Abstract
A thiol-acrylate photopolymerization was used to incorporate enzymatically cleavable peptide sequences into PEG hydrogels to induce chondrogenic differentiation of encapsulated human mesenchymal stem cells (hMSCs). An adhesive sequence, RGD, was designed with an MMP-13 specific cleavable linker. RGD promotes survival of hMSCs encapsulated in PEG gels and has shown to induce early stages of chondrogenesis, while its persistence can limit complete differentiation. Therefore, an MMP-13 cleavage site was incorporated into the peptide sequence to release RGD mimicking the native differentiation timeline. Active MMP-13 production of encapsulated hMSCs was seen to increase from days 9–14 and only in chondrogenic differentiating cultures. Seeded hMSCs attached to the material prior to enzymatic cleavage, but a significant population of the cells detach after cleavage and release of RGD. Finally, hMSCs encapsulated in RGD-releasing gels produce 10 times as much glycosaminoglycan as cells with uncleavable RGD functionalities, by day 21 of culture. Furthermore, 75% of the cells stain positive for collagen type II deposition where RGD is cleavable, as compared to 19% for cultures where RGD persists. Collectively, this data provides evidence that temporal regulation of integrin-binding peptides is important in the design of niches in differentiating hMSCs to chondrocytes.
Keywords: human mesenchymal stem cells, RGD, collagenase-3, chondrogenesis
1 Introduction
Hydrogels synthesized from the chain polymerization of di(meth)/acrylated block copolymers of poly(ethylene glycol) (PEG) and poly(lactic acid) (PLA) are widely used for cell encapsulation, where the hydrolytic degradation is often tuned to allow for cell proliferation and extracellular matrix (ECM) deposition[1–3]. While hydrolytically degradable sequences show some promise with their degree of tunability, the time line for degradation does not always coincide with cellular function. Therefore, enyzmatically degradable materials are quickly evolving to exploit the activity of the cell to alter material properties.
Enzymatically cleavable materials exploit the upregulation and downregulation of cell-secreted enzymes, such as matrix-metalloproteases (MMPs) or other proteases like plasmin(4), to dictate material degradation. These gels are often tailored with a peptide sequence containing an MMP specific cleavage site. Hydrogels are often crosslinked with the MMP-containing peptide sequence, such that the cell-secreted enzyme degrades the gel on a local and cellular timescale[4, 5]. Cellularly-remodeled materials provide specific advantages for the synthesis of highly functional materials that begin to capture aspects of the temporally regulated cues found in development, would healing, and even stem cell niches.
In this regard, studies have shown that human mesenchymal stem cells (hMSCs) upon differentiation into chondrocytes require induction of cell-signaling pathways achieved through their binding to fibronectin[6, 7]. Gene expression profiles suggest that within the first week of differentiation, hMSCs upregulate fibronectin production, which provides an adhesive site for cell condensation, cell-signaling and eventually the early stages of chondrogenic differentiation [7, 8]. After one week, this high level of fibronectin is downregulated, and the cells are free to conform into a spherical shape, which is necessary for complete chondrogenic differentiation[6]. Therefore, the presence of fibronectin or its major binding site, RGD (arginine-lysine-aspartic acid), plays an important role in initiating chondrogenic differentiation[6, 7, 9], as well as maintaining hMSC viability in PEG and other types of hydrogels[9, 10]. While previous results demonstrate that RGD interacts with hMSCs and initiates chondrogenic differentiation, persistence of this molecule inhibits the chondrogenic differentiation of these cells[11]. Thus, we hypothesized that a cellularly dictated mechanism to temporally regulate the persistence of the RGD functionality with time would facilitate chondrogenic differentiation and extracellular matrix deposition.
Studies show that the upregulation of matrix metalloprotease 13 (MMP-13), also known as collagenase-3, occurs within days 7–12 of hMSC chondrogenesis[12]. Further, the upregulation of this enzyme coincides with the cell’s downregulation of fibronectin as seen in vivo(7, 12)]. MMP-13 is a highly active enzyme often seen in patients suffering from osteoarthritis, which is known to degrade collagen, decorin, biglycan, aggrecan and various components of the cartilage ECM[13–15]. A major MMP-13 cleavage site found on aggrecan, a cartilage ECM component, is PENFF (proline-glutamic acid-asparagine-phenylalanine-phenylalanine)[16, 17].
In this work we designed a peptide sequence, incorporating the MMP-13 major cleavage site found on aggrecan (PENFF) and the major fibronectin binding site (RGD). This peptide sequence was then covalently bound to a PEG gel via a thiol-acrylate polymerization mechanism[18]. Specifically, hMSCs were encapsulated in PEG-diacrylate gels copolymerized with cysteine containing peptides, either a persistent, uncleavable RGD tether, CRGDSG, or the cleavable RGD tether, CPENFFGGRGDSG. Cell/gel constructs were cultured in either control stem cell media or chondrogenic differentiation media, and cell viability, chondrogenic differentiation assays and immunostaining techniques were employed to study the effects of RGD release from the system on hMSC function and differentiation.
2 Materials and Methods
2.1 Cell Culture
Adult human mesenchymal stem cells (hMSCs) were obtained from Cambrex Bio Science and plated at 5,000 cells/cm2 in 10 cm tissue culture polystyrene dishes. The hMSCs were cultured in stem cell media (10% fetal bovine serum, 1µg/ml amphotericin B, 50 U/ml penicillin, 50 µg/ml streptomycin, and 20 µg/ml gentamicin in Dulbecco’s modified Eagle medium containing 1 g/L glucose (DMEM-LG)) and grown under standard cell culture conditions (i.e., in a 37°C incubator with 5% CO2). Cells were grown to confluency with media changes twice a week and either passaged, seeded or encapsulated into polymer constructs.
