and an average cDNA size of 1.5 kbp.
Two-Hybrid Screen.
Yeast strain YD2 was generated by transforming strain Y153 (21) with pGBTD2. It was determined that pGBTD2 alone was insufficient for transcriptional activation in the two-hybrid system. The Arabidopsis cDNA library in pGAD424 was then transformed into strain YD2 and positive clones were selected on medium containing 25 mM 3-aminotriazole and confirmed by the β-galactosidase assay as described (21). False positives were eliminated using the unrelated bait plasmids pLAM5 (containing a fragment of the human lamin C gene) and pVA1 (containing a fragment of the murine p53 gene) (18), i.e., the isolated positive library plasmids were tested for interaction with pLAM5 and the interacting clones were eliminated. The largest positive cDNA clone was used as probe to screen the PRL2 Arabidopsis cDNA library (22) in λZipLox (GIBCO/BRL). Approximately 5 × 106 plaques were screened with the digoxigenin-labeled probe immunodetected using the Genius Hybridization system (Boehringer Mannheim). The positive cDNA clones were retrieved from the λZipLox vector according to the manufacturer’s instructions and their sequence was determined using internal oligonucleotide primers and Sequenase 2.0 (United States Biochemical).
Coimmunoprecipitation of VirD2 and AtKAPα.
The pGEMAtKAPα and pGEMD2 plasmids were linearized with BstXI and used as templates for in vitro transcription. Messenger RNAs were transcribed from the templates using the T7 RNA polymerase (Boehringer Mannheim) and 7-methyl-guanosine cap (Pharmacia) as described in the manufacturer’s instructions and ref. 23. The AtKAPα and VirD2 mRNAs were translated in vitro using a wheat germ translation system (Promega) as described (23). Briefly, 10 μCi (1 Ci = 37 gBq) of [35S]methionine (Amersham) was incubated for 1 hr at 25°C with 25 μl wheat germ extract and 2 μg RNA in the reaction volume of 50 μl. Before coimmunoprecipitation assays, 10 μl of the translation reaction were diluted with 90 μl of HBS (10 mM Hepes, pH 7.5/150 mM NaCl) and centrifuged for 5 min at 15,000 × g to sediment insoluble protein aggregates. Immediately after centrifugation, 50 μl of each translation mixture were combined, mixed, and incubated for 2 hr at 4°C. Rabbit preimmune or anti-VirD2 immune serum (1 μl) was added and the incubation continued for 2 hr at 4°C followed by addition of 20 μl of protein A bead slurry and further incubation for 1 hr at 4°C. Beads were washed five times with 1 ml of Hepes-buffered saline and proteins were recovered by boiling in sample buffer and analyzed by gel electrophoresis on a SDS/10% polyacrylamide gel (24) followed by autoradiography.
Expression of AtKAPα in the Yeast srp1-31 Mutant Strain and In vitro Assay for Nuclear Import.
The srp1-31 yeast strain containing a temperature-sensitive mutation in SRP1 (25) was a kind gift from Gerald Fink (Whitehead Institute, Massachusetts Institute of Technology). The pGADΔG, pAtKAPα, and pSRP1 plasmids were introduced into the srp1-31 strain and the resulting transformed strains were grown for 2 days at the permissive (25°C) or restrictive temperatures (37°C). For in vitro assays, srp1-31 cells harboring pGADΔG, pAtKAPα, or pSRP1 were grown for 24 hr at 25°C, and then shifted for 6 hr to 37°C; this period of growth at the restrictive temperature has been shown to inactivate >95% of the mutant SRP1 protein (25). After that, the cells were permeabilized and their cytosolic fractions prepared as described (26). For permeabilization, 200 ml of 107 cells per ml culture was pelleted (800 × g for 10 min at 25°C), resuspended in 20 ml of 100 mM Pipes (pH 9.4), 10 mM DTT, and incubated at 30°C for 10 min with gentle shaking. Cells were then centrifuged, resuspended in 5 ml yeast extract/peptone, 0.2% glucose, 10 mM Pipes (pH 7.5), 0.6 M sorbitol, and 2,000 units of oxalyticase (Enzogenetics, Corvallis, OR), and incubated at 30°C for 15 min. The cells were then collected by centrifugation, washed 5 times in 10 volumes of cold permeabilization buffer (0.4 M sorbitol/20 mM Pipes, pH 6.8/150 mM NaCl/5 mM Mg acetate), resuspended in 2 ml of the same buffer, slowly frozen above liquid nitrogen, and stored at −80°C. For preparation of the cytosol, 20 ml of 107 cells per ml culture was washed with 10 volumes of buffer N (0.25 M sorbitol/20 mM Pipes, pH 6.8/150 mM NaCl/5 mM Mg acetate) and resuspended in 0.25 ml of the same buffer, containing 1 mM DTT, 1 mM phenyl methane sulfonyl fluoride, 5 μg/ml leupeptin, aprotinin, chymostatin, and pepstatin. Cells were lysed by bead-beating with 100 μl of acid-washed 0.5 mm glass beads, followed by centrifugation at 100,000 × g for 1 hr at 4°C. The supernatant was then aliquoted, frozen in liquid nitrogen, and stored at −80°C until use. The final protein concentration of the cytosolic fraction was 2.5 mg/ml.
