Abstract
Background
The ParA/Soj and ParB/Spo0J proteins, and the cis-acting parS site, participate actively in chromosome segregation and cell cycle progression. Genes homologous to parA and parB, and two putative parS copies, have been identified in the Mycobacterium bovis BCG and Mycobacterium smegmatis chromosomes. As in Mycobacterium tuberculosis, the parA and parB genes in these two non-pathogenic mycobacteria are located near the chromosomal origin of replication. The present work focused on the determination of the transcriptional organisation of the ~6 Kb orf60K-parB region of M. bovis BCG and M. smegmatis by primer extension, transcriptional fusions to the green fluorescence protein (GFP) and quantitative RT-PCR.
Results
The parAB genes were arranged in an operon. However, we also found promoters upstream of each one of these genes. Seven putative promoter sequences were identified in the orf60K-parB region of M. bovis BCG, whilst four were identified in the homologous region of M. smegmatis, one upstream of each open reading frame (ORF).
Real-time PCR assays showed that in M. smegmatis, mRNA-parA and mRNA-parB levels decreased between the exponential and stationary phases. In M. bovis BCG, mRNA-parA levels also decreased between the exponential and stationary phases. However, parB expression was higher than parA expression and remained almost unchanged along the growth curve.
Conclusion
The majority of the proposed promoter regions had features characteristic of Mycobacterium promoters previously denoted as Group D. The -10 hexamer of a strong E. coli σ70-like promoter, located upstream of gidB of M. bovis BCG, overlapped with a putative parS sequence, suggesting that the transcription from this promoter might be regulated by the binding of ParB to parS.
Background
Partitioning systems were first characterised in low copy number plasmids of Escherichia coli. In general, plasmid partition modules encode two trans-acting proteins and a cis-acting, centromere-like DNA sequence required for partitioning [1]. E. coli plasmid P1 and F factor partitioning systems encode: i) homologous ATPases (ParA/SopA), characterised by a conserved 'deviant' Walker A motif [2]; and ii) site-specific DNA-binding proteins containing helix-turn-helix (HTH) motifs (ParB/SopB) [3]. The centromere-like sites, parS and sopC, are located downstream of the genes encoding the trans-acting proteins [4,5]. Chromosomal homologues of parA and parB (sometimes denoted as soj and spo0J, because of their involvement in sporulation), as well as parS, have been identified in a wide range of Gram-negative and Gram-positive bacteria, with the exception of certain γ-proteobacteria, including E. coli and Haemophilus influenzae [3,6]. The par genes are commonly arranged in an operon, whose expression is autoregulated by par-encoded proteins [7-9]. In numerous bacteria, chromosomal par genes are located upstream of the dnaA-oriC region [10].
Two or more 16-bp parS inverted repeats, with a consensus sequence 5'-TGTTNCACGTGAAACA-3, are clustered near the origin of chromosome replication (oriC) region [11]. In Bacillus subtilis, Spo0J binds to 8 of these 10 pseudo-palindromic 16-bp invert repeats in vivo. Furthermore, the presence of one of such site on an otherwise unstable plasmid stabilizes it in a Soj- and Spo0J dependent manner [11]. In Streptomyces coelicolor, 20 of the 24 parS sequences are packed around oriC, and ParB binds to many of them in vitro and in vivo [12]. Although the precise function of ParA and ParB is still unclear, it has been proposed that the recruitment of these proteins to parS sites may lead to the positioning of replicated chromosomal origins at opposite poles of the cell [11]. The parAB genes are essential for the viability of Caulobacter crescentus [13], whereas in B. subtilis [14], Streptomyces coelicolor [15] and Pseudomonas putida [16], deletion of soj/parA and spo0J/parB is not lethal. spo0J mutants of B. subtilis display defects in chromosome segregation in both vegetative and sporulating cells [14,17]. Deletion of parAB in S. coelicolor results in the production of significant numbers of anucleate spores, although no detectable defect is visible in vegetatively growing cells [15]. In P. putida, whose cellular division occurs only by binary fission, anucleated-cells are only observed when mutants in these genes are grown in minimal medium or as they enter into stationary phase [16,18]. The Par proteins are involved in other processes, such as chromosome replication, transcription, and a cell-cycle checkpoint that links chromosome segregation to cell division [13,19,20].
New insights about the role of Par proteins in chromosome segregation are emerging with the recent discovery of the bacterial cytoskeleton. A bacterial actin homolog, MreB, has been implicated in chromosome segregation. In the bacterial cells that have MreB, a membrane-associated coiled structure extends along the cell length [21]. In C. crescentus, this structure may be used for transporting oriC rapidly towards the cell poles. MreB may bind to DNA via ParB forming a kinetocore-like complex, which might connect the oriC region to the MreB coil at the membrane, and thus may actively move this region toward the cell poles [22].
Tuberculosis (TB) is a major public health problem with one-third of the world's population infected by its etiologic agent, Mycobacterium tuberculosis. Over two million people die from TB each year [23]. The tubercle bacilli can lie dormant for years, only to rise again when the immune system weakens due to old age, malnutrition or AIDS. M. tuberculosis is a non-capsulate and non-spore forming bacterium with a relatively simple life cycle. Despite the medical importance of this human pathogen, very little is known about the molecular mechanisms controlling its cell cycle.
An interesting problem in M. tuberculosis biology is therefore to understand how this intracellular pathogen regulates progression of its cell cycle during the stages of TB infection, including the dormant state. The dormant state may be considered in some ways analogous to sporulation, and some genes related to sporulation in B. subtilis and S. coelicolor are found in the genome of M. tuberculosis [24]. Nevertheless, the dormant state may also be considered a special physiological state during which mycobacteria grow slowly, but are not sporulated.
Studies based on experimentally-mapped transcriptional start sites have provided a consensus sequence for several mycobacterial promoters [25-27]. Group A includes the σA and σB Mycobacterium promoters, which share homology to the E. coli σ70 consensus sequence. The Group D or "SigGC" Mycobacterium promoters, with -10 (C90R70C50C50M70S90) and -35 (T90G50S80C50S90T30) GC rich-hexamers, are likely to be unique to mycobacteria [27,28]. However, it is still unknown which of the 13 sigma factors described in Mycobacterium actually drive transcription from these promoters [26,27].
In order to understand their possible role in mycobacterial cell cycle, in this work we examined the genetic regulation of the parA and parB partitioning genes, by analysing the transcription of these genes in Mycobacterium bovis BCG and Mycobacterium smegmatis, two non-pathogenic mycobacteria, belonging respectively to the slow and fast-growing groups of the Mycobacterium genus.