2.2 Peptide and Macromer Synthesis
A 12-mer peptide containing both a MMP-13 specific aggrecan binding domain and an RGDS sequence was synthesized using a peptide synthesizer (Applied Biosystems, model 433A). The peptide sequence, CPEN↓FFGRDGSG, (cysteine, proline, glutamic acid, asparagine, phenylalanine, phenylalanine, glycine, arginine, aspartic acid, glycine, serine, glycine) (as seen in Figure 1) was synthesized, cleaved and deprotected using trifluoro acetic acid (TFA) (Sigma), phenol, and triisopropylsilane (TIPS) (Sigma). The final product was ether precipitated, lyophilized, and the sequence verified using matrix-assisted laser desorption/ionization (MALDI). Similar methods were used to synthesize an uncleavable peptide sequence, CRGDSG.
Figure 1.
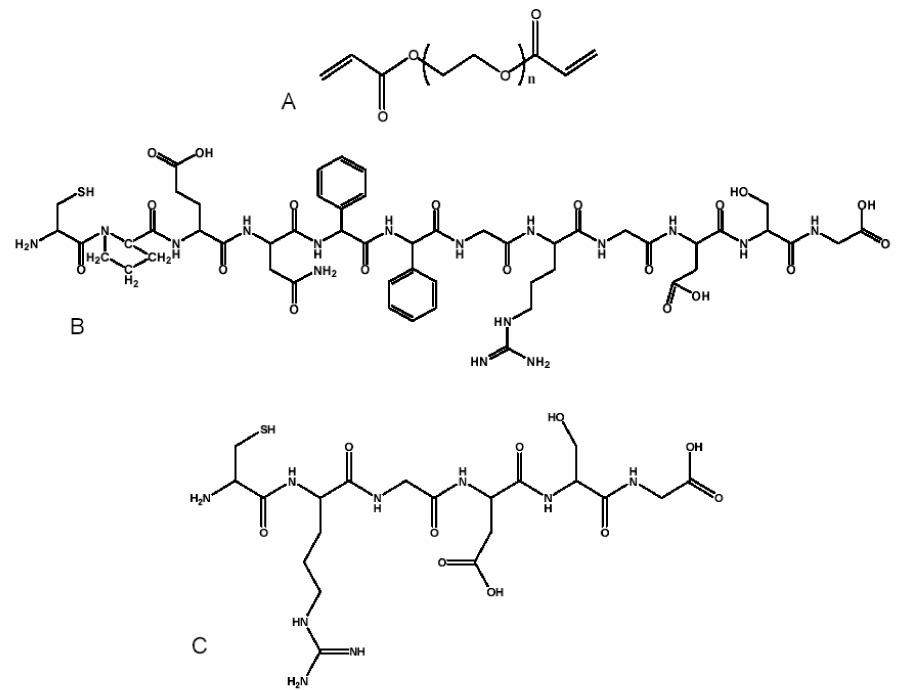
Chemical structures for PEGDA (A), CPENFFGRGDSG (B) and CRGDSG (C).
Poly(ethylene) glycol of M̄n 4600 Da (PEG4600, Sigma) was purchased and modified upon receiving. Twenty grams of PEG was dissolved in a methylene chloride/triethylamine solution before a dropwise addition of the methylene chloride/acrolyl chloride mixture. The reaction mixture was allowed to react overnight and equilibrate to room temperature. Excess TEA salts were removed through rotovapping and filtering the PEGDA in cold ethyl ether. The precipitate was dried overnight in a vacuum oven, redissolved in DI H2O, dialyzed and lyophilized before use. H-NMR was used to verify ~88% acrylation of the final product (PEGDA).
The peptide sequences were incorporated into the PEG macromer networks through photoinitiated mixed-mode thiol-acrylate addition as described previously[18]. All chemical components used in these studies can be seen in Figure 1. Briefly, a photoinitiated radical activates both the thiol and acrylate groups leading to a combination of step and chain growth polymerization resulting in network formation. Peptides were incorporated into the gels at concentrations of 0, 5 and 10mM. The reaction proceeded as a specific volume of PEG-peptide macromer solution containing 0.05% of 4-(2-hydroxyethoxy)phenyl-(2-hydroxy-2-propyl)ketone (I2959) photoinitiator, and was allowed to react under 365nm light at an intensity of ~5mW/cm2 for 10 minutes at room temperature.
2.3 Encapsulation of hMSCs and their MMP-13 Production Profile
Adult human mesenchymal stem cells were encapsulated in pure PEG hydrogels, without peptide functionality, at a concentration of 2×106 cells/ml. The hydrogels were polymerized as stated above using 40 µl of the pure PEGDA solution containing 0.05% 4-(2-hydroxyethoxy)phenyl-(2-hydroxy-2-propyl)ketone (I2959) in the tips of 1 ml syringes, creating 5×1 mm disks. Three hydrogels per composition were then cultured in either control stem cell media or chondrogenic media [Dulbecco’s high glucose modified Eagle medium, ITS+ premix (6.25 µg/ml bovine insulin, 6.25 µg/ml transferrin, 6.25 µg/ml selenous acid, 5.33 µg/ml linoleic acid, 1.25 µg/ml bovine serum albumin), 100 nM dexamethasone, 50 µg/ml ascorbic acid 2-phosphate, 100 µg/ml sodium pyruvate, 100 µg/ml penicillin-streptomycin, 1 µg/ml amphotericin B] and supplemented with 5 ng/ml TGFβ1. The cell/polymer constructs were cultured for 3 weeks at 37°C and 5% CO2 with media changes twice weekly.