Nuclear import assays were performed in buffer N in the total volume of 25 μl, containing 107 permeabilized cells/ml, 1 mM ATP, 0.1 mg/ml creatine kinase, 10 mM creatine phosphate, 1.5 mg/ml cytosolic fraction, and 50 μg/ml of the fluorescently labeled VirD2 or VirD2ΔNLS proteins. Reactions were incubated for 30 min at 30°C, stained with 0.4 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and viewed under a fluorescent microscope. Each experiment was repeated three times.
Fluorescent Labeling of VirD2.
VirD2 and its NLS-lacking mutant, VirD2ΔNLS were overexpressed in E. coli, purified, and labeled with fluorescein-5-maleimide (Molecular Probes) as described (9). The labeled protein was separated from the unincorporated dye on a SDS/12.5% polyacrylamide gel and electroeluted as described (27). The eluted protein was adjusted to 4 mg/ml concentration, aliquoted, and stored at −80°C until use.
RESULTS
AtKAPα–VirD2 Interaction.
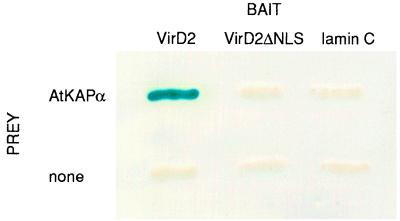
We used the yeast two-hybrid screen (28) with an Arabidopsis cDNA library and the Agrobacterium VirD2 protein as the bait. One positive clone was identified after eliminating the false positives using two nonspecific baits (18) (data not shown). The identified clone was purified and used to isolate the full-length cDNA. First, the direct interaction between the protein product of the isolated cDNA (designated AtKAPα) and VirD2 was tested in vivo using the two-hybrid assay. Fig. 1 shows the positive interaction between AtKAPα and VirD2 in the two-hybrid system. When an unrelated protein C-lamin, known as a nonspecific activator in the two-hybrid system (18), was used as bait, no interaction was observed. Furthermore, AtKAPα also did not interact with VirD2 lacking the NLS (VirD2ΔNLS) (Fig. 1). These results suggest that AtKAPα–VirD2 interaction is likely specific and mediated by the VirD2 NLS.
Figure 1.
VirD2–AtKAPα interaction in the two-hybrid system. The indicated combinations bait/prey were achieved by introducing into the yeast strain Y153 the combinations of the following plasmids: VirD2/AtKAPα, pGBTD2 and pGADAtKAPα; VirD2ΔNLS/AtKAPα, pGBTD2ΔNLS and pGADAtKAPα; lamin C/AtKAPα, pLAM5 and pGADAtKAPα. No prey combinations included the corresponding bait and pGAD424 plasmid without insert. Protein–protein interaction was determined by the β-galactosidase assay on a nitrocellulose filter as described (21). All other experimental conditions were as described for the two-hybrid screen of the cDNA library.