Results
Nucleotide sequence of the jag-parB region and conservation of the parS sites near the chromosomal origin of replication
Analysis of the complete genome sequence indicates that the ParA and ParB proteins of M. tuberculosis H37Rv have high sequence identity (50–60%) with the chromosomal partitioning Soj/ParA and SpoJ/ParB proteins of S. coelicolor, P. putida and C. crescentus [29]. Genes homologous to parA and parB were also identified in the close relatives Mycobacterium leprae [29], Mycobacterium bovis [29] and M. smegmatis [30] and like in M. tuberculosis they are located near the chromosomal origin of replication (oriC).
Eight ORFs could be identified in the 6 Kb region upstream of the dnaA gene in M. tuberculosis, M. bovis BCG and M. smegmatis (see Additional file 1). All eight ORFs were divergently oriented in relation to the dnaA gene and included the parA and parB genes along with several other conserved genes, following a similar gene order to that found in other Gram-positive and -negative bacteria [10].
M. tuberculosis ParA and ParB proteins had sequences that were 99% and 100% identical to the homologous proteins in M. bovis BCG, and 77% and 71% identical to the homologous proteins in M. smegmatis, respectively. In M. tuberculosis and M. bovis BCG, the stop and start codons of gidB, parA and parB genes overlapped, suggesting that these genes could be part of a single operon. In M. smegmatis, the stop and start codons of gidB and parA genes overlapped, while the parA and parB genes were separated by 59 nucleotides, suggesting that promoters localized in the parA-parB intergenic region could initiate the transcription of the M. smegmatis parB gene. Lin and Grossman [8] identified a 16 bp perfect palindrome (5'-TGTTTCACGTGAAACA-3') identical to the parS sequence of B. subtilis, at two sites in the M. tuberculosis chromosome, located at ~1.1 Kb and ~2 Kb upstream of the parB gene. A Blast search of this sequence revealed that two putative parS sites seemed to be conserved in M. bovis BCG and M. smegmatis genomes at similar positions, 1.761 Kb and 0.9 Kb upstream of the start codon of parB for M. bovis BCG, and 1.749 Kb and 0.984 Kb upstream of the start codon of parB for M. smegmatis. No additional parS sequences were found in these mycobacterial chromosomes.
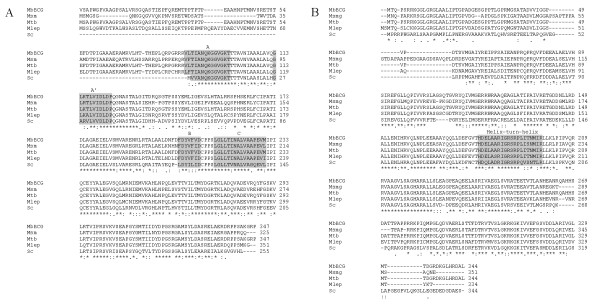
ParA and ParB proteins alignments were performed using the translated par sequences proposed for M. bovis BCG strain Pasteur 1173P2 [29], M. smegmatis mc2155 [30], M. tuberculosis H37Rv [29] and M. leprae [29]. Multiple amino acid sequence alignments showed that all the motifs identified in the chromosomal-coding Par proteins were conserved in the mycobacterial ParA and ParB proteins (Figure 1). The high aa sequence homology at the N-terminal region of the mycobacterial ParAs – and the fact that possible RBS sequences were not identified further downstream of the proposed parA start codons – suggest that in contrast to other chromosome-encoded ParA proteins, mycobacterial ParAs begin far upstream of the Walker A-box motif. Therefore, the mycobacterial ParA proteins may have an unusually long N-terminal domain. However, the helix-turn-helix (HTH) DNA-binding motif present in this region of some plasmid ParA proteins homologues was not present [31].
Figure 1.
Alignment of the ParA and ParB proteins. Comparison of the ParA (left) and ParB (right) aminoacid sequences of M. bovis BCG (MbBCG), M. smegmatis (Msm), M. tuberculosis (Mtb), M. leprae (Mlep) and S. coelicolor (Sc). The alignment was carried out using CLUSTAL W (1.83) http://www.ebi.ac.uk/clustalw/. Conserved amino acids are indicated with asterisk below the alignment; "-" represents gaps, ":" indicate conserved substitutions and "." semi-conserved substitutions. The A (VL/FTIANQKGGVGKT), A' (GLKTLVIDLDP) and B (FDYVFV/ID) boxes typical of the Walker-ATPases and the C motif (LGLLTINALVAAPEVM/L) of ParA proteins are highlighted on grey. The Helix-turn-Helix motif (HDELAARIGRSRPLITNMIR) involved in DNA-protein interactions of ParB is also highlighted on grey [3]. As noted, mycobacterial ParA have a longer N-terminal domain (between 67 to 91 aa) than other bacterial-ParA proteins.
Promoter activity in the parA and parB regulatory regions
In order to locate the promoters responsible for the transcription of the parA and parB genes, we cloned fragments of the orf60K-parB region of M. bovis BCG and M. smegmatis in the promoterless vector pFPV27, upstream of the gfp reporter gene (Table 1). GFP stability produced the accumulation of the fluorescent protein inside the cell and therefore the fluorescence at stationary phase always was higher than at exponential phase. In addition, the absence of a transcriptional terminator upstream of the cloning site in pFPV27 resulted in a relatively high and almost constant fluorescence background during the different growth phases studied, ranging from 175 to 178 RFU. Hence, the GFP fusions performed were not to evaluate cell growth-related expression, but to identify the promoter of each gene under study. Fluorescence > 18–20 % of the background was considered to be indicative of activity of the cloned promoter(s).
Table 1.