Cell/polymer constructs were removed from culture at specific time points and analyzed for production of MMP-13 via the EnzoLyte Plus™ 520 MMP-13 Assay Kit (AnaSpec). Following manufacturer’s protocols, the gels were treated with a buffer solution containing 0.1% (v/v) Triton-X 100, centrifuged and the supernatant was collected. Both the treated gels and the media supernatant were analyzed for MMP-13 release. MMP-13 standards and samples were added to an anti-MMP-13 coated microplate and incubated for 2 hours. The wells were then washed and treated with p-aminophenylmercuric acetate (APMA) at 37°C for 40 minutes to activate the enzyme. Finally, the presence and activity of the MMP-13 enzyme was detected by a 5-FAM/QXL™520 peptide substrate. For normalization of the MMP-13 concentration to cell number, DNA was measured through a PicoGreen assay (Pierce).
2.4 Peptide Cleavage and Release through Exogenously Delivered MMP-13
Hydrogels were synthesized as described, except a peptide sequence CPEN↓FFWGGGG was incorporated for investigation of the accessibility and specificity of the cleavage site. A scrambled sequence, CGEFGPNGGWF, was used as a control. A tryptophan amino acid was used in these sequences for detection purposes. Three hydrogels of each composition were placed in PBS overnight, to allow for extraction of any sol fraction. Swollen gels were then removed from the PBS and placed in a new 48-well plate and enzymatically degraded. The MMP-13 enzyme was activated using 1mM p-Aminophenylmercuric Acetate (APMA) at 37°C for 40 minutes. This activated enzyme was then concentrated using Millipore Centricon Plus-20 centrifugation tubes to remove the remaining APMA and finally diluted to concentrations of 0, 50, 100 and 500 ng/ml. Active MMP-13 reacted with the gels overnight for complete cleavage and release of the FFWGGG portion of the peptide sequence. The peptide concentration remaining in the gel, as well as the peptide released into the solution, were then analyzed with a modified ELISA technique described below.
Measurement of a tryptophan residue in the CPEN↓FFGWGGGG sequence and its cleaved fraction were taken before and after enzymatic treatment. The tryptophan release after treatment was normalized to that prior to exposure to the enzyme. Further, investigation into the sequence released was monitored through a modified-indirect ELISA (enzyme-linked immunosorbent assay). Gels were exposed to 0 or 500ng/ml exogenously delivered MMP-13 and allowed to react overnight. Samples were then blocked in 1% BSA for two hours at room temperature. The samples were washed four times with PBS +1% Tween 20 between each of the incubation steps. The primary antibody (anti-fibronectin binding fragment) was diluted 1:50 in blocking solution and allowed to incubate with the samples for two hours at 37°C. A secondary antibody (horseradishperoxidase (HRP) labeled goat-anti-mouse) reacted with the samples for two hours at room temperature. HRP was then quantified using a substrate solution, 1-Step™ Ultra TMB-ELISA as per manufacturer’s protocol (Pierce). RGD was quantified through a comparison to a standard curve produced via the same reaction scheme against known RGD concentrations adsorbed to high binding Immulon plates.
2.5 hMSC Seeding, Attachment and Enzymatic Degradation of Polymer Constructs
PEG-peptide gels (N=5) were synthesized as described above, but with 100 µl of PEG-peptide macromer solution. The gels were cultured in a 48-well plate, and were swollen overnight in PBS. hMSCs were carefully seeded on the center of each gel at a concentration of 2,000 cells/cm2, fed with hMSC basal media and allowed to attach overnight under proper cell culture conditions. Images were collected for all gels prior to enzymatic degradation. The MMP-13 enzyme was activated as described above, exogenously added and allowed to react on the gels for 5 hours before a second image collection. MMP-13 concentrations used in this study were 0, 100 or 500ng/ml. The images were analyzed to determine the initial amount of attachment to the gels compared to controls and final attachment after treatment with enzyme.
2.6 hMSC Encapsulation and Degradation of PEG-Peptide Hydrogels
Human mesenchymal stem cells were encapsulated at a density of 2 million cells/ml into PEG-peptide constructs containing 0 or 10mM concentrations of either uncleavable CRGDSG or cleavable CPEN↓FFGRGDSG. The cell/polymer-peptide constructs were cultured for up to 4 weeks in either control stem cell or chondrogenic differentiation media with media changes twice weekly. At specific time points during the study, hydrogels were removed and analyzed for DNA concentration, glycosaminoglycan production, and hMSC and chondrocyte markers with immunostaining.
Cell/polymer constructs were collected at each time point and digested in a papain solution overnight. The amount of double stranded DNA was quantified through the use of a PicoGreen assay (Pierce). These same constructs were analyzed via the dimethylmethylene blue[19] (DMMB) assay to determine the amount of glycosaminoglycans deposited. Constructs were also collected, fixed and cryosectioned at 40µm for immunostaining purposes. Briefly, the sections were incubated in blocking solution (1% BSA) for two hours prior to treatment with a primary antibody. Some of the sections were treated with anti-αvβ3 or anti-α5β1 primary antibodies raised in mouse to investigate cell-surface integrins. Other sections were treated with anti-CD105 (mouse) together with anti-collagen type II (rabbit). All primary antibodies used in this study were diluted 1:100 and left to react with the sections for two hours in a humidity chamber at 37°C. The samples were washed four times with PBS buffer between each incubation step. The samples stained for integrins were treated with a biotinylated secondary antibody (anti-mouse IgG) for 1 hour, followed by a 30 minute incubation with a HRP complex, and finally a 30 minute treatment with a peroxidase substrate (Vector NovaRed or Vector VIP). The CD105 and collagen samples were treated with fluorescently labeled secondary antibodies (goat anti-rabbit AlexaFluor 633 and goat-anti-mouse AlexaFluor 488) for 1 hour. All sections were then treated with a DAPI counterstain for 10 minutes and imaged using confocal microscopy with a Zeiss Axioplan 2 with LSM 5 Pascal laser and related software. A total of three images (with an average of 50 cells per image) were taken for each gel system at each time point and analyzed for positively stained cells. Cells staining positive were counted and normalized to the total number of cells in each image to determine a percentage of positive cells.