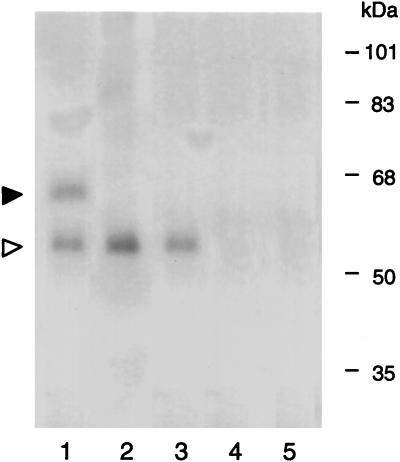
The specificity of the AtKAPa protein interaction was further established in vitro by coimmunoprecipitation. When in vitro-translated AtKAPα and VirD2 were incubated and immunoprecipitated with anti-VirD2 antiserum, coprecipitation of both proteins was observed (Fig. 2, lane 1). Incubation of AtKAPα with VirD2ΔNLS resulted only in precipitation of the VirD2ΔNLS mutant protein (lane 2). No AtKAPα band was detected when VirD2 was incubated alone (lane 3). Fig. 2 also shows that AtKAPα, when incubated alone, was not precipitated using the anti-VirD2 antiserum (lane 4). Finally, following coincubation, neither AtKAPα nor VirD2 was precipitated by the preimmune serum (Fig. 2, lane 5). Thus, AtKAPα most likely directly interacts with the NLS signal of VirD2. An alternative possibility of nonspecific AtKAPα binding to the positively charged NLS sequence is unlikely because several internal regions of VirD2 (e.g., amino acid residues in positions between 289 and 327) also contain numerous basic amino acids that do not promote interaction with AtKAPα and do not function as an NLS.
Figure 2.
VirD2–AtKAPα interaction in vitro. Protein–protein interaction was assayed by coimmunoprecipitation with anti-VirD2 antibody as described. Lane 1, VirD2+AtKAPα; lane 2, VirD2ΔNLS+AtKAPα; lane 3, VirD2 alone; lane 4, AtKAPα alone; lane 5, VirD2+AtKAPα+preimmune serum. Solid and open arrowheads indicate the positions of the radioactively labeled AtKAPα and VirD2 proteins, respectively. Protein molecular mass standards are indicated on right in kDa. Note that the electrophoretic mobility of VirD2 is 56 kDa as described (39), although its molecular mass calculated on the basis of the known DNA sequence is 49,579 Da (40).
Complementation of a Yeast SRP1 Mutation by AtKAPα.
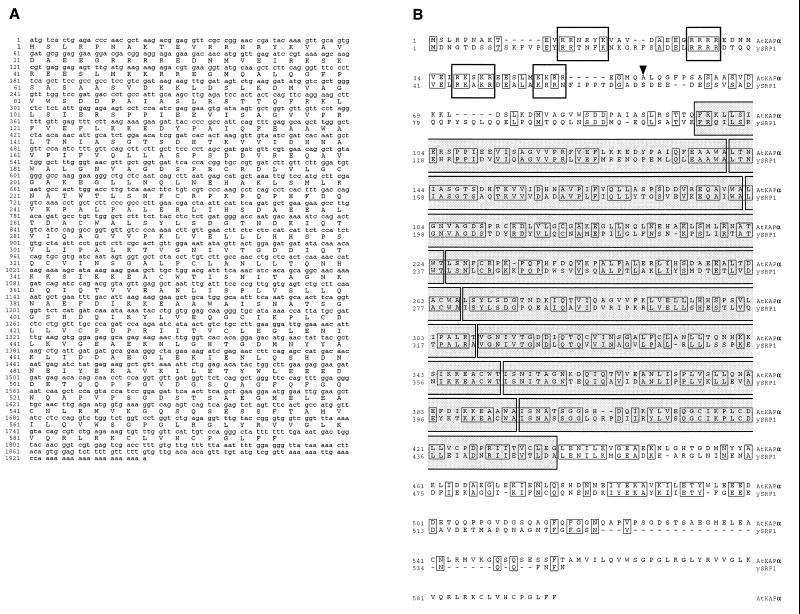
Sequence analysis of the AtKAPα cDNA showed that it contained a single ORF encoding a protein of 65.6 kDa (Fig. 3A). This predicted molecular mass was consistent with the electrophoretic migration of the in vitro translated AtKAPα at 66 kDa (Fig. 2, lane 1). The deduced amino acid sequence of AtKAPα was homologous to animal karyopherin α (data not shown) and yeast SRP1 proteins (Fig. 3B) that function as NLS receptors (refs. 29 and 30 and references therein). Specifically, AtKAPα (Arabidopsis thaliana karyopherin α) contained two features characteristic of this protein family. It carried eight contiguous repeats of the “arm motif” first identified in the Drosophila armadillo protein (31) and then in yeast SRP1 (32) (Fig. 3B). AtKAPα also contained four clusters of basic amino acids in its amino-terminal region (Fig. 3B).