Plasmids used in this work
| Plasmid | Relevant features | Reference or source |
| pFPV27 | Kmr, shuttle vector for operon and gene fusion to gfp gene | [46] |
| pD19B | 261 bp PCR fragment from M. bovis BCG containing the upstream region of the gene jag | This work |
| pJ1B | 148 bp PCR fragment from M. bovis BCG containing part of the coding region of the orf60K | This work |
| pA3B | 114 bp PCR fragment from M. bovis BCG containing the upstream region of the gene jag | This work |
| pB5B | 205 bp PCR fragment from M. bovis BCG containing the upstream region of the gene gidB | This work |
| pA15B | 116 bp PCR fragment from M. bovis BCG containing the coding region of the gene jag | This work |
| pB3B | 113 bp PCR fragment from M. bovis BCG containing the upstream region of the gene gidB | This work |
| pA2B | 214 bp PCR fragment from M. bovis BCG containing the upstream region of the gene parA | This work |
| pC5B | 113 bp PCR fragment from M. bovis BCG containing part of the coding region of the gene parA | This work |
| pE1B | 229 bp PCR fragment from M. bovis BCG containing the upstream region of the gene parB | This work |
| pJ3M | 320 bp PCR fragment from M. smegmatis containing the upstream region of the gene jag | This work |
| pD1M | 159 bp PCR fragment from M. smegmatis containing the upstream region of the gene jag | This work |
| pG2M | 256 bp PCR fragment from M. smegmatis containing part of the coding region of the gene jag | This work |
| pC18M | 217 bp PCR fragment from M. smegmatis containing part of the coding region of the gene parA cloned in the direction of parA gene | This work |
| pC11M | 217 bp PCR fragment from M. smegmatis containing part of the coding region of the gene parA cloned in the reverse direction of parA gene | This work |
| pA1M | 120 bp PCR fragment from M. smegmatis containing part of the coding region of the gene gidB | This work |
| pB1M | 200 bp PCR fragment from M. smegmatis containing part of the coding region of the gene gidB | This work |
| pB16M | 475 bp PCR fragment from M. smegmatis containing the upstream region of the gene parB | This work |
| pC1M | 122 bp PCR fragment from M. smegmatis containing the upstream region of the gene parB | This work |
All the constructs were tested for fluorescence emission in M. smegmatis mc2155. The Figures 2B and 3B show the fluorescence obtained during the stationary phase of growth for each transcriptional fusion corrected by subtracting the fluorescence emission of M. smegmatis bearing the plasmid pFPV27. We found that M. bovis BCG promoter activities were well expressed in the heterologous host M. smegmatis.
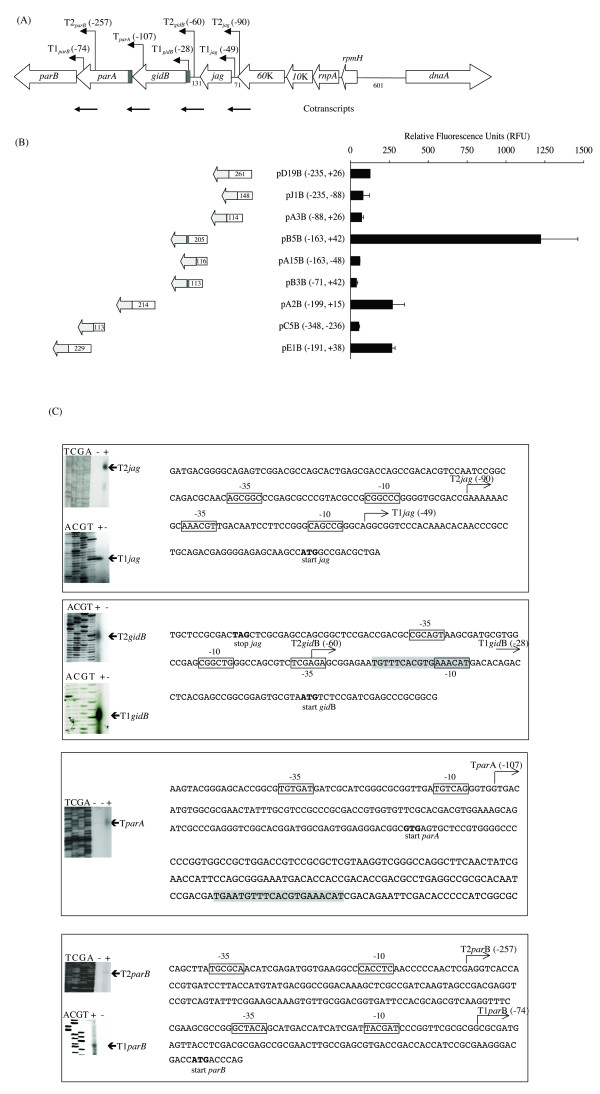
Figure 2.
Transcriptional pattern of the M. bovis BCG orf60K-parB region. (A): Schematic representation of the M. bovis BCG orf60K-parB region showing the position of the transcriptional start sites (TSSs). The parS sequences are represented by solid grey rectangles. Cotranscripts identified by RT-qPCR are shown as horizontal bold arrows. TSSs are showed as bent arrows. The position of the TSSs mapped are in parenthesis and it localization is related to the start of the gene immediately downstream. (B): Transcriptional fusions to gfp and measurement of the fluorescence emission. Recombinant plasmids were obtained by cloning of PCR fragments (white rectangles) upstream of the gfp. The coordinates (5' and 3' ends with respect to the start codon of the gene being evaluated), of the cloned fragments are shown in parenthesis together with the plasmid name. The length (in bp) of the cloned fragments is indicated within the white rectangles and the grey arrows represent the cloning direction and the gfp gene. Promoter activity was measured by fluorimetry as Relative Fluorescent Units (RFU) in M. smegmatis corrected by subtracting pFPV27 mediated background fluorescence.The bars on the graphic represent RFU (means ± SE of at least three independently experiments) during stationary phase of growth. (C): Mapping of the mRNA 5' termini on the jag-gidB-parA-parB region of M. bovis BCG by primer extension. The mRNA 5'-ends or TSSs using specific oligos are indicated (T1jag, transcription start site for the promoter 1 of gene jag, etc.). Sequencing reaction with the same primers is shown alongside. The ParA1B primer was annealed to total RNA at 48°C. The highlighted boxed region defines the -35 and -10 promoter sequences identified upstream of each TSS; the numbers in parenthesis indicate the position to the TSS according to the start codon of the gene locate immediately downstream. Start codon for jag, gidB, parA and parB is shown in bold and the putative parS sequence located upstream gidB is highlighted with grey.