2.7 Statistics
Data collected throughout this study is presented as a mean ± standard deviation of five data samples. A student’s t-test was employed to compare data sets using p values of less than 0.05 in order to determine significance.
3 Results
An enzymatically degradable MMP-13 peptide sequence containing an RGD motif was incorporated as a pendant tether into a PEG network as a means to mimic the native upregulation and downregulation of fibronenctin in the extracellular matrix (ECM) of a differentiating human mesenchymal stem cell (hMSC). Here, we exploit the timeline at which the hMSC secretes MMP-13 to degrade the peptide sequence and release RGD from the network. Theoretically, hMSCs will initially respond to the RGD present in the PEG network; promoting their survival and initiating early stages of chondrogenic differentiation. Once the differentiation process begins, the cells will start secreting MMP-13, which will degrade the peptide sequences in the local environment of the cell, release RGD from the network and allow the hMSCs to complete their chondrogenic differentiation process.
3.1 hMSC Production of Active MMP-13
hMSCs were encapsulated in PEG gels, without the presence of a peptide sequence, and cultured in basal stem cell media or chondrogenic differentiation media. Active MMP-13 enzyme production levels were monitored over a two week time course via a FRET modified ELISA technique based upon a MMP-13 specific kit (AnaSpec). Both the gels and supernatant were analyzed for active MMP-13 production. As seen in Figure 2, those cells cultured in control stem cell media retained basal levels of MMP-13 production over the course of the study. Those cells cultured in chondrogenic media showed an increase in MMP-13 production by ~day 9, which remained constant until day 14. The highest level of active MMP-13 released from the encapsulated cells was 640 ± 10 ng/ml. The upregulation and eventual downregulation of this enzyme follows nicely with the gene expression data published previously[12], where MMP-13 levels increased within 7–12 days of culture for differentiating hMSCs.
Figure 2.
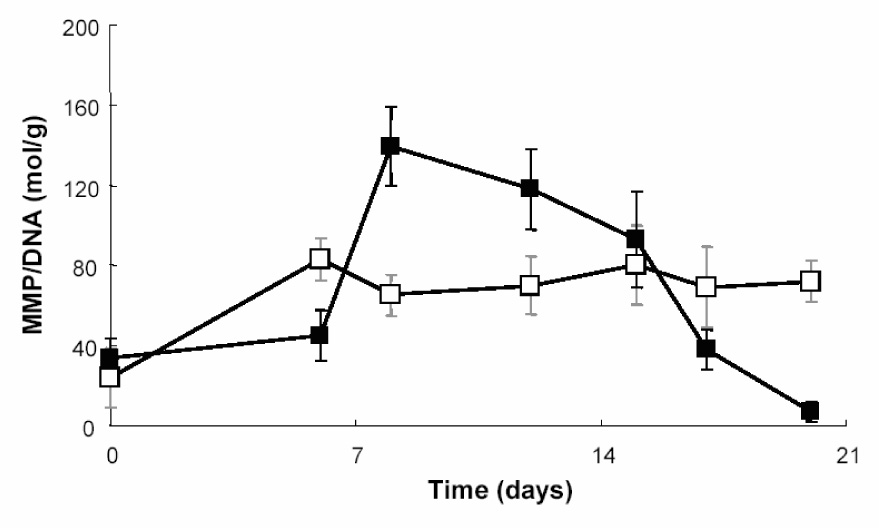
Measurement of active MMP-13 produced by hMSCs encapsulated in plain PEG gels and cultured in either control stem cell media (□) or chondrogenic differentiation media (■). MMP-13 production was normalized to DNA content to better quantify MMP production per cell basis.
3.2 Peptide Characterization
The cleavable peptide sequence, CPEN↓FFWGGGG, containing a tryptophan residue, was characterized first to determine the cleavage specificity and release of the peptide fragment. PEG gels were copolymerized with the cleavable sequence at varying concentrations of 0, 5 and 10mM or a scramble sequence (CGEFGPNGGWF) at 10mM. The amount of tryptophan in the swelling solution prior to enzymatic treatment was determined via UV-Vis measurements at 280nm. The gels were then exogenously treated with 0, 10, 50, 100 and 500ng/ml MMP-13 and left to react overnight. Peptide release was monitored through tryptophan release in the soluble fraction and normalized to initial values (Figure 3A). A loading concentration of 10mM and treatment with 100ng/ml of MMP-13, resulted in ~23% cleavage and release of the peptide fragment, while treatment of the same gel with 500ng/ml MMP-13, reached a release of ~49% in the same time. No release was observed with the pure PEG gel or with the scrambled sequence.
Figure 3.