Figure 3.
Nucleotide sequence of the AtKAPα cDNA (A) and alignment of its deduced amino acid sequence with the yeast SRP1 protein (B). The GenBank accession numbers for AtKAPα and SRP1 are U69533 and M75849, respectively. Alignment was performed by the clustal algorithm (41). Regions of identity are indicated by narrow boxes, gaps introduced for alignment are indicated by dashes. Wide open boxes indicate the amino-terminal basic regions and wide shaded boxes indicate eight “arm” motifs (32). Arrowhead indicates the position of the cdc2 kinase site absent in AtKAPα and SRP1 but found in animal karyopherin α proteins (38).
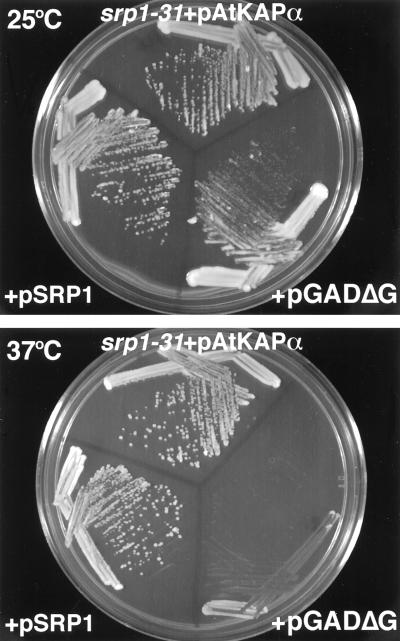
Amino acid sequence alignment of AtKAPα with yeast SRP1 (Fig. 3B) showed 44% identity between these proteins. It is possible, therefore, that AtKAPα functions similarly to SRP1 that is required for nuclear import and mitosis in yeast cells (25). To test this hypothesis, we expressed AtKAPα in a temperature sensitive srp1-31 yeast mutant. In this strain, the SRP1 protein is inactivated when the cells are grown at the restrictive temperature (37°C), resulting in arrest at the G2/M phase of the cell cycle (25). Fig. 4 shows that while the srp1-31 cells transformed with the expression vector alone (pGADΔG) grew normally at 25°C, their growth was dramatically inhibited at 37°C. This conditional lethal phenotype was completely reversed by constitutive expression of AtKAPα (Fig. 4). The degree of complementation was very high since the cell growth at 37°C following expression of AtKAPα was identical to that after expression of the wild-type SRP1 protein (Fig. 4).
Figure 4.
Complementation of the srp1-31 mutation by AtKAPα. The yeast srp1-31 strain was transformed with either pAtKAPα, pGADΔG, or pSRP1. The transformants were plated on selective medium and grown for 2 days at either 25°C (Upper) or 37°C (Lower).
AtKAPα Transports VirD2 into the Nuclei of Permeabilized Yeast Cells.
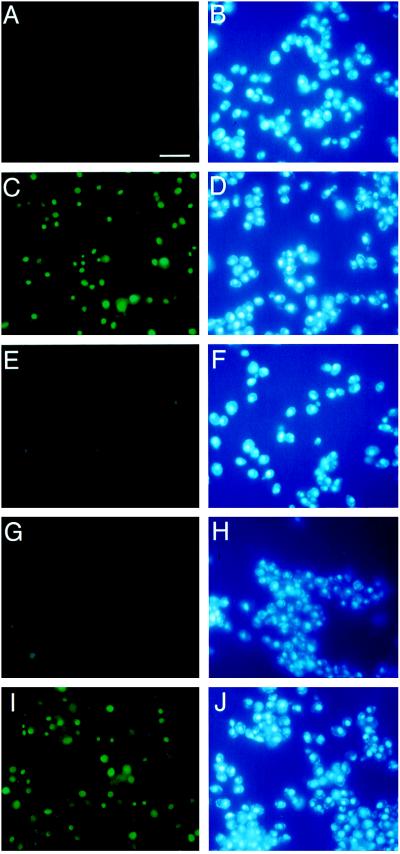
Because AtKAPα may function as the host cell receptor for VirD2 during the T-DNA transport, it was essential to test whether this protein is directly involved in VirD2 nuclear import. Presently, no plant experimental system exists in which the role of isolated components of protein nuclear import machinery can be tested in vitro. Complementation of the yeast srp1-31 phenotype by AtKAPα suggested that this protein may also function in an in vitro permeabilized yeast cell assay (26). Permeabilized cells were prepared from the srp1-31 strain grown at 25°C and shifted to 37°C for 6 hr. Under these conditions, the srp1-31 mutant loses >95% of its nuclear import ability (25). Cytosolic fractions for the in vitro assay were prepared from the srp1-31 cells that were shifted to 37°C while expressing AtKAPα, wild-type SRP1, or harboring the expression vector alone. When cytosol from cells containing vector alone was mixed with permeabilized cells and the fluorescently labeled VirD2, the latter was excluded from the cell nuclei (Fig. 5 A and B). However, when cytosol from cells expressing AtKAPα was used in the same type of experiment, the fluorescently labeled VirD2 was imported into the yeast nuclei (Fig. 5C), colocalizing with the nucleus-specific stain, 4′,6-diamidino-2-phenylindole (Fig. 5D).