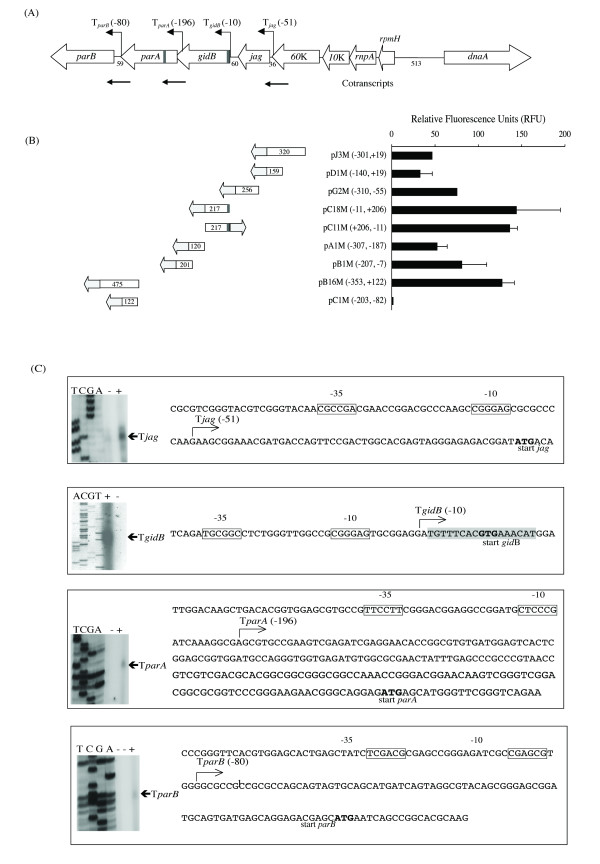
Figure 3.
Transcriptional pattern of the M. smegmatis orf60K-parB region. (A): Schematic representation of the M. smegmatis orf60K-parB region showing the position of the transcriptional start sites (TSSs). The parS sequences are represented by solid grey rectangles. Cotranscripts identified by RT-qPCR are shown as horizontal bold arrows. TSSs are showed as bent arrows. The position of the TSSs mapped are in parenthesis and it localisation is related to the start of the gene immediately downstream. (B): Transcriptional fusions to gfp and measurement of the fluorescence emission. Recombinant plasmids were obtained by cloning of PCR fragments (white rectangles) upstream of the gfp. The coordinates (5' and 3' ends with respect to the start codon of the gene being evaluated), of the cloned fragments are shown in parenthesis together with the plasmid name. The length (in bp) of the cloned fragments is indicated within the white rectangles and the grey arrows represent the cloning direction and the gfp gene. Promoter activity was measured by fluorimetry as Relative Fluorescent Units (RFU) in M. smegmatis corrected by subtracting pFPV27 mediated background fluorescence. The bars on the graphic represent RFU (means ± SE of at least three independently experiments) during stationary phase of growth. (C): Mapping of the mRNA 5' termini on the jag-gidB-parA-parB region of M. smegmatis by primer extension. The mRNA 5'-ends or TSSs using specific oligos are indicated (T1jag, transcription start site for the promoter 1 of gene jag, etc.). Sequencing reactions with the same primers is shown alongside. The highlighted boxed region defines the -35 and -10 promoter sequences identified upstream of each TSS; the numbers in parenthesis indicate the position to the TSS according to the start codon of the gene locate immediately downstream. Start codon for jag, gidB, parA and parB is shown in bold and the putative parS sequence located upstream gidB is highlighted with grey.
M. smegmatis cells emitted fluorescence when they bore plasmids containing the orf60K-jag (pD19B) and jag-gidB (pB5B) intergenic regions, as well as plasmids containing the 3'-end coding region of the gidB (pA2B) and parA (pE1B) genes of M. bovis BCG (Figure 2B), suggesting that jag, gidB, parA and parB genes of M. bovis BCG may be transcribed from promoters localised immediately upstream of each one of these genes. The parA and parB genes of M. smegmatis could also be transcribed from their own promoters, because substantial fluorescence was detected when the cells had the GFP transcriptional fusion to the orf60K-jag (pJ3M), jag-gidB (pG2M plasmid) and parA-parB (pB16Ms plasmid) intergenic regions as well as to the 3'-end of the gidB gene (pB1M plasmid) (Figure 3B). Unexpectedly, we found that a 217 bp fragment containing the parS motif localised in the 5'-end of the gidB gene of M. smegmatis (pC18Ms and pC11Ms plasmids) showed fluorescence emission independently of the clone direction, suggesting divergent promoter activity in this region.
When we deleted 89 bp of the 3'-end (pA15B) or 92 bp of the 5'-end (pB3B) from pB5B, the fluorescence emission was practically abolished, showing that the entire 205 bp region of pB5B was necessary in order to have the activity observed with this transcriptional fusion (Figure 2B). Finally, the fluorescence of M. smegmatis bearing some constructs (pA3B, pA15B, pB3B, pC5B, pJ3M and pG2M) was not detectable during the exponential phase of growth (data not shown), suggesting that the promoters contained in these fragments were weak and their expression could be detected only after enough GFP have accumulated during growth.
Mapping the transcription start sites in the jag-parB region
In an attempt to precisely localise the transcriptional start sites (TSSs) in the jag-parB region of M. bovis BCG and M. smegmatis, primer extension experiments were carried out using several specific primers and total RNA isolated from exponentially growing mycobacteria (Figure 2C and 3C). Analysis of the nucleotide sequence upstream of the identified TSSs was performed in order to identify potential promoters. Published consensus promoter sequences as well as the distance between the -10 hexamer and the TSS and the length of the spacer between the -10 and -35 regions were considered. Promoter sequences proposed according to our results are shown in Table 2. All but one of the TSSs of all genes corresponded to a purine (A or G) and each one was very well associated to a recognised promoter sequence. All the identified promoters in both M. smegmatis and M. bovis BCG belonged to the Group D of Mycobacterium promoter recognition sequences, with the exception of two possible E. coli σ70- like promoters located upstream of gidB (P1gidB) and parB (P1parB) in M. bovis BCG. We found two TSSs upstream of the jag, gid and parB genes in M. bovis BCG (Figure 2C). They were close to each other, suggesting that two promoters may drive the expression of each one of these genes. Fragments containing only one of the proposed promoters for jag (pJ1B and pA3B), gid (pJA15 and pB3B) and parB (pC5B and pE1B) genes of M. bovis BCG showed fluorescence activity (Figure 2B) corroborating the presence of two promoters upstream of each one of these genes.
Table 2.
Promoter sequences for jag, gidB, parA and parB genes of M. bovis BCG (Mb) and M. smegmatis (Ms)
| Promoter | -35 | Spacer† | -10 | Spacer‡ | TSS§ | Group |
| P1jag (Mb) | aaaCGT | 16 | CAgCCG | 03 | A | D |
| P2jag (Mb) | aGCgGc | 18 | CGGCCC | 11 | G | D |
| P1gidB (Mb) | TcGAgA | 19 | aAacAT | 04 | C | A |
| P2gidB (Mb) | cGCaGT | 18 | CGgCtG | 13 | A | D |
| PparA (Mb) | TGtgaT | 21 | tGtCAG | 04 | G | D |
| P1parB (Mb) | gctACA | 17 | TAcgAT | 12 | G | A |
| P2parB (Mb) | TGCgCa | 20 | CACCtC | 12 | G | D |
| Pjag (Ms) | cGCCGa | 18 | CGGGAG | 10 | G | D |
| PgidB (Ms) | TGCgGc | 14 | CGggAG | 07 | G | D |
| PparA (Ms) | TtCCtT | 18 | CtCCCG | 10 | A | D |
| PparB (Ms) | TcGaCg | 16 | CGagCG | 04 | G | D |
(†) Length of the spacer between the -35 and -10 hexamers. (‡) Length of the spacer between the -10 hexamer and TSS. (§) Transcription start site (TSS) determined by primer extension. Consensus nucleotides are shown with capital letters.