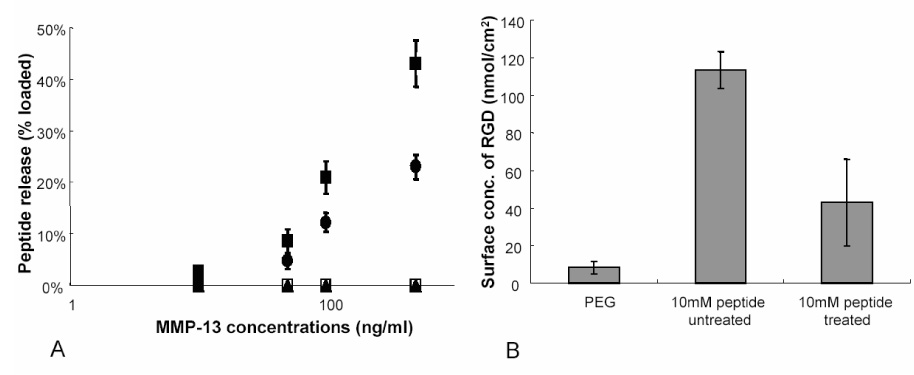
Characterization of peptide release from PEG gels. Varying concentrations of tryptophan containing peptide sequence were copolymerized with PEG and treated with 0, 10, 50, 100 and 500ng/ml exogenously delivered MMP-13. PEG-peptide concentrations were as follows; 10mM (■), 5mM (●), 0mM (□), 10mM scramble sequence (▲). UV-Vis measurements for released tryptophan residue (A) were normalized to loaded concentrations to determine a release profile. (B) RGD ELISA was performed on 10mM peptide containing PEG gels treated with either 0 or 500ng/ml exogenously delivered MMP-13. Surface concentration determining RGD release was fit to a standard curve of known RGD concentrations.
The specific release of an RGD containing fraction was then determined through a modified ELISA technique. Here, the gels were either treated with 500ng/ml of exogenously delivered MMP-13 or left untreated. Known concentrations of peptide were adsorbed onto high binding plates and a standard curve was generated via the same ELISA method. The gels were treated with a primary antibody against the major binding region in FN encompassing RGD. As Figure 3B shows, the control PEG gel shows negligible amounts of RGD background staining. The untreated, 10mM sample had an RGD surface concentration of ~114 nmol/cm2. After enzymatic treatment, the RGD was cleaved and released from the surface of the gel as can be seen with a >50% reduction in RGD measured on the surface. Both release studies verify that the peptide designed with the aggrecan MMP-13 cleavage site and an RGD binding motif can be cleaved on an appropriate time scale and with specificity to the targeted site, altering release of RGD.
hMSCs were seeded on the surface of PEG gels containing 0 or 10mM CPENFFGRGDSG (cleavable sequence) to determine their affinity to the RGD portion of this designed peptide sequence. Cells were allowed to attach to the surfaces overnight, were imaged and counted to calculate an initial cell attachment population. hMSCs seeded on a tissue culture polystyrene plate (TCPS) adhere to the surface with an inherent, elongated and spindly morphology indicative of an undifferentiated, well-maintained hMSC. When hMSCs are seeded on a pure PEG hydrogel, without the presence of an adhesive site the cells ball up and are left to undergo apoptosis due to lack of attachment to the surface. Approximately 74% of hMSCs initially seeded on the PEG gels, containing 10mM of the cleavable peptide, (prior to enzymatic treatment) remained attached and spread on the surface of these gels. After treatment with 100ng/ml MMP-13, 27% ± 4% of the adhered hMSCs detached. When treated with 500ng/ml, 63% ± 6% of the hMSCs had detached from the gel, as compared to the number of cells initially attached. These simple cell attachment studies demonstrate that the hMSCs respond to the presence and absence or removal of RGD from these gels through initial adhesion and ultimately a large portion of the cells detaching from the surface.
3.4 hMSC Chondrogenic Differentiation Via MMP-13 Cleavable Sites
hMSCs were encapsulated in PEG gels containing either an uncleavable RGD tether (CRGDSG) or a cleavable RGD tether (CPENFFGRGDSG) at 0 or 10mM concentrations. The cells were analyzed for viability over the course of the study (Figure 4A). DNA was measured and normalized to day 0. Those hMSCs cultured in the presence of uncleavable RGD in both control and chondrogenic media show a slight decrease in viability reaching ~68% after 24 days. hMSCs in chondrogenic media encapsulated in cleavable RGD platforms maintain their viability through day 11, just as the hMSCs in uncleavable RGD cultures; however, once MMP-13 production ensues and RGD is cleaved, there is a decrease in viable cells reaching an ~34% survival rate after day 24. Interestingly, those hMSCs in cleavable cultures with control media lose viability considerably after day 11 and are found to have ~9% of their population viable after 24 days.
Figure 4.