Figure 5.
AtKAPα promotes nuclear import of VirD2 in permeabilized yeast cells. Nuclear import assays were performed as described in Materials and Methods. The following combinations of cytosolic fractions and the fluorescently labeled protein were added to the permeabilized srp1-31 cells: A and B, VirD2+cytosol from cells expressing pGADΔG; C and D, VirD2+cytosol from cells expressing pAtKAPα; E and F, VirD2ΔNLS+cytosol from cells expressing pAtKAPα; G and H, VirD2+cytosol from cells expressing pAtKAPα+ 2 μg of a synthetic VirD2 NLS peptide; I and J, VirD2+cytosol from cells expressing pSRP1. A, C, E, G, and I, fluorescein-labeled protein; B, D, F, H, and J, 4′,6-diamidino-2-phenylindole staining. The one-letter code amino acid sequence of the VirD2 NLS is KRpredddgepseRKReR with basic residues of the first and second domains of the bipartite signal indicated in uppercase (11). (Bar = 50 μm.)
To demonstrate the specificity of the AtKAPα function in nuclear import, fluorescently labeled VirD2 lacking the NLS (VirD2ΔNLS) was mixed with cytosol from cells expressing AtKAPα and permeabilized cells. The VirD2ΔNLS protein remained cytoplasmic (Fig. 5 E and F), indicating that AtKAPα promotes nuclear import of VirD2 through an NLS-dependent pathway. Consistent with this idea, a synthetic peptide corresponding to the VirD2 NLS efficiently blocked nuclear import, probably competing with the fluorescently labeled VirD2 for interaction with AtKAPα (Fig. 5 G and H). As in the in vivo complementation experiments (Fig. 4), the activity of AtKAPα in vitro (number of permeabilized cells showing nuclear import of VirD2) was comparable to that of the wild-type SRP1 (Fig. 5 I and J).
DISCUSSION
Agrobacterium–plant cell interaction is a fascinating process comprising a wide variety of biological reactions that ultimately result in genetic transformation of the host plant cell. One of the critical steps in the transformation event involves transport of the T-DNA molecule into the host cell nucleus. Presumably, the T-DNA is imported into the nucleus as a protein–nucleic acid T-complex composed of a T-strand DNA molecule, and two bacterial proteins, VirD2 and VirE2, which protect the T-strand, shape it into a transferable form, and supply the targeting NLS sequences (reviewed in ref. 5).
VirD2 performs perhaps the most numerous functions among all Agrobacterium proteins involved in T-DNA transport. In addition to its role in the T-complex nuclear import, VirD2 is a part of the endonuclease activity directly involved in cleavage of the T-DNA borders and production of the T-strand. During this process, VirD2 covalently attaches to the 5′ end of the T-strand molecule, becoming an integral part of the T-complex. Finally, VirD2 has a ligase activity, potentially involved in the T-DNA integration into the host cell genome (reviewed in ref. 5). Because VirD2 is a central player in the process of Agrobacterium–plant cell interaction, it was important to identify its cellular receptor. Using the yeast two-hybrid system to probe protein–protein interaction (28), we have isolated an Arabidopsis cDNA that encodes a VirD2-interacting protein.