In contrast, we found just a single TSS upstream of the jag, gid, parA and parB genes in M. smegmatis (Figures 3A and 3C) and upstream of the parA gene in M. bovis BCG (Figure 2A and 2C). This implied the presence of only one promoter for each one of these genes.
The -10 (AAACAT) hexamer associated to the T1gidB of M. bovis BCG overlapped with a putative parS sequence (Figure 2C), suggesting that ParB could be regulating the transcription from P1gidB by competing for the same region with the RNA polymerase.
Dicistronic transcripts in the jag-parB region
The primer extension, transcriptional fusions to gfp, and nucleotide sequence analysis together indicated that the gid, parA and parB genes of both M. bovis BCG and M. smegmatis, seem to be transcribed independently from their own promoters. However, the short or missing intergenic regions found in this study do not eliminate the possibility that gid and the two par genes can be part of a single transcript. To ascertain whether the par genes had a dicistronic arrangement, RT-qPCR was performed using M. bovis BCG and M. smegmatis RNAs. Specific primers were designed in order to obtain products encompassing from the 3'-end to the 5'-start of the orf60K-jag, jag-gidB, gidB-parA and parA-parB pair genes (Table 3), which always excluded the contribution of the promoters located immediately upstream of each evaluated gene. Although the possible presence of transcriptional termination signals into the downstream gene cannot be discarded, our results suggested that all the transcripts, except the one for jag gene of M. smegmatis, were at least dicistronic (Table 4, Figures 2A and 3A).
Table 3.
Sequences of PCR primers used for RT-qPCR¶
| parAB expression | ||||
| Gene | Forward (5'→3') | Reverse (5'→3') | Amplicon (bp) | Coordinates (5', 3') |
| parA (Mb) | aagtgttgcggacggtgattc | ggtcacgctcggcaagttc | 140 | +874, +1014 |
| parB (Mb) | gcgtaagccgattcagatgcc | ccgagccgaactccaccac | 122 | +833, +954 |
| parA (Ms) | acgacggccgcaccaagct | gtcgagatagctcagtgctcc | 177 | +754, +930 |
| parB (Ms) | cgtaagccgatccagatgcca | tcgttctgggcgctcatcag | 171 | +882, +1052 |
| Co-transcription | ||||
| Region | Forward (5'→3') | Reverse (5'→3') | Amplicon (bp) | Coordinates (5', 3') |
| orf60K-jag (Mb) | aatgcggcagccccaacag | tcggtggtgtcagcgtcg | 256 | -233, +23 |
| jag-gidB (Mb) | ccagaacgccgagtcgttgtgc | gtccgaagatcgcagacgc | 204 | -164, +40 |
| gidB-parA (Mb) | gcggttgatgtcagggtggtg | cgtcggtgtcggtggtgtc | 236 | -124, +112 |
| parA-parB (Mb) | gcgttggagggtgtgtcg | ccctttctgcgtgacggc | 352 | -326, +26 |
| orf60K-jag (Ms) | gctccgccaccgaactgac | gcgtccgcagcgagagtg | 187 | -184, +3 |
| jag-gidB (Ms) | ttccgccgcctcaagcc | cacgccctgtcctttgttctg | 199 | -124, +75 |
| gidB-parA (Ms) | atgctcccgatcaaaggc | cgaacccatgctcatctcc | 230 | -215, +15 |
| parA-parB (Ms) | cctcgcagtgtgaaggtctcg | cggctgattcatgctcgtctcc | 212 | -200, +12 |
¶Normalization was performed using primers for 16S rRNA amplification previously published [50]
Table 4.
Co-transcription in the jag-parB region
| Cotranscription region (cDNA copies/16S × 10-6) | ||||
| orf60K-jag | jag-gidB | gidB-parA | parA-parB | |
| M. bovis BCG | ||||
| Exponential (7 days) | 9.16 ± 5.4 | 16.40 ± 1.4 | 17.92 ± 2.2 | 1.42 ± 0.1 |
| Stationary (14 days) | 43.45 ± 5.9 | 108.70 ± 20.4 | 65.85 ± 23.9 | 10.39 ± 0.1 |
| M. smegmatis | ||||
| Early Exponential (OD585nm = 0.6) | 58.27 ± 5.6 | 0 | 4.29 ± 1.2 | 14.95 ± 0.1 |
| Late Exponential (OD585nm = 1.2) | 35.61 ± 4.5 | 0 | 3.39 ± 0.5 | 12.36 ± 1.9 |
| Stationary (OD585nm = 2.0) | 0 | 0 | 0.28 ± 0.0 | 1.65 ± 0.1 |
Quantification of parA and parB mRNA levels during mycobacterial growth
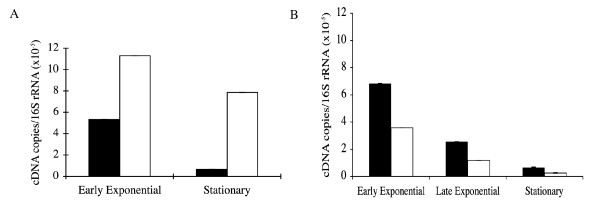
The levels of parA and parB genes mRNAs in M. bovis BCG and M. smegmatis were quantified by real-time RT-PCR (RT-qPCR) in exponential as well as in the stationary growth phase. Quantitative PCRs for parA, parB and 16S-rRNA were performed using the cDNAs obtained from the same RT reaction. The amount of mRNA for each par gene was calculated and expressed in relation to the total RNA and normalized by the 16S-rRNA levels. We detected mRNA-parA that was double of mRNA-parB levels in M. smegmatis, although the mRNAs of both genes decreased between the exponential and stationary phases. In contrast, the mRNA-parB levels in M. bovis BCG were very similar between the exponential and stationary phases, but mRNA-parA levels showed an important reduction in the stationary growth phase. Additionally, unlike the transcriptional pattern observed in M. smegmatis, the mRNA-parB levels were higher that mRNA-parA in M. bovis BCG (Figure 4).