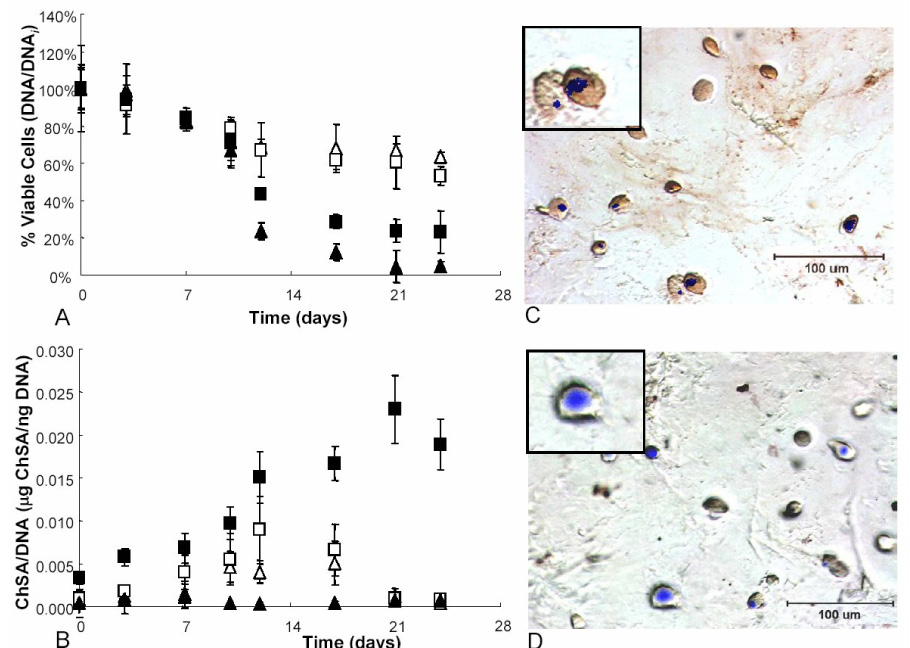
hMSCs were entrapped in PEG-peptide gels in either control (▲) or chondrogenic (■) media. The composition of the gels was either PEG with 10mM CRGDSG (open symbols) or PEG with 10mM CPENFFGRGDSG (closed symbols). The cell constructs were analyzed for viability as measured via DNA amounts and normalized to the initial time point (A). These cell/gel materials were also analyzed for glycosaminoglycan deposition (B). Finally the cell surface integrin (α5β1) was stained in the cleavable RGD chondrogenic culture on day 3 (C) and day 24 (D) to determine the regulation of this adhesion binding complex. Heavy red staining on day 3 is indicative of integrin surface markers, where little to no red staining on day 24 indicates that this marker has been downregulated by the cell.
Further investigation into chondrogenic differentiation of these hMSCs when exposed to environments which aim to mimic the inherent upregulation and downregulation of fibronectin (or RGD) in culture was also conducted. As seen in Figure 4B, glycosaminoglycan (GAG) production was monitored for all culture platforms. hMSCs in both uncleavable RGD and cleavable RGD platforms cultured in control media show basal level GAG deposition of 0.003 µg ChSA/ng DNA. The hMSCs in uncleavable RGD gels in chondrogenic culture show an increasing trend in GAG production reaching ~0.01 µg ChSA/ng DNA by day 10. This trend is similar to that of cells in chondrogenic cleavable RGD cultures until day 13, where the hMSCs in cleavable gels continues to increase to a level of 0.025 µg ChSA/ng DNA, through day 21. By day 21 of culture, cells in cleavable RGD gels in chondrogenic culture produce 10 times more GAG than those cells in uncleavable RGD chondrogenic culture. The GAG deposition as seen by cells in cleavable RGD chondrogenic cultures slightly decreases to 0.002 µg ChSA/ng DNA by day 24. Furthermore, cell/gel constructs were immunostained to determine the population of undifferentiated hMSCs (via the CD105 integrin) versus the population of cells which deposited collagen type II, indicative of chondrogenic differentiation (Table 1). Sections on day 3 show 100% ± 1% CD105 staining for all culture platforms. By day 24 of culture, those cells in uncleavable control culture stained 97% ± 3% positive for CD105, whereas those in uncleavable chondrogenic culture stained 54% ± 4% positive for CD105 and 46% ± 6% positive for collagen type II deposition. hMSCs in cleavable platforms cultured in control media stained 98% ± 2% positive for CD105, of the remaining viable cells, whereas the cells in chondrogenic culture stained 24% ± 4% positive for CD105 and 75% ± 7% for collagen type II.
Table 1.
Percentage of cells stained for undifferentiated stem cell marker, CD105, or chondrogenic differentiation marker, collagen type II. Cells were encapsulated in cleavable RGD and uncleavable RGD platforms cultured in control stem cell or chondrogenic differentiation media
| Composition | Media | CD105 positive (%) | collagen type II positive (%) |
|---|---|---|---|
| uncleavable RGD | control | 97±3 | n/a |
| chondrogenic | 54±4 | 46±6 | |
| cleavable RGD | control | 98±2 | n/a |
| chondrogenic | 24±4 | 75±7 |
Finally, cell surface integrins responsible for binding to the RGD motif were also stained and analyzed to determine their upregulation and downregulation in response to the presence or absence of RGD in culture. As seen in Figure 4C & D, hMSCs encapsulated in cleavable RGD chondrogenic platforms were stained for α5β1 integrin expression on day 3 and day 24 of culture. On day 3, there is a heavy red staining of the cells indicative of α5β1 integrin positive cells or a high degree of α5β1 integrin expression. By day 24 of culture, ~12% of the cells continue to stain red for α5β1 integrin expression, whereas the majority of the cells show no staining (Figure 4D). The αvβ3 integrins stained in a similar fashion (data not shown). This demonstrates the cells internal and external response to the presence or absence of RGD in culture.
4 Discussion
Design of a 3D cell culture niche, which incorporates specific and necessary components for the induction and maintenance of cell differentiation, survival and extracellular matrix production, requires an understanding of cell response in vivo. The development of a tissue involves numerous cell-signaling pathways and the constant control of cell secreted extracellular matrix (ECM) components and enzymes[6, 20–23]. Specifically, for the chondrogenic differentiation of human mesenchymal stem cells, studies suggest that there are important cues needed to induce differentiation, namely fibronectin or its adhesive component, RGD[7]. Furthermore, investigations conclude that the persistence of RGD in culture with differentiating hMSCs will hinder the extent of chondrogenesis[11]. Yet, other studies pinpoint an enzyme, MMP-13, which is upregulated for a short period, coinciding with the downregulation and ultimate disappearance of fibroenctin in native cultures[8, 12, 24]. Therefore, a specific gel microenvrionment designed for entrapment of hMSCs for purposes of chondrogenic differentiation and cartilage tissue development must incorporate mimics such as these.