The deduced amino acid sequence of this protein, designated AtKAPα, was homologous to the yeast NLS-binding protein SRP1 and other members of the growing family of karyopherin α-like proteins (29, 30). Karyopherin α acts as a subunit of an NLS receptor complex in which it (i) binds NLS-containing proteins to be transported into the cell nucleus, and (ii) associates with the second subunit, karyopherin β, which mediates docking at the nuclear pore (33, 34). So far, none of the functional nuclear transport components have been identified in plants. Recently, a cDNA encoding an Arabidopsis homolog (aIMPα) of vertebrate and yeast NLS receptors has been isolated (35); however, the biological activity of aIMPα has not been demonstrated. Furthermore, although most of the amino acid sequence of AtKAPα is almost identical to that of aIMPα (35), the carboxyl-terminal part of AtKAPα has a 64 amino acid-long domain not found in aIMPα (amino acid residues in positions 533 to 596; see Fig. 3 and ref. 35). Thus, AtKAPα likely represents a new member of the karyopherin α/importin α family in Arabidopsis. AtKAPα bears two hallmarks of this protein family: (i) It contains eight contiguous repeats of the “arm motif” (31, 32) that may function as a binding site for NLS sequences (33). (ii) The amino terminus of AtKAPα contains four clusters of basic amino acids that are involved in binding to the karyopherin β subunit (33). The presence of these functional domains in AtKAPα suggests that it may mediate nuclear import of VirD2 during the T-DNA transport process.
Direct interaction of AtKAPα with VirD2 was tested in vivo and in vitro. AtKAPα-VirD2 binding was examined using the yeast two-hybrid system as well as coimmunoprecipitation. In both approaches, AtKAPα bound VirD2 specifically. Furthermore, the binding was absolutely dependent on the presence of the bipartite NLS sequence in VirD2, indicating that AtKAPα recognized this signal. Interestingly, the presence of a basic region homologous to a monopartite NLS in the amino-terminal part of the protein (36, 37) was not sufficient for interaction with AtKAPα, consistent with the previous observation that only the carboxyl-terminal bipartite NLS is functionally active (11).
Involvement of AtKAPα in nuclear import was also demonstrated both in vivo and in vitro. First, this Arabidopsis protein was shown to complement a yeast mutant with a defective SRP1. Complementation with AtKAPα was very efficient and indistinguishable from that by the wild-type SRP1. Interestingly, AtKAPα is the first nonyeast member of the karyopherin α family that complemented a yeast SRP1 mutation. The absence of the potential cdc2 kinase (p34) site in both AtKAPα and SRP1 proteins may underlie this complementation ability. Phosphorylation of animal karyopherins α at a conserved p34 phosphorylation site has been proposed to regulate their nuclear entry in a cell cycle-dependent fashion (30, 38). In contrast, SRP1 that lacks the p34 recognition sequence is located in the yeast cell nucleus throughout the cell cycle (38). Thus, AtKAPα that also does not carry this conserved phosphorylation site may be correctly localized within the yeast cell, allowing complementation of the SRP1 function.
That AtKAPα functioned in yeast allowed us to use permeabilized yeast cells (26) as an in vitro system to examine nuclear import of VirD2 by AtKAPα. Our results show that AtKAPα actively imported VirD2 into the yeast cell nuclei and that this transport was mediated by the VirD2 NLS. Based on these observations, we suggest that the Arabidopsis protein AtKAPα is a cellular receptor for Agrobacterium VirD2. During Agrobacterium infection, AtKAPα probably recognizes VirD2 and promotes its nuclear import. Because VirD2 is tightly associated with the invading T-strand (reviewed in ref. 5), AtKAPα likely mediates nuclear import of the Agrobacterium T-DNA as well. Unlike VirD2, VirE2, which is the second NLS-containing protein component of the T-complex (reviewed in ref. 5), did not interact with AtKAPα in the two-hybrid assay (data not shown). Thus, another, as yet unidentified, plant protein that recognizes VirE2 NLSs may be involved in the T-complex nuclear import. In uninfected plants, AtKAPα may function as an NLS-binding protein mediating transport of the nuclear proteins of the cell. Amino acid homology between the plant NLS receptor AtKAPα and animal and yeast karyopherins α suggests evolutional conservation of the nuclear import pathway among these organisms.
Acknowledgments
We thank Nancy Hollingsworth for help with the design of complementation experiments. This work was supported by grants from National Institutes of Health (Grant R01-GM50224), U.S. Department of Agriculture (Grant 94-02564), U.S.–Israel Binational Research and Development Fund (Grant US-2247-93), and Center for Biotechnology at Stony Brook, funded by the New York State Science and Technology Foundation, to V.C.