Figure 4.
parA and parB mRNA synthesis during growth of Mycobacteria in broth culture. (A): Total RNA isolated from exponential (7 days) and stationary (21 days) cultures of M. bovis BCG. (B): Total RNA isolated from exponential (OD595 nm = 0.9), late exponential (OD595 nm = 1.2) and stationary (OD595 nm = 3.0) cultures of M. smegmatis. At the indicated time, bacterial RNA was extracted and transcript levels of parA (black bars) and parB (white bars) were analysed by real-time PCR; 16S rRNA levels were used for normalization. The error bars show the mean (± SD) of at least two separate determinations made with different batches of total RNA.
Discussion
We found evidence that the chromosomal parA and parB genes of M. bovis BCG and M. smegmatis are expressed from multiple promoters. To identify the promoter sequences that regulate the expression of the par genes, we mapped the transcription start sites of the par-mRNAs by primer extension and confirmed the activity of the identified promoters by transcriptional fusions to a fluorescent reporter. We also demonstrated that in M. bovis BCG the parA and parB genes are differentially expressed during the exponential and stationary growth phases.
In all microorganisms studied thus far, plasmid and chromosome-encoded partitioning genes are arranged in an operon. Transcription of the par genes is driven by one (in F and R1 plasmids, P1 prophage and C. crescentus) or two (in S. coelicolor) promoters located upstream of the gene encoding the ATPase (parA or sopA) [5,7,13,15,32]. The jag, gidB, parA and parB genes of M. bovis BCG and M. smegmatis shared orientation and close spacing, suggesting that they may be co-transcribed. However, we identified at least one promoter sequence for each of these genes (Figures 2 and 3 and Table 2). RT-qPCR (Table 4) and Northern blot hybridisation (data not shown) demonstrated that the parA-parB, gidB-parA and orf60-jag gene pairs were also transcribed as dicistronic operons; however, co-transcription between the jag-gidB region was only detected in M. bovis BCG (Table 3).
Most of the putative promoter sequences identified (Table 2) had features of the Mycobacterium promoters denoted as Group D. Only two of the promoter sequences found belonged to Group A Mycobacterium promoters. We were unable to identify promoter sequences for σ factors different from σA (or σB) and "SigGC" in the jag-parB region of both mycobacterial species, probably due to the exiguous data accumulated regarding DNA sequences recognized by RNA polymerases containing other σ factors. Nevertheless, no variation in the parA and parB gene expression has been observed in M. tuberculosis knockout mutants of σE [33], σH [34], σF [35], σC [36], σD [37], σL [38] or σM[39], suggesting that none of these σ factors were involved in the parAB expression.
Based on our results, we propose that in both M. bovis BCG and M. smegmatis, the parA and parB genes comprise an operon. Therefore, the expression of parB may be derived from three promoters in M. bovis BCG – two Group D and one Group A promoters – whereas parB transcription in M. smegmatis seems to be driven from only two promoters, both belonging to the Group D of Mycobacterium promoters (Figures 2 and 3 and Tables 2 and 4).
Results also indicated that the parA and parB genes in M. bovis BCG and M. smegmatis were differentially expressed (Figure 4), possibly due to the differential quantity and activity that each promoter contributed to transcribe the gidB parA and parB genes in each mycobacteria. It has been suggested that mycobacterial promoters homologous to E. coli σ70 have a higher activity than the Group D Mycobacterium promoters [27]. In agreement with these observations, we found that the TSSs in Group D mycobacterial promoter sequences (T2gidB and T2parB) showed weaker signals in comparison with those preceded by Group A (T1gidB and T1parB) of Mycobacterium promoters (Figure 2C).
The decrease of the mRNAs for parA and parB observed during the transition from exponential to stationary phase in M. smegmatis (Figure 3B) may be in agreement with the assumption that genes involved in replication and cell division must be down regulated during the stationary phase. In keeping with this interpretation, the expression of these genes decreases when M. tuberculosis is cultured under starvation [40]. The parB gene expression in M. bovis BCG seems to be differently regulated, because one Group A Mycobacterium promoter as well as two "SigGC" promoters appeared to contribute to parB expression in this mycobacterial species (Figure 2 and Table 2). The expression of E. coli σ70-like promoters (P1parB) appears to be particularly important for parB, because the transcription from P2parB (T2parB in Figure 2C) as well as from parA (Table 4) did not account for the mRNA-parB levels observed at the stationary growth phase (Figure 4A). Since during stationary growth, the levels of σA decrease [41] whilst σB expression increases [41,42], we proposed that transcription from P1parB may be driven by σB, the principal-like sigma factor.
On the other hand, it has been suggested that the correct stoichiometry of the Par proteins is important for partition of plasmids [43,44] and the bacterial chromosome [9,45], and that therefore the par loci must be under strict regulation. Recently, it has been suggested that modulation of the chromosomal parAB expression may be mediated by the binding of ParB to parS sites located near promoter sequences [9]. Here, one putative parS site was identified in the regulatory region of the gidB gene of M. bovis BCG, which overlapped with the -10 sequence of one Group A promoter (Figure 2C), suggesting that the binding of the ParB protein to the parS sequence may obstruct the access of the RNA Polymerase and negatively regulate the gidB expression. The other putative parS sequence identified was located within the coding region of the parA gene (Figure 2A). This suggests that ParB protein may also affect the expression of the parA gene in M. bovis BCG by blocking transcription initiated from TparA or the translation of the mRNA-parA. Thus, the regulation of the gidBparA genes and the parA expression by ParB binding to the parS sequences might contribute to maintain appropriate levels of the Par proteins.
Conclusion
Transcriptional analysis demonstrated that the par genes in M. bovis BCG and M. smegmatis had a dicistronic arrangement in which parA and parB were mainly expressed from weak "SigGC" promoters. However, additional Group A promoters were found upstream of parB and gidB in M. bovis BCG. Furthermore, the presence of multiple promoters for genes related to cell cycle as parAB, which may be regulated by different sigma factors, might be responsible of the differential regulation of these genes.
Methods
Media, bacterial strains and growth conditions
E. coli XL1-blue cultures were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C. M. smegmatis mc2155 [46] and M. bovis BCG Pasteur (ATCC 35734) were grown at 37°C using Middlebrook 7H9 broth or 7H10 agar supplemented with 0.5 % (v/v) glycerol and 10 % (v/v) Middlebrook OADC (Difco). To avoid clumping, Tween 80 (0.05 %) was added to liquid media. The following concentrations of antibiotics were added when appropriate: Carbenicillin (Cb, 50 μg ml-1) or Kanamycin (Km, 50 μg ml-1 for E. coli, 25 μg ml-1 for mycobacteria).