The work presented here studied the effectiveness of MMP-13 release by hMSCs encapsulated in PEG hydrogels when forced down the chondrogenic pathway and the role of RGD release on hMSC chondrogenic differentiation. Initial studies determined the timeline for active MMP-13 production by hMSCs entrapped in pure PEG gels cultured in chondrogenic differentiation, which followed the previous reports regarding upregulated gene expression levels of chondrogenic differentiating hMSCs[12]. Further characterization into the peptide sequence confirmed that RGD was being released from the system through MMP-13 cleavage. Although 100% release of RGD was not attained in this system, this result can be explained through the fact that the half-life of MMP-13 is relatively short, ~1 hour and the size of the enzyme limits its diffusion through the covalently crosslinked PEG network. Therefore, the majority of RGD release was seen from cleavage of this peptide on the surface, where the half-life limitations lessened the degree of release seen (≤50% release with 500ng/ml of MMP-13).
Ultimately, hMSCs were encapsulated in PEG gels containing an uncleavable RGD tether and the designed cleavable RGD sequence to analyze hMSC response to the persistence or eventual absence of RGD in PEG cultures. As other studies suggest, RGD is an important added cue in permissive PEG gels for encapsulated hMSCs, which provides the cells with an adhesive ligand to maintain survival and perhaps initiate chondrogenic differentiation[9, 10]. hMSC viability was analyzed in both culture platforms, and it was seen that the viability of cells in cleavable systems decreased, after the cleavage and release of RGD from the gel. Here, the RGD plays an important role in maintaining high levels of hMSC survival in PEG gels. Once the cells begin producing MMP-13 to cleave and release RGD from their environment, around day 9, a drop in viability ensues. However, those cells in chondrogenic cultures with cleavable RGD maintain a higher rate of cell survival, indicating that these cells may have progressed far enough down the chondrogenic pathway that they are no longer in need of an adhesive RGD motif. While the cells progressing down a chondrogenic pathway are no longer in need of the RGD ligand, a greater number of cells must differentiate to avoid apoptosis. In this system, the RGD and chondrogenic media are the only means to initiate chondrogenic differentiation. To expand the number of cells differentiating, incorporation of other cartilage-specific ECM molecules can be employed such as deocrin[25] or other growth factors. The presentation of these other molecules should aid in furthering chondrogenic differentiation of these cells to ensure that a larger population will survive after the release of RGD from the system.
While this system provides temporal delivery of ECM cues for the chondrogenic differentiation of hMSCs, it has broad applications for the design of many cell delivery niches. Delivery of cells in permissive environments, such as PEG, requires that the cells are provided with much needed extracellular matrix cues, often in the form of peptides[26]. Cell function requires the upregulation and downregulation of numerous signals within the cellular environment[6]. In creating an environment to sustain cell survival and function while delivering cells to an area of tissue damage, a cell’s native environment is often mimicked in these systems. However, the persistence of cues in these gels will often lead to cell signaling cascades that are not regulated as they should be in proper cell differentiation and function. Therefore, it is of great importance to mimic these environments with specific degrees of regulated cues. Here, we present an easy method to introduce cues in a way that can then be specifically adjusted by the cells. The thiol-acrylate polymerization method allows a facile approach to incorporate cysteine-containing peptides into PEG gels at various concentrations and conformations. MMP-degradable peptide sequences have often been used for gel degradation or delivery of ECM cues in the form of growth factors[27]. Knowledge of native cell environments and the rise and fall of cell cues throughout their cellular function timeline is essential for designing a system that can better mimic the environment of a cell. The method for altering cell function in this system combines the use of cell secreted enzymes with the known specific cues, which are upregulated and downregulated. As the cell sees fit, the gel environment and specific cues are released, thereby mimicking a native timeline for directed cell progression.
5 Conclusion
Incorporation of a cell-mediated, enzymatically cleavable peptide sequence into a PEG hydrogel shows promise for enhancing the chondrogenic differentiation of encapsulated hMSCs. The production of MMP-13 by the encapsulated hMSCs reached levels high enough to completely degrade the designed peptide sequence. Once the hMSCs were exposed to the cleavable RGD, early stage chondrogenic differentiation ensued, as seen through a 10-fold increase in GAG production over uncleavable RGD cultures. Furthermore, those hMSCs cultured in gels with the cleavable RGD stained positive for a higher population of chondrogenic differentiated cells than cultures with uncleavable RGD. Therefore, introducing RGD in a cleavable and releasable fashion maintains high levels of hMSC survival and initiates extensive chondrogenic differentiation.