ABBREVIATIONS
- Ti
tumor inducing
- T-DNA
the portion of the Ti plasmid that is transferred to plant cells
- NLS
nuclear localization signal
- T
transport
- T-strand
transported single-stranded copy of the T-DNA
- aIMPα
Arabidopsis import in α
- AtKAPα
Arabidopsis thaliana karyopherin α
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U69533).
References
- 1.Tinland B, Hohn B, Puchta H. Proc Natl Acad Sci USA. 1994;91:8000–8004. doi: 10.1073/pnas.91.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yusibov V M, Steck T R, Gupta V, Gelvin S B. Proc Natl Acad Sci USA. 1994;91:2994–2998. doi: 10.1073/pnas.91.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stachel S E, Timmerman B, Zambryski P. Nature (London) 1986;322:706–712. [Google Scholar]
- 4.Hooykaas P J J, Beijersbergen A G M. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 5.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hainfeld J F, Wall J S, Desmond E J. Ultramicroscopy. 1982;8:263–270. doi: 10.1016/0304-3991(82)90242-x. [DOI] [PubMed] [Google Scholar]
- 7.Howard E A, Citovsky V. BioEssays. 1990;12:103–108. [Google Scholar]
- 8.Howard E A, Citovsky V, Zambryski P. UCLA Symp Mol Cell Biol. 1990;129:1–11. [Google Scholar]
- 9.Guralnick B, Thomsen G, Citovsky V. Plant Cell. 1996;8:363–373. doi: 10.1105/tpc.8.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zupan J, Citovsky V, Zambryski P. Proc Natl Acad Sci USA. 1996;93:2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard E, Zupan J, Citovsky V, Zambryski P. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 12.Citovsky V, Warnick D, Zambryski P. Proc Natl Acad Sci USA. 1994;91:3210–3214. doi: 10.1073/pnas.91.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1803–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 14.Rossi L, Hohn B, Tinland B. Mol Gen Genet. 1993;239:345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 15.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 16.Howard E A, Winsor B A, De Vos G, Zambryski P. Proc Natl Acad Sci USA. 1989;86:4017–4021. doi: 10.1073/pnas.86.11.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraft R, Tardiff J, Kranter K S, Leinwand L A. BioTechniques. 1988;6:544–547. [PubMed] [Google Scholar]
- 18.Bartel P, Chien C, Sternglanz R, Fields S. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 19.McClary J A, Witney F, Geisselsoder J. BioTechniques. 1989;7:282–289. [PubMed] [Google Scholar]
- 20.Yano R, Oakes M, Yamigishi M, Dodd J A, Nomura M. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 22.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Tomashow M, Retzel E, Sommerville C. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean B G, Zupan J, Zambryski P. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Loeb J D J, Schlenstedt G, Pellman D, Kornitzer D, Silver A P, Fink G R. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlenstedt G, Hurt E, Doye V, Silver P. J Cell Biol. 1993;123:785–798. doi: 10.1083/jcb.123.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Citovsky V, Wong M L, Zambryski P. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields S, Song O-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 29.Weis K, Mattaj I W, Lamond A I. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 30.Kussel P, Frasch M. J Cell Biol. 1995;129:1491–1507. doi: 10.1083/jcb.129.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riggleman R, Wieschaus E, Schedl P. Genes Dev. 1989;3:96–113. doi: 10.1101/gad.3.1.96. [DOI] [PubMed] [Google Scholar]
- 32.Peifer M, Berg S, Reynolds A B. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 33.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 34.Gorlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 35.Hicks G R, Smith H M S, Lobreaux S, Raikhel N V. Plant Cell. 1996;8:1337–1352. doi: 10.1105/tpc.8.8.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tinland B, Koukolikova-Nicola Z, Hall M N, Hohn B. Proc Natl Acad Sci USA. 1992;89:7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrera-Estrella A, Van Montagu M, Wang K. Proc Natl Acad Sci USA. 1990;87:9534–9537. doi: 10.1073/pnas.87.24.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kussel P, Frasch M. Mol Gen Genet. 1995;248:351–363. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]
- 39.Yanofsky M F, Porter S G, Young C, Albright L M, Gordon M P, Nester E W. Cell. 1986;47:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- 40.Rogowsky P M, Powel B S, Shirasu K, Lin T-S, Morel P, Zyprian E M, Steck T R, Kado C I. Plasmid. 1990;23:85–106. doi: 10.1016/0147-619x(90)90028-b. [DOI] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]