Transcriptional fusion to gfp and fluorescence measurement
The nucleotide sequences of the orf60k-parB regions were obtained in a Blast search [29,30]. Fragments of variable length containing the upstream region of the genes parA and parB from M. smegmatis and M. bovis BCG were inserted into the shuttle plasmid pFPV27 [47] to obtain the transcriptional fusions to gfp. The fragments were the products of PCR amplification using specific primers and chromosomal DNA as template. Plasmids digestions with restriction endonucleases and sequencing confirmed the direction of the inserts. The plasmids generated (Table 1) were electroporated in M. smegmatis mc2155 and grown at 37°C in 7H9 medium containing Km. Aliquots of the cultures were taken at exponential (OD595 nm = 0.8 – 1.3) and stationary (OD595 nm > 1.6) growth phases for fluorescence measurements. Fluorescence was determined from 150 μl of culture using a fluorimeter (Tecan GENius) and the appropriate filter combinations for GFP. The specific promoter activities were expressed as relative fluorescence units (RFU) corrected by subtracting the fluorescence emission of M. smegmatis bearing the promoterless plasmid pFPV27.
RNA extraction and primer extension analysis
RNA was isolated from M. smegmatis and M. bovis BCG by cell disruption as previously described [48]. For primer extension experiments, at least six synthetic oligonucleotides complementary to the mRNA strand of the upstream jag-gidB-parA-parB sequences were 5' end labeled with [γ-32P] ATP and T4 polynucleotide kinase. Each labeled primer (100 fmol) was annealed to 5–20 μg of total RNA at 52°C for 30 min. After cooling at room temperature, the primer extension reactions were carried out with AMV reverse transcriptase (Promega) at 42°C for 45 min. The extension products were separated on an 8% polyacrylamide/urea gel, alongside the sequencing reaction generated using the PCR fragments corresponding to the analysed sequence and the oligonucleotide used in the primer extension reaction as primer [49].
Detection of mRNA by quantitative RT-PCR
Total RNA was treated with DNAseI (Promega) during 45 min at 37°C and the absence of DNA was checked before reverse transcription by PCR amplification. The number of amplicons was measured by real-time PCR using gene-specific primers and SYBR Green. A standard curve was obtained for each set of primers by performing four different PCRs in parallel, using 10-fold dilutions of known amounts of M. bovis BCG or M. smegmatis chromosomal DNA (1,000, 10,000, 100,000, and 1,000,000 theoretical copies) alongside the uncharacterized samples. The melting curve of each amplicon was determined at the end of each experiment. Each measurement was performed at least in duplicate and repeated twice using independent RNA preparations from different cultures. In each sample 500 ng (or as indicated) of RNA and 0.5 μg of random hexamers (total concentration of 1 μM) were mixed in a total volume of 12 μl, heated to 65°C for 10 min and immediately chilled in ice-water for at least 5 min. Subsequently, 1 × PCR Buffer (10 mM Tris-Cl pH 8.3; 50 mM KCl), 5 mM MgCl2, 40 U of RNase inhibitor (RNasin Plus, Promega), 200 U of M-MLV (Moloney murine leukemia virus; Invitrogen) or AMV (Avian myeloblastosis virus; Promega) reverse transcriptase (RT) and all four deoxynucleoside triphosphates (final concentration of 1 mM each) were added. The reverse transcription reaction was performed at 42°C for 60 min. In all cases, a duplicate sample was prepared without RT as a control to measure DNA carryover. The enzyme was inactivated by heating at 99°C for 5 minutes.
Amplifications were performed in the DNA Engine Opticon (MJ Research) with sampling during elongation. Reactions were performed in 20 μl volume consisting of 0.25 μM concentration of each primer (Table 3), 10 μl of 2 × SG1Master mix (DyNAmo SYBR Green qPCR Kit. FINNZYMES) and 2 μl of the cDNA previously obtained. A control without RT was included in each run. The samples were subjected to 40 cycles of amplification (96°C denaturation for 10 s, specific annealing temperature for 15 s and 72°C extension for 20 s) in sealed strip tubes with optical caps; followed by incubation at 72°C for 5 min. To ensure that the fluorescent levels detected were due to the amplification of a specific product, a melting curve followed the final extension step, from 60°C to 95°C, with readings every 0.2°C.
Other molecular techniques
Digestions, ligations, filling of protruding ends and plasmid DNA isolation were performed according to standard procedures. Amplified fragments and plasmid DNAs were sequenced with USB Sequenase 2.0 (USB, Amersham) and [α-35S]dATP or with a dye terminator cycle sequencing kit and an ABI377 sequencer (PE Biosystem), using the appropriate primers.
Authors' contributions
LS and YC conceived the work and participated in its design. LS and JG-M coordinated the study. YC carried out the majority of the experiments while that EG and SR-G conducted some of the transcriptional fusion and primer extension assays. LS drafted the manuscript. LS and YC edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Gene organization in the parB-dnaN region of mycobacterial chromosome. The chromosomal gene organization is shown for Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. Arrows indicate gene orientations. Numbers inside of the boxes denote the size in amino acids of the predicted proteins. Numbers in bold denote the length in bp of the intergenic regions. The perpendicular black lines indicate the putative parS motifs. An asterisk shows the oriC region.
Acknowledgments
Acknowledgements
This work was supported by grants from the Fondo Nacional de Investigaciones Científicas y Tecnológicas – Venezuela (FONACIT-S1-2001000706), and the European Union through its INCO program (ICA4-CT-2002-10063). JG-M received support from COFAA, EDI and SIP-20071141, IPN, Mexico, and CONACyT (Grant SEP-2004-C01-46404). We are grateful to A. Sánchez for technical support. We thank K. Rodriguez-Clark and I. Beacham for reading the manuscript and making helpful suggestions.
Contributor Information
Yveth Casart, Email: ycasart@ivic.ve.
Elida Gamero, Email: egamero@ivic.ve.
Sandra Rivera-Gutierrez, Email: leiriasalazar@gmail.com.
Jorge A González-y-Merchand, Email: jgonzal1212@yahoo.com.mx.
Leiria Salazar, Email: leiriasalazar@gmail.com.