Acknowledgements
Funding for this project was provided by National Institutes of Health (DE12998), as well as a grant from the Graduate Assistantship in Areas of National Need fellowship to CNS. Thanks to Greg Rocheleau for assistance in supplementary work regarding this paper, and to Chuck Cheung for careful review and suggested edits to this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rice MA, Anseth KS. Encapsulating chondrocytes in copolymer gels: bimodal degradation kinetics influence cell phenotype and extracellular matrix development. J Biomed Mater Res A. 2004;70(4):560–568. doi: 10.1002/jbm.a.30106. [DOI] [PubMed] [Google Scholar]
- 2.Nuttelman CR, Henry SM, Anseth KS. Synthesis and characterization of photocrosslinkable, degradable poly(vinyl alcohol)-based tissue engineering scaffolds. Biomaterials. 2002;23(17):3617–3626. doi: 10.1016/s0142-9612(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 3.Bryant SJ, Anseth KS. Controlling the spatial distribution of ECM components in degradable PEG hydrogels for tissue engineering cartilage. J Biomed Mater Res A. 2003;64(1):70–79. doi: 10.1002/jbm.a.10319. [DOI] [PubMed] [Google Scholar]
- 4.Levesque SG, Shoichet MS. Synthesis of enzyme-degradable, peptide-cross-linked dextran hydrogels. Bioconjug Chem. 2007;18(3):874–885. doi: 10.1021/bc0602127. [DOI] [PubMed] [Google Scholar]
- 5.Rizzi SC, Hubbell JA. Recombinant protein-co-PEG networks as cell-adhesive and proteolytically degradable hydrogel matrixes. Part I: Development and physicochemical characteristics. Biomacromolecules. 2005;6(3):1226–1238. doi: 10.1021/bm049614c. [DOI] [PubMed] [Google Scholar]
- 6.DeLise AMF L, Tuan RS. Cellular interactions and signaling in cartilage development. Osteoarthritis and cartilage. 2000;8(5):309–334. doi: 10.1053/joca.1999.0306. [DOI] [PubMed] [Google Scholar]
- 7.Tavella S, Bellese G, Castagnola P, Martin I, Piccini D, Doliana R, et al. Regulated expression of fibronectin, laminin and related integrin receptors during the early chondrocyte differentiation. J Cell Sci. 1997;110(Pt 18):2261–2270. doi: 10.1242/jcs.110.18.2261. [DOI] [PubMed] [Google Scholar]
- 8.Yamada KM. Integrin signaling. Matrix Biol. 1997;16(4):137–141. doi: 10.1016/s0945-053x(97)90001-9. [DOI] [PubMed] [Google Scholar]
- 9.Salinas CN, Cole BB, Kasko AM, Anseth KS. Chondrogenic differentiation potential of human mesenchymal stem cells photoencapsulated within poly(ethylene glycol)-arginine-glycine-aspartic acid-serine thiol-methacrylate mixed-mode networks. Tissue Eng. 2007;13(5):1025–1034. doi: 10.1089/ten.2006.0126. [DOI] [PubMed] [Google Scholar]
- 10.Nuttelman CR, Tripodi MC, Anseth KS. Synthetic hydrogel niches that promote hMSC viability. Matrix Biology. 2005;24:208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Connelly JT, Garcia AJ, Levenston ME. Inhibition of in vitro chondrogenesis in RGD-modified three-dimensional alginate gels. Biomaterials. 2007;28(6):1071–1083. doi: 10.1016/j.biomaterials.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Sekiya I, Vuoristo JT, Larson BL, Prockop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci U S A. 2002;99(7):4397–4402. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monfort J, Tardif G, Reboul P, Mineau F, Roughley P, Pelletier JP, et al. Degradation of small leucine-rich repeat proteoglycans by matrix metalloprotease-13: identification of a new biglycan cleavage site. Arthritis Res Ther. 2006;8(1):R26. doi: 10.1186/ar1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reboul P, Pelletier JP, Tardif G, Cloutier JM, Martel-Pelletier J. The new collagenase, collagenase-3, is expressed and synthesized by human chondrocytes but not by synoviocytes. A role in osteoarthritis. J Clin Invest. 1996;97(9):2011–2019. doi: 10.1172/JCI118636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tardif G, Reboul P, Pelletier JP, Martel-Pelletier J. Ten years in the life of an enzyme: the story of the human MMP-13 (collagenase-3) Mod Rheumatol. 2004;14(3):197–204. doi: 10.1007/s10165-004-0292-7. [DOI] [PubMed] [Google Scholar]
- 16.Deng S, Bickett D, Mitchell J, Lambert M, Blackburn R, Carter H, et al. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. Jounral of Biological Chemistry. 2000;275(40):31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 17.Fosang AJ, Last K, Knauper V, Murphy G, Neame PJ. Degradation of cartilage aggrecan by collagenase-3 (MMP-13) FEBS Lett. 1996;380(1–2):17–20. doi: 10.1016/0014-5793(95)01539-6. [DOI] [PubMed] [Google Scholar]
- 18.Salinas CN, Anseth KS. Characterization of Thiol-Acrylate Mixed-Mode Photopolymerization for the incorporation of Peptides into a PEG gel. submitted Macromolecues. 2008 [Google Scholar]
- 19.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883(2):173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 20.Albelda SB CA. Integrins and other cell adhesion molecules. Faseb Journal. 1990;4:2868–2880. [PubMed] [Google Scholar]
- 21.Alliston T, Derynck R. Transforming growth factor-b in skeletal development and maintenance. Skeletal Growth Factors. 2000:233–249. [Google Scholar]
- 22.Frenz DA, Jaikaria NS, Newman SA. The mechanism of precartilage mesenchymal condensation: a major role for interaction of the cell surface with the amino-terminal heparin-binding domain of fibronectin. Dev Biol. 1989;136(1):97–103. doi: 10.1016/0012-1606(89)90133-4. [DOI] [PubMed] [Google Scholar]
- 23.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee GML RF. Interactions of the Chondrocyte with its Pericellular Matrix. Cells and Materials. 1998;8:135–149. [Google Scholar]
- 25.Salinas CN, Anseth KS. Decorin moieties tethered into PEG networks induce chondrogenesis of human mesenchymal stem cells. submitted J Biomed Mater Res. 2008 doi: 10.1002/jbm.a.32112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang SL K-F, Du A, Chua C-K. The design of scaffolds for use in tissue engineering, Part I. Traditional factors. Tissue Engineering. 2001;7:679–689. doi: 10.1089/107632701753337645. [DOI] [PubMed] [Google Scholar]
- 27.Lutolf MP, Weber FE, Schmoekel HG, Schense JC, Kohler T, Muller R, et al. Repair of bone defects using synthetic mimetics of collagenous extracellular matrices. Nat Biotechnol. 2003;21(5):513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]