References
- Hiraga S. Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Kondo A, Ohshima A, Ogura T, Hiraga S. Structure and function of the F ÿplasmid genes essential for partitioning. J Mol Biol. 1986;192:1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- Austin S, Abeles AL. Partitioning of unit copy miniplasmids to daughter cells. II. The partititon regions of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Bugge-Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Friedman SA, Austin SJ. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988;19:103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- ÿDam Mikkelsen N, Gerdes K. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- Dubarry N, Pasta F, Lane D. ParABS systems of the four replicons of Burkholderia cenocepacia : new chromosome centromeres confer partition specificity. J Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O, Borovok I, Av-Gay Y, Cohen G, Aharonowitz Y. Gene organization in the trxA/B-oriC region of the Streptomyces coelicolor chromosome and comparison with other eubacteria. Gene. 1998;217:83–90. doi: 10.1016/s0378-1119(98)00357-6. [DOI] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Jakimowicz D, Chater K, Zakrzewska-Czerwinska J. The ParB protein of Streptomyces coleicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the lineal chromosome. Mol Microbiol. 2002;45:1365–1377. doi: 10.1046/j.1365-2958.2002.03102.x. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crecentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Ireton K, Gunther NW, IV, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involved an oriC-linked parAB locus. J Bacteriol. 2000;182:1313–1320. doi: 10.1128/jb.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology. 2002;148:537–548. doi: 10.1099/00221287-148-2-537. [DOI] [PubMed] [Google Scholar]
- Sharpe ME, Errington J. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- Godfrin-Estevenon AM, Pasta F, Lane D. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol Microbiol. 2002;43:39–49. doi: 10.1046/j.1365-2958.2002.02735.x. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Easter J, Jr, Gober JW. The chromosome partitioning protein, ParB, is required for citokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Ogasawara N, Harry EJ, Moriya S. Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J Bacteriol. 2003;185:6316–6324. doi: 10.1128/JB.185.21.6316-6324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann PL. Cytoskeletal elements in bacteria. Curr Opin Microbiol. 2004;7:565–571. doi: 10.1016/j.mib.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) Tuberculosis (TB) http://www.who.int/tb/en/
- Cole ST, Brosh R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, other authors. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Gomez M, Smith I. Determinants of mycobacterial gene expression. In: Hatfull GF, Jacobs WR, Jr, editor. Molecular genetics of Mycobacteria. Washington, DC. American Society of Microbiology; 2000. pp. 111–129. [Google Scholar]
- Smith I, Bishai WR, Nagaraja V. Control of mycobacterial transcription. In: Cole ST, Eisenach D, McMurray DN, Jacobs WR, Jr, editor. Tuberculosis and the tubercle bacillus. Washington, DC. American Society of Microbiology; 2005. pp. 219–231. [Google Scholar]
- Unniraman S, Chatterji M, Nagaraja V. DNA gyrase genes in Mycobacterium tuberculosis : a single operon driven by multiple promoters. J Bacteriol. 2002;184:5449–5456. doi: 10.1128/JB.184.19.5449-5456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannatine JP, Barletta RG, Thoen CO, Andrews RE., Jr Identification of Mycobacterium paratuberculosis gene expression signals. Microbiology. 1997;143:921–928. doi: 10.1099/00221287-143-3-921. [DOI] [PubMed] [Google Scholar]
- The new multi-microbial genome browser (GenoList) GenoList genome browser http://genolist.pasteur.fr/
- J. Craig Venter Institute Comprehensive microbial resource: Mycobacterium smegmatis mc2 genome page http://cmr.tigr.org/cgi-bin/CMR/GenomePage.cgi?org=gms
- Hayes F, Barilla D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nature Rev. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- Hayes F, Radnedge L, Davis MA, Austin SJ. The homologous operons for P1 and P7 plasmid partition are autoregulated from dissimilar operator sites. Mol Microbiol. 1994;11:249–260. doi: 10.1111/j.1365-2958.1994.tb00305.x. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Voskuil MI, Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Role of the extracytoplasmic-function sigma factor sigma (H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- Geiman DE, Kaushal D, Ko C, Tyagi S, Manabe YC, Schroeder BG, Fleischmann RD, Morrison NE, Converse PJ, Chen P, Bishai WR. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect Immun. 2004;72:1733–1745. doi: 10.1128/IAI.72.3.1733-1745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Converse PJ, Ko C, Tyagi S, Morrison NE, Bishai WR. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol. 2004;52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
- Raman S, Hazra R, Dascher CC, Husson RN. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palù G, Manganelli R. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor σ L and roles in virulence and in global regulation of gene expression. Infect Immun. 2006;74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal N, Woolwine SC, Tyagi S, Bishai WR. Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes. Infect Immun. 2007;75:452–461. doi: 10.1128/IAI.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis. 2004;84:218. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Funnel BE. Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J Bacteriol. 1988;170:954–960. doi: 10.1128/jb.170.2.954-960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusukawa N, Mori H, Kondo A, Hiraga S. Partitioning of the F plasmid: overproduction of an essential protein for partition inhibits plasmid maintenance. Mol Gen Genet. 1987;208:365–372. doi: 10.1007/BF00328125. [DOI] [PubMed] [Google Scholar]
- Figge RM, Easter J, Jr, Gober JW. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crecentus. Mol Microbiol. 2003;47:1225–1237. doi: 10.1046/j.1365-2958.2003.03367.x. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustapha S, Kieser T, Jacobs WR. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Hromockyj AE, Monarck D, Ramakrishnan L, Falkow S. Applications for the green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- Salazar L, Guerrero E, Casart Y, Turcios L, Bartoli F. Transcription analysis of the dnaA gene and oriC region of the chromosome of Mycobacterium smegmatis and Mycobacterium bovis BCG, and its regulation by the DnaA protein. Microbiology. 2003;149:773–784. doi: 10.1099/mic.0.25832-0. [DOI] [PubMed] [Google Scholar]
- Movahedzadeh F, Gonzalez-Y-Merchand JA, Cox RA. Transcription start-site mapping. In: Parish T, Stoker NG, editor. Mycobacterium tuberculosis protocols, Methods in Molecular Medicine. Vol. 54. Humana Press, Totowa, NJ; 2001. pp. 105–124. [DOI] [PubMed] [Google Scholar]
- Shi L, Jung YJ, Tyagi S, Gennaro ML, North RJ. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc Natl Acad Sci. 2003;100:241–246. doi: 10.1073/pnas.0136863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene organization in the parB-dnaN region of mycobacterial chromosome. The chromosomal gene organization is shown for Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. Arrows indicate gene orientations. Numbers inside of the boxes denote the size in amino acids of the predicted proteins. Numbers in bold denote the length in bp of the intergenic regions. The perpendicular black lines indicate the putative parS motifs. An asterisk shows the oriC region.